Abstract
Nanoparticles can encapsulate a range of therapeutics, from small molecule drugs to sensitive biologics, to significantly improve their biodistribution and biostability. Whilst the regulatory approval of several of these nanoformulations has proven their translatability, there remain several hurdles to the translation of future nanoformulations, leading to a high rate of candidate nanoformulations failing during the drug development process. One barrier is that the difficulty in tightly controlling nanoscale particle synthesis leads to particle-to-particle heterogeneity, which hinders manufacturing and quality control, and regulatory quality checks. To understand and mitigate this heterogeneity requires advancements in nanoformulation characterisation beyond traditional bulk methods to more precise, single particle techniques. In this review, we compare commercially available single particle techniques, with a particular focus on single particle Raman spectroscopy, to provide a guide to adoption of these methods into development workflows, to ultimately reduce barriers to the translation of future nanoformulations.
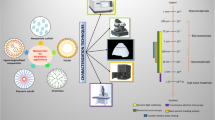
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nanoformulations, the encapsulation of active compounds in nanoparticles for delivery, are at the forefront of pharmaceutical development as a versatile modality from vaccines to gene therapy to cancer therapeutics (1,2,3). Encapsulating these compounds into nanoparticles allows access to unique physiochemical properties to overcome the low solubility, poor biostability, and fast clearance suffered by many bioactive compounds (4,5,6,7). Nanoformulation translation to the clinic began with FDA approval of Doxil in 1995 to treat cancers including breast cancer and solid tumours. Doxil offers an improved toxicity profile and reduced side effects compared to free doxorubicin by encapsulating the hydrophobic drug in crystallised form within the aqueous core of a spherical lipid bilayer, or liposome (8). Liposomes have been further utilised to solubilise hydrophobic drugs and reduce toxicity in several approved nanoformulations, including the chemotherapies Onivyde and Marqibo, and the anti-fungal treatment AmBisome (9, 10). As well as liposomes, solid lipid nanoparticles (SLNs) have found success in the clinic. The hereditary transthyretin-induced amyloidosis treatment Onpattro encapsulates short interfering ribonucleic acid (siRNA) in SLNs to protect the sensitive cargo from rapid hydrolysis and degradation in the body (11,12,13). Nanoformulations are now receiving greater attention due to the success of the Pfizer/BioNTech and Moderna SARS-Cov-2 lipid nanoparticle (LNP) vaccines, with over 3 billion Pfizer/BioNTech vaccines manufactured in 2021 (14). As well as liposomes and LNPs, there are many other clinical or experimental nanoparticle morphologies which include polymer micelles, nanogels, dendrimersomes, polymersomes, and hybrid lipid-polymer and polymer-inorganic particles (2, 15,16,17,18,19).
The self-assembly mechanisms employed to prepare nanoformulations, through film rehydration, emulsification, solvent injection, or microfluidics-assisted assembly, cause inherent particle-to-particle heterogeneity within batches and reduced batch-to-batch reproducibility (6, 20). This heterogeneity impacts clinical outcomes, because nanoparticle-biological interactions are affected significantly by their physicochemical properties such as size, surface charge, shape, and cargo loading (21,22,23). Therefore, nanoformulation heterogeneity hinders both safe manufacturing according to good manufacturing practice (GMP) and the definition of critical quality attributes (CQA) by regulators to identify acceptable ranges of drug product properties (24). Furthermore, the development of structure-function relationships is impeded, which prevents rational design of nanomedicines. A key step to understanding the impact of heterogeneity on clinical outcomes and devising novel strategies to mitigate heterogeneity is comprehensive characterisation.
Standard nanoformulation characterisation techniques to measure nanocarrier composition and cargo loading amount include liquid chromatography-mass spectrometry (LC-MS) and high-performance liquid chromatography (HPLC). However, these techniques are bulk methods, so can only measure ensemble average properties. To characterise heterogeneity in full, it is necessary to characterise nanoformulations at the single particle level. Several single particle characterisation techniques, such as cryogenic transmission electron microscopy (cryo-TEM) and atomic force microscopy (AFM), are low throughput either through long measurement times per particle or extensive sample preparation. Although these techniques can capture single particle-level detail, they cannot currently measure enough particles in a reasonable timeframe to be representative of population statistics. Therefore, there has been significant work in recent years to develop widely applicable, high throughput, single particle techniques which can fully characterise nanoformulation heterogeneity (25,26,27).
Many particle properties are essential to understand in order to predict nanoformulation behaviour in vivo. Morphological characteristics, such as size, shape, and rigidity can determine circulation time, cellular uptake, and biodistribution (18, 28, 29). These have been covered in excellent reviews previously (6, 30, 31). Here, we focus on chemical properties, specifically nanocarrier composition, cargo loading and surface chemistry and interactions. These properties impact biological behaviour: for example increasing the amount of a targeting ligand can increase cellular uptake; however, too high a targeting ligand density can reduce antifouling behaviour, thus increasing off-target accumulation (32). Additionally, liposome composition affects protein corona identity and cellular uptake (33). Despite their influence on clinical outcomes, these properties are often more challenging to characterise than morphological features, so require careful selection of characterisation technique. We focus on lipid and polymer-based nanoformulations for cargo delivery, which represent the majority of FDA-approved nanoformulations for delivery (34). Whilst there are several clinically approved formulations based on inorganic nanoparticles, these are not covered here as they often have different requirements for measuring chemical properties, which are detailed in recent reviews (35, 36). We aim in this review to provide an overview of available techniques to measure single particle chemical properties, with a particular focus on single particle automated Raman trapping analysis (SPARTA®). We place this method in the context of commercially available single particle characterisation techniques, by first giving a technical description of relevant techniques, followed by practical considerations for selecting a characterisation method. We then give examples of how chemical properties, specifically nanocarrier composition, cargo loading/release, and surface functionalisation/interactions have been characterised using the techniques detailed.
Single Particle Characterisation Techniques
Here, we present a brief technical overview of single particle characterisation techniques used to study nanoformulation chemical properties (Fig. 1). We have particularly focused on techniques which are commercially available and for which their utility for studying the chemical properties of nanoformulations has been demonstrated with published examples. We have excluded several single particle characterisation techniques, such as AFM with functionalised tips or cryo-TEM with staining, as these provide limited quantitative information on chemical properties and are rendered low throughput through laborious sample preparation and/or measurement protocols (37,38,39). There are also several techniques, such as nanoparticle tracking analysis (NTA) which are traditionally used to measure single nanoparticle size and/or concentration, but where to the best of our knowledge there are no published examples of application to study chemical properties. By describing how each technique experimentally measures nanoformulations and what data they output, we aim to provide context for later sections on how each technique can be used to study specific nanoformulation properties.
Overview of single particle techniques for characterising nanoformulation chemical properties. Single particle Raman trapping: a particle is stably held in an optical trap whilst its Raman spectrum is captured to give label-free size, nanocarrier composition, cargo loading, and surface chemistry information. Fluorescence correlation spectroscopy: the time correlation between fluorescence intensity fluctuations, caused by fluorescent species diffusing through a small volume, is analysed to give particle size, concentration, and surface and loading properties. Tunable resistive pulse sensing: ions and particles flow through a small pore under the influence of an applied electric field. The magnitude and duration of ionic current impedance whilst particles are resident in the pore can be used to calculate nanoparticle size, concentration and zeta potential. Nano-flow cytometry: the single particle scattering and fluorescence from a stream of nanoparticles in a flow channel can be used to quantify particle size, concentration, functionalisation, and loading. Single molecule localisation microscopy: blinking fluorophores can be precisely located from a time series of images where different fluorophores are in different on/off states in each frame. This allows determination of nanoparticle size, composition, surface functionalisation/interactions, and cargo loading
Nano-Flow Cytometry
In flow cytometry (FCM), particles are hydrodynamically focused in a flow channel to form a single particle stream that passes through a laser beam. This irradiation induces light scattering and fluorescence from each particle. Fluorescent labelling enables characterisation of surface functionalisation and cargo loading, as well as particle concentration. Conventional flow cytometers were designed for cell biology, where scattered light is used to gauge cell size and concentration. However, these instruments are usually unable to detect light scattered from submicron size particles with a refractive index similar to water, such as liposomes and polymersomes. Therefore, nano-flow cytometry (nFCM) uses more recently developed instruments that enable highly sensitive light scattering detection from single nanoparticles as small as 40 nm for determining their size and concentration without the need for fluorescent labelling (40, 41). Aside from advancements in laser power and detector performance employed, the enhanced sensitivity of nFCM over conventional FCM is brought about mainly by prolonging the transit time of each particle through the focused laser beam (ms compared to ns-µs) for increased photon generation, and by reducing the sample stream diameter (∼1.4 μm from typically 10 μm) resulting in a ∼10 fL detection volume to reduce scattering emission from the sample background significantly (40). Combined with fluorescent labelling, nFCM has been demonstrated to provide high throughput (10,000 particles/min), multiparameter (functional, size and concentration) analyses of single nanoparticles, including viruses, liposomes, and extracellular vesicles (31, 42,43,44,45).
Super Resolution Microscopy
Fluorescence microscopy can provide quantitative and localisation information on labelled biochemical constituents. However, the resolution of conventional fluorescence microscopy is diffraction limited (∼250 nm lateral direction, ∼500 nm in the axial direction) (46) and therefore insufficient to study individual nanoparticles. To overcome the diffraction limit, super resolution microscopy (SRM) techniques have been developed. SRM includes stimulated emission depletion (STED), structured illumination microscopy (SIM), and single-molecule localisation microscopy (SMLM). Compared to other SRM techniques, SMLM achieves the highest resolution (∼20–50 nm lateral direction, ∼50–60 nm in the axial direction) (47, 48). Therefore, SMLM is particularly useful for elucidating the spatial distribution of fluorescently-labelled nanoformulation constituents, such as cargoes, ligands, polymer coatings, and corona proteins, on individual nanoparticles as well as the inter-particle heterogeneity in these parameters (47, 49,50,51). SMLM includes photoactivated localisation microscopy (PALM) and stochastic optical reconstruction microscopy (STORM). SMLM techniques exploit fluorophores capable of switching stochastically between ON and OFF states to differentiate the position of individual fluorophores in time rather than space, which would not be possible with all the fluorophores fluorescing at the same time. When a sample is imaged, a time-series of images, in which different fluorophores are in the ON state in different frames, is acquired. Although the light from single emitting fluorophores remains diffraction limited, the distribution of its emission photons enables its precise centroid position to be calculated. A super-resolution image is then reconstructed with sub-diffraction sized pixels by combining the calculated localisation information from individual fluorophores in each of the image frames. This enables the distribution and heterogeneity of multi-colour labelled single molecules inside or on the surface of nanoparticles to be visualised with high spatial resolution.
Fluorescence Correlation Spectroscopy
Fluorescence correlation spectroscopy (FCS) enables calculation of particle size, concentration, cargo loading and release, and surface interactions (52, 53). The Brownian motion of fluorescent molecules or particles through a tightly focused, often confocal volume of ~0.5 fL causes fluorescence intensity fluctuations (54). These fluctuations are represented with an autocorrelation function, which describes how closely the signal resembles itself at different points in time. Fitting the autocorrelation function gives the average number of molecules in the detection volume, the molecular brightness, and the diffusion coefficient, which can be used to calculate particle hydrodynamic diameter. Since FCS also allows measurement of the fluorescent label alone or the fluorescent cargo as a reference, the number of fluorescent molecules per nanoparticle can be estimated (55). Furthermore, an extension to FCS named fluorescence cross-correlation spectroscopy (FCCS) uses two spectrally distinguished fluorophores, which are excited and detected separately, to compare the autocorrelation and cross-correlation of two species to measure their interactions.
Tunable Resistive Pulse Sensing
Tunable resistive pulse sensing (TRPS) measures single nanoparticle size and zeta potential and nanoparticle concentration, simultaneously. A TRPS setup uses two fluid cells separated by a non-conductive, nano-porous membrane and nanoparticles suspended in an electrolyte. Applying a voltage across the cells causes a flow of ions through the pore, which establishes a baseline current. As particles also move through the pore, they temporarily block the baseline current with a magnitude that is proportional to particle size and at a rate which denotes particle concentration. TRPS represents an advance over traditional resistive pulse sensing as the nanopore can be stretched to control fluid and particle flow (56). TRPS also measures electrophoretic mobility by analysing the durations of the resistive pulses under varying driving forces compared to a calibration standard (57,58,59,60,61). This can be used to measure single particle zeta potential, which is the potential difference between the particle with a layer of ions adsorbed to its surface and the dispersant, that is often used to infer surface charge (62).
Single Particle Raman Trapping
Raman spectroscopy is a label-free, non-destructive analysis method that can be used to characterise the chemical make-up of materials (63). When a sample is irradiated with monochromatic light, inelastic scattering can occur, where energy is transferred between the sample and incoming light. Different chemical bonds cause different amounts of energy transfer, which is represented as peaks in a Raman spectrum, where the peak position can be used to identify chemical species and peak intensity can indicate concentration. When Raman spectroscopy is combined with optical trapping, where a single particle is stably held in a tightly focused laser beam by gradient forces, the Raman spectrum of an individual nanoparticle can be generated. Using this technique, the user can assess the biochemical composition of single nanoparticles, and particle-to-particle variability of nanoparticle composition, loading and/or functionalisation steps (64, 65).
Initial Raman single particle trapping methods (65,66,67,68) were limited by inefficient manual optical trapping which resulted in only a handful of nanoparticle spectra per hour. By automating the nanoparticle trapping as described in Penders et al. (27), a platform, SPARTA®, has been developed that allows for generating statistically significant and sample-representative spectral data sets (>200 nanoparticle spectral acquisitions per hour) in a short time period. SPARTA® has been used to study single nanoparticle chemistry of liposomes (17, 27), LNPs (69), mixed lipid-polymer vesicles (16), polymersomes (27, 70, 71), dendrimersomes (17), and extracellular vesicles (72).
Considerations for Selecting a Characterisation Technique
There are several considerations when selecting a characterisation technique, such as labelling requirements, temporal/spatial resolution, the need to measure in physiologically relevant conditions, data analysis required, and the capability to simultaneously measure multiple parameters. We compare each of these considerations for the single particle techniques introduced above, and discuss their impact on the measurement of nanoformulation chemical properties (Table I).
Labelling
Many single particle techniques, including nFCM, FCS, and SRM, measure fluorescence signal. The benefits of fluorescence measurements include simple readout, high signal to noise ratio (when employing strategies in instrumentation design, measurement parameters selection, analysis, and sample preparation), and up to single molecule sensitivity (73,74,75,76,77). This high sensitivity allows for short measurement times (~mins) and low sample volumes (~μL). Moreover, high sensitivity allows accurate detection of cargo loading (78, 79) and surface functionalisation, where the functionalisation molecules often make up a small fraction of the whole nanoparticle, so high fluorescence sensitivity enables detection of very small changes in the nanoparticle chemical properties, such as single ligand binding or cleavage on the particle surface (51, 80).
However, whilst some drug and nanocarrier materials are inherently fluorescent, most materials require fluorophore labelling, either through covalent conjugation or encapsulation. These fluorophores can exhibit their own behaviours such as photobleaching or phase separation, thus inadvertently influencing the properties of the labelled molecule (81). This a particular concern for low molecular weight cargo and functionalisation molecules, where the fluorophore makes up a significant proportion of the molecule. Furthermore, labelling requires optimisation of staining protocols, which can be laborious and not necessarily applicable between different nanoformulations, as well as careful selection of fluorophores. These issues with labelling can be avoided by using label-free techniques, such as TRPS or SPARTA®. Although these label-free techniques can display reduced sensitivity, their ability to holistically measure nanoparticle chemical information without additional sample preparation time or the potential for non-specific signal and label-altered behaviour is a significant advantage.
In addition to the benefits and disadvantages of labelling, it is also necessary to consider whether a characterisation technique has suitable spatial and temporal resolution to sufficiently characterise a nanoformulation.
Spatial and Temporal Resolution
Spatial resolution is an important consideration when spatially mapping components within a particle, for example to characterise phase separation or surface interactions, whilst temporal resolution is a key factor for dynamic reaction monitoring, such as protein corona formation. The only technique considered here which is capable of sub-particle spatial resolution is SMLM. For example, SMLM provides spatially localised quantification of the number of corona proteins on the surface of each nanoparticle (82, 83). This can provide essential information on which factors (e.g., particle geometry) influence the particle corona composition for rational formulation design. Although SMLM can to some extent measure corona formation kinetics, the temporal resolution is low (on the scale of mins) (84).
Although the other techniques we consider in this review measure the whole nanoparticle, there is still a wealth of chemical information that can be gained from single particle resolution, especially when combined with high temporal resolution for monitoring surface interactions. The high temporal (∼ns) resolution of FCS has been applied to study in situ protein interactions and corona formation on various nanoparticles (85, 86) including lipid (87) and polymer-based drug carriers (52, 70, 71, 78) formulated with and without polymeric stabilisers. The temporal resolution of TRPS (~ms) also allows real-time analysis of nanoparticle reactions or interactions with biomolecules such as monitoring the kinetics and thickness of protein corona formation on a particle by monitoring changes in nanoparticle surface charge and size (58,59,60).
There are also many surface reactions that take place on the ~minute-hour timescale, such as enzymatic conversion or synthetic functional group interconversions. Thus, although SPARTA® measures one particle on the ~s timescale, so cannot monitor sub-second reactions, SPARTA® has been used for label-free, real-time monitoring of several surface interactions including click chemistry reactions and lipid headgroup interconversion (27, 69).
Physiologically Relevant Conditions
Another consideration when comparing characterisation techniques is whether measurements can be performed at physiologically relevant conditions. In vivo environments are complex mixtures containing proteins and electrolytes which can affect nanoparticle behaviour. For example, the proteins present can destabilise nanoparticles, leading to different rates of cargo release (88). Often these environments are challenging to characterise due to their high background signal, strong scattering, and different refractive index and viscosity to water. Despite this, FCS has been used to measure cargo release in blood using near-infrared light or by incorporating a cell-impermeable membrane in the measurement setup (78, 86). SPARTA® has been used to characterise doxorubicin and liposome signals simultaneously in the commercial nanoformulation Doxil in 90% v/v foetal bovine serum (FBS), although additional analysis is required due to high background signal from the serum (89). Nevertheless, the release of doxorubicin from Doxil over 48 hours was successfully measured in 10% v/v FBS (89).
Measurement and Data Analysis Complexity
An important practical consideration between different characterisation techniques is the complexity of performing measurements, such as required equipment and data analysis. SRM requires complex, high-quality optics and detectors. Both FCS and SPARTA® use a confocal optical arrangement to produce a small sampling volume to ensure single particle information (27, 53), and also to create sufficient optical trapping forces in the case of SPARTA®. SPARTA®, SRM, FCS, and nFCM utilise lasers to induce scattering and/or fluorescence, although SPARTA® only requires one wavelength of laser, whilst multi-colour FCS, SRM, and nFCM employ multiple lasers. TRPS is a laser-free technique; however, disposable cartridges are required for each measurement, which are prone to clogging. TRPS and nFCM also require frequent calibration for absolute determination of properties. For sample preparation, SRM is intensive due to required optimisation of sample immobilisation as well as staining protocols. FCS and nFCM have limited sample preparation requirements after labelling optimisation, as nanoformulations can be characterised in suspension. As TRPS and SPARTA® are both label-free techniques performed on nanoparticle suspensions, they can be performed with minimal sample preparation.
In addition to required equipment and sample preparation, it is also important to consider data analysis complexity. SMLM requires extensive data analysis to map the intensities of blinking fluorophores, and to combine data from the time series to reconstruct an image accounting for artefacts such as sample drift. FCS and nFCM comparatively require limited data analysis. However, each of these techniques also has specific considerations with analysis. FCS signal can be prone to domination by slow-diffusing species and requires fitting of autocorrelation curves, but this can often be automated. In nFCM, setting an accurate background threshold is prone to user error, because even a small excess in free fluorophores results in high fluorescence noise leading to significant overestimations in number of labelled species and masking of subtle signals differences when comparing different nanoformulations (90). SPARTA® gives extensive chemical information which can require thorough data analysis to extract. However, much of this process can be automated, and there are an increasing number of libraries for identification of chemical signatures.
Multiplexing Parameters
Nanoformulations are complex, with several important, inter-related properties that determine biological behaviour. To reduce time and cost, it is preferable to use the fewest techniques to gain the most information, particularly as different methods often have different volume and concentration requirements. Furthermore, measuring multiple parameters can reveal relationships between different nanoformulation properties. It can often be instructive to combine size measurements with chemical information, as is possible with FCS, nFCM, SPARTA®, SRM, and TRPS (43, 59, 61, 78, 91), for example to compare the variation in surface functionalisation with concentration, or to relate drug release with nanocarrier disintegration (51, 78). Furthermore, multiplexing with multiple fluorophores in FCCS, nFCM, and SMLM allows the detection of colocalization of different cargoes, distinct nanocarrier components, or reporter molecules (17, 92, 93). However, it is key to carefully select fluorophores to minimise cross-talk, and the inclusion of multiple labels increases the complexity of labelling optimisation and fluorophore selection. Furthermore, multi-colour labelling measurements are often limited to 2–4 colours (94).
SPARTA® displays an inherent multiplexing ability because Raman spectroscopy holistically measures all chemical components, above the limit of detection, of a nanoformulation in a label-free manner. This ability to agnostically and simultaneously characterise nanoformulation chemical properties offers an advantage in characterising the heterogeneity of nanoformulations, to ultimately understand their biological behaviour.
Nanoformulation Chemical Properties
Nanoformulations are multi-component systems, which exhibit a complex range of chemical properties. We now present key chemical properties, namely nanocarrier composition, cargo loading and release, and surface chemistry and interactions (Fig. 2). We demonstrate why these properties are essential to characterise with single particle detail; and we give examples of how these properties have been characterised on the single particle level.
Nanocarrier Composition
Many nanoformulations are prepared from a mixture of nanocarrier components, including phospholipids, cholesterol, polysaccharides, and/or polymers, which encapsulate a pharmaceutic agent (e.g., drug molecule, protein, nucleic acid). However, the initial reagent ratios do not always correlate with the final particle composition due to the difficulties in controlling self-assembly mechanisms and reagent ratios at the nanoscale, and differences in solubility potentially triggering phase separation during formulation or storage. Current evidence suggests that these compositional differences affect biological activity (95,96,97,98). For example, Zhang et al. found that even though two LNP batches displayed similar bulk properties, one batch was homogenous in composition whilst the other was heterogenous. After fractionation, the two batches displayed significantly different in vitro behaviour (98). However, there are limited studies on the impact of particle-to-particle compositional heterogeneity on biological behaviour, highlighting the need for single particle characterisation of nanoformulations to understand the impact of heterogeneity on biological behaviour.
Nanocarrier composition has been studied by FCS/FCCS and nFCM. The multiplexing ability of FCCS was used to confirm that nanoparticles prepared from block polymers contained three orthogonal reactive sites: one for cross-linking to ensure particle formation and two additional sites for the covalent addition of different chemical moieties (Fig. 3a) (99). nFCM was used to characterise hybrid nanoparticles prepared from thermosensitive liposomes and extracellular vesicles that were genetically engineered to overexpress a CD47 marker, which is a potent marker that signals to macrophages not to consume that object. Labelling either the liposomes or CD47 allowed determination that the fusion efficiency was 95.7% of particles (Fig. 3b) or that 56.8% hybrid particles prepared from genetically engineered extracellular vesicles contained CD47 compared to only 5.4% of nanoparticles fused with wild-type extracellular vesicles respectively (100). Although there has been some work using nFCM to study extracellular vesicle composition, there has been relatively limited application to nanoformulations, despite these methods being applicable (101). However, a key limitation is that nFCM is currently limited to two dye colours.
Single particle characterisation of nanocarrier composition. a (i) Auto-correlation and cross-correlation curves from FCS analysis of polymer (PSar-b-PCys(SO2Et)) self-assembly into micelles, followed by cross-linking at one site and covalent labelling of 2 additional orthogonal reactive sites with DBCO-CR110 and Cy5-NHS labels respectively. Reproduced with permission from Schäfer et al. (99). Copyright 2017 American Chemical Society. b nFCM analysis of hybrid liposome-exosome particles. Comparing the side scatter (SS-H) and fluorescence (FITC-H) plots for (i) labelled liposomes alone and (ii) hybrid particles where only the liposomes were labelled before fusion confirms that fusion efficiency was 95.7%. Reproduced from Lv et al. (100). c STORM images of temperature-responsive, core-shell nanogels. Labelling of (i) both core and shell, (ii) only core region, and (iii) only shell region. (iv) Correlation between core radius and shell thickness for each particle from STORM analysis. Selectivity was achieved by conjugating Alexa 647 dye derivatives to either primary amines in the core or disulfide moieties in the shell. Diffraction-limited images are shown in the top left corner. Scale bars 500 nm. Adapted with permission from Gelissen et al. (91). Copyright 2016 American Chemical Society. d SPARTA® analysis of polymer-lipid hybrid particles. (i) Schematic representation of PDLLA-b-PAA nanoparticle modification with aminomethanesulfonic acid (AMSA) and 5-amino-2-methoxybenzenesulfonic acid (AMBS). (ii) Mean Raman spectra with standard deviation of single particle traps. Green bands highlight polymer-specific peaks at 1032 and 1613 cm−1, and grey band denotes PEG-specific band at 851 cm−1. (iii) Comparison of single particle polymer and lipid intensity ratios for polymer-containing samples (PEG-0.3 and noPEG), PEGylated and lipid-only particles (PEGonly and POPC-Chol). Adapted from Najer et al. (16)
The high spatial resolution of SMLM (~20 nm) (47) has been used to study phase separation in polymeric cylindrical micelles and core/shell thermo-responsive nanogels (Fig. 3c) (51, 91, 92, 102). However, investigations into nanocarrier composition with SMLM are currently limited by available fluorophores. Many fluorophores traditionally used for SMLM are optimised for specific aqueous buffers (103). However, studying nanocarrier composition often requires staining of hydrophobic components, which can change key fluorophore behaviours such as quantum yield and blinking on/off ratio. Therefore, there has recently been increased focus on developing new dyes suitable for staining hydrophobic environments, including spiropyrans and inorganic nanoparticles (92, 103). Whilst this has allowed some studies of nanocarrier composition with SMLM (51, 91, 92, 102), progress to measure a wider range of systems and to obtain component stoichiometry is currently limited by further development and understanding of fluorophore behaviour in hydrophobic environments.
SPARTA® provides a label-free alternative to study nanocarrier composition. The inherent multiplexing ability of SPARTA® allowed simultaneous characterisation of the variation in both polyethylene glycol (PEG) stabiliser and pathogen-binding polymer components in different hybrid polymer-lipid nanoformulations (16). These results confirmed homogeneity in particle composition with the solvent injection preparation method used, which led to longer circulation time for particles formulated with PEG compared to without PEG, for the purpose of malaria parasite and virus inhibition (Fig. 3d). SPARTA® was also used to compare polymersomes made from a binary mixture of triblock copolymers where one copolymer had a much longer stabiliser chain (71). SPARTA® revealed that there was only one population of particles formed for all mixing ratios of the copolymers, and that the stabiliser signal increased with increasing initial ratio, indicating good incorporation into the particles. The proportion of stabiliser directly influenced biological behaviour, with a higher proportion of stabiliser decreasing cellular uptake and prolonging circulation time.
Cargo Loading and Release
A key parameter to characterise in discovery and regulatory settings is cargo loading and release. Bulk characterisation methods, such as HPLC or UV-Vis spectroscopy, typically measure either encapsulation efficiency (the proportion of drug loaded into the particle), or drug loading capacity (the proportion of drug component relative to the whole particle by weight). Bulk-level release studies are typically performed with a dialysis method, where released material is sampled and quantified (104). However, for optimising nanoformulations, it is also key to characterise how homogenous the drug loading per particle is. This includes the proportion of unloaded particles and the variation in release kinetics across the particle population. These parameters have direct impact on nanoformulation performance, because a high percentage of unloaded particles could reduce nanoformulation efficacy and contribute to immunogenic load, whilst a high proportion of particles experiencing burst release could increase off-target effects. Furthermore, single particle analysis has revealed significant cargo loading heterogeneity in diverse systems from anticancer-loaded polymeric particles to proteoliposomes, where only a small percentage of particles have drug loading properties represented by the “average” value (105,106,107). Therefore, only certain fractions of heterogenous nanoformulations will have the desired therapeutic properties, which could impact on toxicity and efficacy. It is therefore crucial to characterise cargo loading and release on the single particle level, also to allow next-generation rational design of improved nanoformulations.
nFCM, FCS, and SMLM are well suited to study cargo loading and release due to their high sensitivity. nFCM was used to determine that close to 100% of liposomes were loaded with doxorubicin in Doxoves, which is a research-grade, PEGylated liposomal nanoformulation (44). This information was combined with nanoparticle size information to determine the distribution of drug loading with particle size. The multiplexing ability of nFCM was used to measure the release of the fluorescent drug mitoxantrone from responsive liposomes in the presence of a peroxidation reporter molecule (Fig. 4a) (93). FCCS also has multiplexing ability to measure multiple fluorescent species, which was used to confirm co-loading of two different enzymes in a dendrimersome cascade nanoreactor (17), whilst FCS was used to simultaneously measure the decrease in nanoparticle size and drug loading (78). FCS and FCCS release studies have been performed in complex environments including 10% v/v FBS and whole blood (Fig. 4b) (78, 88). Furthermore, SMLM has been applied to study the intracellular trafficking and release of nucleic acid-loaded polyplexes in cells (108, 109).
Characterisation of cargo loading and release using single particle techniques. a nFCM analysis of liposomes functionalised with a ROS sensor and loaded with the fluorescent drug mitoxantrone. Upon addition of hydrogen peroxide, ROS sensing and cargo release could simultaneously be detected on a single particle level. (i) 3D scatter plot of side scattering, green and red fluorescence before (red dots) and after (green dots) addition of hydrogen peroxide. Change in (ii) green fluorescence from ROS sensor and (iii) red fluorescence from drug release. Reprinted from Chen et al. (93), with permission from Elsevier. b Simultaneous measurement of nanocarrier degradation and cargo release in blood with FCS. (i) Schematic of sample setup which utilises a semi-permeable membrane that allows nanoparticles to permeate, but prevents larger blood components such as cells from entering the probe volume. (ii) Autocorrelation curves of degradable nanocarriers measured between 0 and 72 h after intravenous injection into a mouse. (iii) Resulting calculation of the number of dyes per nanocarrier over time. Reproduced from Schmitt et al. (78). c SPARTA® measurements of cargo loading heterogeneity in polymer vesicles loaded with increasing amounts (0.1, 0.5, 1, 1.5, or 2 mM feed amount) of 2 different model cargo molecules. (i) Single particle scatter plots of polymer signal (x-axis) and cargo signal (y-axis). Top row corresponds to increasing loading from one model cargo whilst bottom row are samples from another cargo. Darker points are outlying particles, as determined by multivariate analysis: at 0.1 mM there is only one loading population which is distinguished by particle size whilst a distinct population appears for both cargoes at higher loading amounts. The proportion of these highly loaded particles is summarised in (ii). Adapted from Saunders et al. (89)
The intrinsic label-free, simultaneous detection of carrier and cargo molecules by Raman spectroscopy is highly suitable to study cargo loading and release. Raman spectroscopy has previously been used to correlate drug signal with increases in lipid disorder to determine the degree of bilayer disruption when incubating different drugs with liposomes (64, 110, 111). In addition to comparing loading of deuterated glucose in leaky dendrimersomes and non-leaky liposomes (17), SPARTA® could distinguish different loading sub-populations in polymersomes, including highly loaded particles (Fig. 4c) (89). Furthermore, SPARTA® could distinguish core and membrane loading in nanovesicles, based on the relationship between the cargo and nanocarrier signals detected simultaneously. This analysis was also shown to be applicable to commercial nanoformulations by measuring drug retention of doxorubicin within Doxil over a 48-hour period (89).
Surface Functionalisation and Interactions
The biodistribution, stability, and biological interactions of therapeutic nanoparticles for controlled delivery strongly depend on nanoparticle surface chemistry, including surface charge and functionalisation (3, 112). Nanoparticles with no or low charge tend to aggregate, whilst highly charged particles maintain inter-particle repulsion allowing for stable particle suspensions (62). Furthermore, nanoparticle surface charge influences electrostatic interaction with proteins and cell surfaces, modulating biostability, cellular uptake, and the accompanied residence time in circulation (3, 70). Generally, there is a trade-off between the two: a negative or neutral charge for circulation and enhanced biostability and positive charge for uptake (113, 114). Adjei et al. found that separate injection of either neutral or anionic polymeric nanoparticles led to accumulation in the kidneys or the liver respectively, whereas simultaneous co-injection altered the biodistribution (115), highlighting the influence of population heterogeneity on biological behaviour. In addition to controlling surface charge, nanoparticle surfaces are commonly functionalised with moieties to induce targeted cellular uptake for therapeutic delivery. These molecules include specific antigen recognition molecules such as antibodies (116), peptides (117), affibodies (118), aptamers (119), or general receptor ligands (120, 121) that target receptors that are overexpressed on the surface of the diseased cells. For example, as folate receptors are commonly overexpressed in over 40% of human cancers, folic acid surface-functionalised nanoparticles have shown improved general targeting towards cancer cells (122).
Single particle surface charge is relatively challenging to characterise; however, TRPS has been used to study single particle zeta potential, which can be used to indicate surface charge (62), as well as to infer changes to surface functionalisation based on changes to zeta potential. TRPS was used to compare liposomes formed from mixing varying ratios of zwitterionic and anionic lipids (Fig. 5a) (58). Compared to the bulk ensemble technique phase analysis light scattering (PALS), TRPS could distinguish multimodal surface charge populations and an increase in variance in zeta potential for DNA-modified particles compared to unmodified particles. TRPS could also identify differences in the distribution of zeta potential amongst unPEGylated and PEGylated liposomes, which were not evident from their respective size distributions (57). These measurements enabled the assessment that all the liposomes incorporated some PEG, but the degree of PEGylation was not homogenous. However, TRPS is limited to measuring nanoparticle size, concentration, and zeta potential, whereas often more detailed nanoformulation characterisation is required.
Characterisation of single particle surface charge, functionalisation, and interactions. a TRPS simultaneous size and zeta potential characterisation of liposomes formed with increasing amount of negatively charged DMPG lipid. Adapted from Vogel et al. (58). b Characterisation of streptavidin-cyanin5 (SA-Cy5) (red) ligand conjugation to rhodamine-green (RhG) labelled biotin-functionalised polymer nanocontainers (NCs) (green) by FCS/FCCS. (i) Autocorrelation curves (red and green) and cross correlation curve (black). (ii) Using the relative correlation amplitudes and the detection volume, the fraction bound SA−Cy5 against the total concentration of NCs was determined. Reprinted with permission from Rigler et al. (55). Copyright 2006 American Chemical Society. c Ligand conjugation density characterisation by nFCM. (i) Bivariate dot-plot and histograms of folate ligand number against particle size for folate-conjugated liposomes. (ii) Distribution of folate ligand density for folate-conjugated liposomes with various folate input concentrations (0.05 to 4.0%). (iii) Quantification of the available transferrin (Tf) ligand density for transferrin-conjugated liposomes with different Tf input concentrations. Reprinted with permission from Chen et al. (80). Copyright 2022 American Chemical Society. d Ligand conjugation density characterisation by SRM. (i) STORM DNA-PAINT image (scale bar 1000 nm) and a small field (upper left, scale bar 100 nm) of PLGA–PEG 30% maleimide nanoparticles conjugated to thiol-DNA 9-mer oligonucleotides labelled using complementary DNA-PAINT (red) and Dil signal (yellow) used for drift correction. Normalised frequency histograms of DNA-PAINT localisations per NP and bar graph depicting the number of ligands per nanoparticle quantified with PAINT (upper right) for PLGA–PEG formulated with (ii) 5% and (iii) 30% maleimide. Adapted from Andrian et al. (51). e (i) Schematic of stimuli-responsive changes in lipid nanoparticle headgroup composition after addition of phospholipase D (PLD). (ii) Corresponding kinetic SPARTA® analysis of headgroup compositional changes from choline to phosphatidic acid after enzyme addition at different concentrations. Each data point represents a single particle measurement and a new trapping event, whilst the dashed lines represent first order kinetics fit of PLD activity, with fitting parameter time to 50% choline conversion shown in (iii). Adapted from Barriga et al. (69)
Single particle surface functionalisation has been characterised using nFCM (80), FCS (55, 123), SRM (51), and TRPS (61). FCS/FCCS can be employed for estimating the number of ligands per nanoparticle and accompanying receptor binding, for example measuring the kinetics of streptavidin binding to biotin-functionalised polymer nanoparticles (Fig. 5b) (55). nFCM has been used to determine the ligand density on folate- (Fig. 5c I, II), transferrin- (Fig. 5c III), and HER2-antibody-conjugated liposomes (80). Using this same method enabled optimisation of the conjugation reaction conditions, including the PEG spacer length and ligand concentration for maximum ligand density and cell uptake. DNA-based Point Accumulation in Nanoscale Topography (DNA-PAINT), an SMLM tool, was combined with STORM to characterise the number of thiol-DNA 9-mer oligonucleotides conjugated to nanoparticles formulated with varying concentrations of maleimide (Fig. 5d) (51).
One key biological interaction is protein fouling, or protein corona formation, where proteins adsorb to the nanoparticle surface upon exposure to serum. This protein corona can cause instability, immune system recognition, or potentially beneficial adsorption of dysopsonins for prolonging circulation time (124). Feiner-Gracia et al. found that within a nanoparticle population there were significant differences in protein corona radius and identity, leading to the conclusion that even small differences in native nanoparticle surface chemistry were amplified upon protein adsorption to yield particles with distinct biological identity (84). Therefore, as was recently recognised, a better understanding and modulation of the complex surface interactions of nanomedicines in biological environments is key for aiding clinical translation (107, 125). Utilising single particle characterisation techniques for these properties enables detailed measurement of population variance, especially arising from surface modification processes and biological interactions for improved control and design of uniform nanoformulations.
Protein corona formation has been characterised using FCS and SMLM. Increases in radius measured by FCS revealed information about the orientation and thickness of corona proteins on nanoparticle surfaces, whilst the equilibrium dissociation coefficients provided information on the protein binding affinity and kinetics (85). An alternative FCS technique is to randomly label plasma and then to mix with unlabelled nanoparticles to detecting protein fouling, which is a highly sensitive method but requires using diluted plasma or studying individually labelled plasma proteins (70). Nevertheless, observed differences between PEG-decorated polymersomes and liposomes provided important links between the copolymer membrane material properties, particle PEG-density and resulting corona formation. Meanwhile, staining specific serum proteins with multiple fluorophores for SMLM visualisation enables quantification of the relative protein content in the corona formed on individual nanoparticle (126).
Besides protein corona formation, there are many other nanoparticle-biological interactions that can be studied on a single particle level, including enzymatic changes to surface chemistry. SPARTA® was used to characterise the kinetics of enzymatic conversion of lipid headgroups from phosphatidylcholine to phosphatidic acid on a single particle level in Barriga et al. (69), including reaction kinetics (Fig. 5e). The ability to characterise stimuli-responsive compositional changes on the surface of nanoparticles provides the opportunity for optimisation of “smart” nanocarriers for targeted delivery prior to in vivo studies. Furthermore, SPARTA® was used to characterise a multi-step surface click chemistry reaction on polystyrene nanoparticles, both by holding a single particle in the optical trap over an extended time (~2 mins) and by sequentially trapping particles for shorter time (~s) to measure kinetics within a population (27). This not only provided a particle-by-particle population distribution of the degree of successful surface functionalisation, but also kinetic information on the rate of functionalisation. In the same study, SPARTA® provided single particle verification of sequential disulfide exchange surface functionalisation of polystyrene nanoparticles.
Conclusions
Nanoformulations offer a versatile platform for drug delivery to improve biodistribution, biostability and targeting potential compared to traditional small molecule drugs. However, their inherent particle-to-particle heterogeneity in key chemical properties such as drug loading and surface chemistry has been found to influence biological activity including protein corona formation, biodistribution and in vitro dose-response behaviour (84, 98, 115). This heterogeneity is not accounted for by bulk analysis methods, which can only return an average value, and where particles with the “average” property may not exist. Therefore, there is a need to introduce more extensive, in-depth single particle analysis to understand and predict nanoformulation in vivo behaviour. When selecting a technique, the requirements must be set out, including required sensitivity, kinetic information, complexity of environment and available labelling methods. Whilst fluorescence-based techniques such as FCS or nFCM offer high sensitivity, often combined with simple data readout and low sample volumes, the need for labelling can increase sample preparation time and limits what features can be characterised. Label-free single particle methods include the sensitive TRPS; however, this is limited to measuring only size, concentration, and zeta potential. Whilst SPARTA® requires more involved data analysis and has lower sensitivity, it offers a label-free, holistic view of all nanoformulation components above the limit of detection. This allows for agnostic, simultaneous, single particle characterisation of cargo loading, nanocarrier composition, and surface chemistry which reveals relationships between properties. This flexibility places SPARTA® as a useful tool in a nanoformulation development workflow.
References
Hou X, Zaks T, Langer R, Dong Y. Lipid nanoparticles for mRNA delivery. Nat Rev Mater. 2021;6:1078–94.
Rezvantalab S, Drude NI, Moraveji MK, Guvener N, Koons EK, Shi Y, et al. PLGA-based nanoparticles in cancer treatment. Front Pharmacol. 2018;9:1260.
Mitchell MJ, Billingsley MM, Haley RM, Wechsler ME, Peppas NA, Langer R. Engineering precision nanoparticles for drug delivery. Nat Rev Drug Discov. 2021;20(2):101–24.
Wais U, Jackson AW, He T, Zhang H. Nanoformulation and encapsulation approaches for poorly water-soluble drug nanoparticles. Nanoscale. 2016;8:1746–69.
Soares S, Sousa J, Pais A, Vitorino C. Nanomedicine: principles, properties, and regulatory issues. Front Chem. 2018;6:360.
Sayes CM, Aquino GV, Hickey AJ. Nanomaterial drug products: manufacturing and analytical perspectives. AAPS J. 2017;19(1):18–25.
Shi J, Kantoff PW, Wooster R, Farokhzad OC. Cancer nanomedicine: progress, challenges and opportunities. Nat Rev Cancer. 2017;17:20–37.
Rafiyath SM, Rasul M, Lee B, Wei G, Lamba G, Liu D. Comparison of safety and toxicity of liposomal doxorubicin vs. conventional anthracyclines: a meta-analysis. Exp Hematol Oncol. 2012;1:10.
Lui P, Chen G, Zhang J. A review of liposomes as a drug delivery system: current status of approved products, regulatory environments, and future perspectives. Molecules. 2022;27:1372.
Stone NRH, Bicanic T, Salim R, Hope W. Liposomal amphotericin B (AmBisome®): a review of the pharmacokinetics, pharmacodynamics, clinical experience and future directions. Drugs. 2016;76(4):485–500.
Akinc A, Maier MA, Manoharan M, Fitzgerald K, Jayaraman M, Barros S, et al. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat Nanotechnol. 2019;14:1084–7.
Schoenmaker L, Witzigmann D, Kulkarni JA, Verbeke R, Kersten G, Jiskoot W, et al. mRNA-lipid nanoparticle COVID-19 vaccines: structure and stability. Int J Pharm. 2021;601: 120586.
Hou X, Zaks T, Langer R, Dong Y. Lipid nanoparticles for mRNA delivery. Nat Rev Mater. 2021;6:1078–94.
Warne N, Ruesch M, Siwik P, Mensah P, Ludwig J, Hripcsak M, et al. Delivering 3 billion doses of Comirnaty in 2021. Nat Biotechnol. 2023;41(2):183–8.
Discher BM, Won Y-Y, Ege DS, Lee JC, Bates FS, Discher DE, et al. Polymersomes: tough vesicles made from diblock copolymers. Science (80- ). 1999;284:1143–7.
Najer A, Blight J, Ducker CB, Gasbarri M, Brown JC, Che J, et al. Potent virustatic polymer−lipid nanomimics block viral entry and inhibit malaria parasites in vivo. ACS Cent Sci. 2022;8(9):1238–57.
Potter M, Najer A, Klöckner A, Zhang S, Holme MN, Nele V, et al. Controlled dendrimersome nanoreactor system for localized hypochlorite-induced killing of bacteria. ACS Nano. 2020;14:17333–53.
Anselmo AC, Zhang M, Kumar S, Vogus DR, Menegatti S, Helgeson ME, et al. Elasticity of nanoparticles influences their blood circulation, phagocytosis, endocytosis, and targeting. ACS Nano. 2015;9(3):3169–77.
Komsthöft T, Bovone G, Bernhard S, Tibbitt MW. Polymer functionalization of inorganic nanoparticles for biomedical applications. Curr Opin Chem Eng. 2022;37: 100849.
Souto EB, Silva GF, Dias-Ferreira J, Zielinska A, Ventura F, Durazzo A, et al. Nanopharmaceutics: Part I- Clinical Trials Legislation and Good Manufacturing Practices (GMP) of nanotherapeutics in the EU. Pharmaceutics. 2020;12:146.
Albanese A, Tang PS, Chan WCW. The effect of nanoparticle size, shape, and surface chemistry on biological systems. Annu Rev Biomed Eng. 2012;14:1–16.
Harush-Frenkel O, Rozentur E, Benita S, Altschuler Y. Surface charge of nanoparticles determines their endocytic and transcytotic pathway in polarized MDCK cells. Biomacromolecules. 2008;9:435–43.
Meng H, Yang S, Li Z, Xia T, Chen J, Ji Z, et al. Aspect ratio determines the quantity of mesoporous silica nanoparticle uptake by a small gtpase-dependent macropinocytosis mechanism. ACS Nano. 2011;5(6):4434–47.
Mühlebach S. Regulatory challenges of nanomedicines and their follow-on versions: a generic or similar approach? Adv Drug Deliv Rev. 2018;131:122–31.
Fraikin J-L, Teesalu T, Mckenney CM, Ruoslahti E, Cleland AN. A high-throughput label-free nanoparticle analyser. Nat Nanotechnol. 2011;6:308–13.
Lignos I, Stavrakis S, Kilaj A, DeMello AJ. Millisecond-timescale monitoring of PbS nanoparticle nucleation and growth using droplet-based microfluidics. Small. 2015;11(32):4009–17.
Penders J, Pence IJ, Horgan CC, Bergholt MS, Wood CS, Najer A, et al. Single particle automated Raman trapping analysis. Nat Commun. 2018;9:4256.
Xu M, Soliman MG, Sun X, Pelaz B, Feliu N, Parak WJ, et al. How entanglement of different physicochemical properties complicates the prediction of in vitro and in vivo interactions of gold nanoparticles. ACS Nano. 2018;12(10):10104–13.
Palomba R, Palange AL, Rizzuti IF, Ferreira M, Cervadoro A, Barbato MG, et al. Modulating phagocytic cell sequestration by tailoring nanoconstruct softness modulating phagocytic cell sequestration by tailoring nanoconstruct softness. ACS Nano. 2018;12(2):1433–44.
Modena MM, Rühle B, Burg TP, Wuttke S. Nanoparticle characterization: what to measure? Adv Mater. 2019;31:1901556.
Chen C, Chen C, Li Y, Gu R, Yan X. Characterization of lipid-based nanomedicines at the single-particle level. Fundam Res. 2023;3(4):488–504
Gu F, Zhang L, Teply BA, Mann N, Wang A, Radovic-Moreno AF, et al. Precise engineering of targeted nanoparticles by using self-assembled biointegrated block copolymers. Proc Natl Acad Sci U S A. 2008;105(7):2586–91.
Caracciolo G, Pozzi D, Capriotti AL, Cavaliere C, Piovesana S, Amenitsch H, et al. Lipid composition: a “key factor” for the rational manipulation of the liposome-protein corona by liposome design. RSC Adv. 2015;5(8):5967–75.
Anselmo AC, Mitragotri S. Nanoparticles in the clinic : an update. Bioeng Transl Med. 2019;4: e10143.
Zhou J, Chizhik AI, Chu S, Jin D. Single-particle spectroscopy for functional nanomaterials. Nature. 2020;579:41–50.
Mourdikoudis S, Pallares RM, Thanh NTK. Characterization techniques for nanoparticles: comparison and complementarity upon studying. Nanoscale. 2018;10:12871–934.
Brader ML, Williams SJ, Banks JM, Hui WH, Zhou ZH, Jin L. Encapsulation state of messenger RNA inside lipid nanoparticles. Biophys J. 2021;120:2766–70.
Ebner A, Wildling L, Gruber J. Functionalization of AFM tips and supports for molecular recognition force spectroscopy and recognition imaging. In: Santos N, Carvalho F, editors. Atomic force microscopy methods in molecular biology, vol 1886. New York, NY: Humana Press; 2019.
Malatesta M. Transmission electron microscopy for nanomedicine: novel applications for long-established techniques Ultrastructural morphology for nanotechnology. Eur J Histochem. 2016;60(4):2751.
Zhu S, Ma L, Wang S, Chen C, Zhang W, Yang L, et al. Light-scattering detection below the level of single fluorescent molecules for high-resolution characterization of functional nanoparticles. ACS Nano. 2014;8(10):10998–1006.
Tian Y, Ma L, Gong M, Su G, Zhu S, Zhang W, et al. Protein profiling and sizing of extracellular vesicles from colorectal cancer patients via flow cytometry. ACS Nano. 2018;12:671–80.
Ma L, Zhu S, Tian Y, Zhang W, Wang S, Chen C, et al. Label-free analysis of single viruses with a resolution comparable to that of electron microscopy and the throughput of flow cytometry. Angew Chemie - Int Ed. 2016;55:10239–43.
Chen C, Zhu S, Wang S, Zhang W, Cheng Y, Yan X. Multiparameter quantification of liposomal nanomedicines at the single-particle level by high-sensitivity flow cytometry. ACS Appl Mater Interfaces. 2017;9:13913–9.
Zhu S, Ma L, Wang S, Chen C, Zhang W, Yang L, et al. Light-scattering detection below the level of single fluorescent molecules for high-resolution characterization of functional nanoparticles. ACS Nano. 2014;8(10):10998–1006.
Dooley K, McConnell RE, Xu K, Lewis ND, Haupt S, Youniss MR, et al. A versatile platform for generating engineered extracellular vesicles with defined therapeutic properties. Mol Ther. 2021;29(5):1729–43.
Chen X, Wang Y, Zhang X, Liu C. Advances in super-resolution fluorescence microscopy for the study of nano–cell interactions. Biomater Sci. 2021;9(16):5484–96.
Lelek M, Gyparaki MT, Beliu G, Schueder F, Griffié J, Manley S, et al. Single-molecule localization microscopy. Nat Rev Methods Prim. 2021;1:39.
Valli J, Garcia-Burgos A, Rooney LM, Vale de Melo B, Duncan RR, Rickman C. Seeing beyond the limit: a guide to choosing the right super-resolution microscopy technique. J Biol Chem. 2021;297(1):100791.
Davis JL, Zhang Y, Yi S, Du F, Song KH, Scott EA, et al. Super-resolution imaging of self-assembled nanocarriers using quantitative spectroscopic analysis for cluster extraction. Langmuir. 2020;36(9):2291–9.
Pujals S, Albertazzi L. Super-resolution microscopy for nanomedicine research. ACS Nano. 2019;13(9):9707–12.
Andrian T, Delcanale P, Pujals S, Albertazzi L. Correlating super-resolution microscopy and transmission electron microscopy reveals multiparametric heterogeneity in nanoparticles. Nano Lett. 2021;21(12):5360–8.
Schmitt S, Nuhn L, Barz M, Butt H, Koynov K. Shining light on polymeric drug nanocarriers with fluorescence correlation spectroscopy. Macromol Rapid Commun. 2022;43(12):e2100892
Yu L, Lei Y, Ma Y, Liu M, Zheng J, Dan D, et al. A comprehensive review of fluorescence correlation spectroscopy. Front Phys. 2021;9: 644450.
Rigler R, Mets U, Widengren J, Kask P. Fluorescence correlation spectroscopy with high count rate and low background: analysis of translational diffusion. Eur Biophys J. 1993;22:169–75.
Rigler P, Meier W. Encapsulation of fluorescent molecules by functionalized polymeric nanocontainers: investigation by confocal fluorescence imaging and fluorescence correlation spectroscopy. J Am Chem Soc. 2006;128:367–73.
Hill AF. Exosomes and microvesicles: methods and protocols [Internet]. 1st ed. New York: Hill, Andrew F; 2017.
Kozak D, Broom M, Vogel R. High resolution particle characterization to expedite development and regulatory acceptance of nanomedicines. Curr Drug Deliv. 2014;12(1):115–20.
Vogel R, Pal AK, Jambhrunkar S, Patel P, Thakur SS, Reátegui E, et al. High-resolution single particle zeta potential characterisation of biological nanoparticles using tunable resistive pulse sensing. Sci Reports. 2017;7(1):1–13.
Pal AK, Aalaei I, Gadde S, Gaines P, Schmidt D, Demokritou P, et al. High resolution characterization of engineered nanomaterial dispersions in complex media using tunable resistive pulse sensing technology. ACS Nano. 2014;8(9):9003–15.
Blundell ELCJ, Healey MJ, Holton E, Sivakumaran M, Manstana S, Platt M. Characterisation of the protein corona using tunable resistive pulse sensing: determining the change and distribution of a particle’s surface charge. Anal Bioanal Chem. 2016;408:5757–68.
Kozak D, Anderson W, Vogel R, Chen S, Antaw F, Trau M. Simultaneous size and ζ-potential measurements of individual nanoparticles in dispersion using size-tunable pore sensors. ACS Nano. 2012;6(8):6990–7.
Bhattacharjee S. DLS and zeta potential – what they are and what they are not? J Control Release. 2016;235:337–51.
Smith E, Dent G. Modern Raman spectroscopy-a practical approach. Chichester (UK): John Wiley and Sons Ltd; 2005.
Fox CB, Harris JM. Confocal Raman microscopy for simultaneous monitoring of partitioning and disordering of tricyclic antidepressants in phospholipid vesicle membranes. J Raman Spectrosc. 2010;41:498–507.
Sanderson JM, Ward AD. Analysis of liposomal membrane composition using Raman tweezers. Chem Commun. 2004;4(9):1120–1.
Cherney DP, Conboy JC, Harris JM. Optical-trapping Raman microscopy detection of single unilamellar lipid vesicles. Anal Chem. 2003;75(23):6621–8.
Cherney DP, Bridges TE, Harris JM. Optical trapping of unilamellar phospholipid vesicles: investigation of the effect of optical forces on the lipid membrane shape by confocal-Raman microscopy. Anal Chem. 2004;76(17):4920–8.
Carney RP, Hazari S, Colquhoun M, Tran D, Hwang B, Mulligan MS, et al. Multispectral optical tweezers for biochemical fingerprinting of CD9-positive exosome subpopulations. Anal Chem. 2017;89(10):5357–63.
Barriga HMG, Pence IJ, Holme MN, Doutch JJ, Penders J, Nele V, et al. Coupling lipid nanoparticle structure and automated single-particle composition analysis to design phospholipase-responsive nanocarriers. Adv Mater. 2022;34(26):2200839.
Najer A, Rifaie-Graham O, Yeow J, Adrianus C, Chami M, Stevens MM. Differences in human plasma protein interactions between various polymersomes and stealth liposomes as observed by fluorescence correlation spectroscopy. Macromol Biosci. 2022;23(8):2200424.
Najer A, Belessiotis-Richards A, Kim H, Saunders C, Fenaroli F, Adrianus C, et al. Block length-dependent protein fouling on poly (2-oxazoline)-based polymersomes: influence on macrophage association and circulation behavior. Small. 2022;18:2201993.
Penders J, Nagelkerke A, Cunnane EM, Pedersen SV, Pence IJ, Coombes RC, et al. Single particle automated Raman trapping analysis of breast cancer cell-derived extracellular vesicles as cancer biomarkers. ACS Nano. 2021;15:18192–205.
Elson EL. Fluorescence correlation spectroscopy: past, present, future. Biophys J. 2011;101:2855–70.
Lian H, He S, Chen C, Yan X. Flow cytometric analysis of nanoscale biological particles and organelles. Annu Rev Anal Chem. 2019;12:389–409.
Diekmann R, Kahnwald M, Schoenit A, Deschamps J, Matti U, Ries J. Optimizing imaging speed and excitation intensity for single molecule localization microscopy. Nat Methods. 2020;17:909–12.
Reismann AWAF, Atanasova L, Schrangl L, Zeilinger S, Schütz GJ. Temporal filtering to improve single molecule identification in high background samples. Molecules. 2018;23:3338.
Fortunato D, Mladenović D, Criscuoli M, Loria F, Veiman K-L, Zocco D, et al. Opportunities and pitfalls of fluorescent labeling methodologies for extracellular vesicle profiling on high-resolution single-particle platforms. Int J Mol Sci. 2021;22:10510.
Schmitt S, Huppertsberg A, Klefenz A, Kaps L, Mailänder V, Schuppan D, et al. Fluorescence correlation spectroscopy monitors the fate of degradable nanocarriers in the blood stream. Biomacromolecules. 2022;23:1065–74.
Dai Y, Bai S, Hu C, Chu K, Shen B, Smith ZJ. Combined morpho-chemical profiling of individual extracellular vesicles and functional nanoparticles without labels. Anal Chem. 2020;92:5585–94.
Chen C, Zhou Y, Chen C, Zhu S, Yan X. Quantification of available ligand density on the surface of targeted liposomal nanomedicines at the single-particle level. ACS Nano. 2022;16(4):6886–97.
Münter R, Kristensen K, Pedersbæk D, Larsen JB, Simonsen JB, Andresen TL. Dissociation of fluorescently labeled lipids from liposomes in biological environments challenges the interpretation of uptake studies. Nanoscale. 2018;10(48):22720–4.
Battaglini M, Feiner N, Tapeinos C, De Pasquale D, Pucci C, Marino A, et al. Combining confocal microscopy, dSTORM, and mass spectroscopy to unveil the evolution of the protein corona associated with nanostructured lipid carriers during blood–brain barrier crossing. Nanoscale. 2022;14:13292–307.
Woythe L, Porciani D, Harzing T, Van Veen S, Burke DH, Albertazzi L. Valency and affinity control of aptamer-conjugated nanoparticles for selective cancer cell targeting. J Control Release. 2023;355:228–37.
Feiner-Gracia N, Beck M, Pujals S, Tosi S, Mandal T, Buske C, et al. Super-resolution microscopy unveils dynamic heterogeneities in nanoparticle protein corona. Small. 2017;13:1701631.
Shang L, Nienhaus GU. In situ characterization of protein adsorption onto nanoparticles by fluorescence correlation spectroscopy. Acc Chem Res. 2017;50(2):387–95
Negwer I, Best A, Schinnerer M, Schäfer O, Capeloa L, Wagner M, et al. Monitoring drug nanocarriers in human blood by near-infrared fluorescence correlation spectroscopy. Nat Commun. 2018;9:5306.
Kristensen K, Urquhart AJ, Thormann E, Andresen TL. Binding of human serum albumin to PEGylated liposomes : insights into binding numbers and dynamics by fluorescence correlation spectroscopy. Nanoscale. 2016;8:19726–36.
Bouchaala R, Richert L, Anton N, Vandamme TF, Djabi S, Mély Y, et al. Quantifying release from lipid nanocarriers by fluorescence correlation spectroscopy. ACS Omega. 2018;3:14333–40.
Saunders C, Foote JEJ, Wojciechowski JP, Cammack A, Pedersen SV, Doutch JJ, et al. Revealing population heterogeneity in vesicle-based nanomedicines using automated, single particle Raman analysis. ACS Nano. 2023;17(12):11713–28.
Tcherniak A, Reznik C, Link S, Landes CF. Fluorescence correlation spectroscopy: criteria for analysis in complex systems. Anal Chem. 2009;81(2):746–54.
Gelissen APH, Oppermann A, Caumanns T, Hebbeker P, Turnhoff SK, Tiwari R, et al. 3D structures of responsive nanocompartmentalized microgels. Nano Lett. 2016;16(11):7295–301.
Yan J, Zhao LX, Li C, Hu Z, Zhang GF, Chen ZQ, et al. Optical nanoimaging for block copolymer self-assembly. J Am Chem Soc. 2015;137(7):2436–9.
Chen C, Gao K, Lian H, Chen C, Yan X. Single-particle characterization of theranostic liposomes with stimulus sensing and controlled drug release properties. Biosens Bioelectron. 2019;131:185–92.
Orth A, Ghosh RN, Wilson ER, Doughney T, Brown H, Reineck P, et al. Super-multiplexed fluorescence microscopy via photostability contrast. Biomed Opt Express. 2018;9(7):2943–54.
Arpicco S, Battaglia L, Brusa P, Cavalli R, Chirio D, Dosio F, et al. Recent studies on the delivery of hydrophilic drugs in nanoparticulate systems. J Drug Deliv Sci Technol. 2016;32:298–312.
Carrasco MJ, Alishetty S, Alameh M, Said H, Wright L, Paige M, et al. Ionization and structural properties of mRNA lipid nanoparticles influence expression in intramuscular and intravascular administration. Commun Biol. 2021;4:956.
Sahay G, Alakhova DY, Kabanov AV. Endocytosis of nanomedicines. J Control Release. 2010;145(3):182–95.
Zhang J, Pei Y, Zhang H, Wang L, Arrington L, Zhang Y, et al. Assessing the heterogeneity level in lipid nanoparticles for sirna delivery: size-based separation, compositional heterogeneity, and impact on bioperformance. Mol Pharm. 2013;10(1):397–405.
Schäfer O, Klinker K, Braun L, Huesmann D, Schultze J, Koynov K, et al. Combining orthogonal reactive groups in block copolymers for functional nanoparticle synthesis in a single step. ACS Macro Lett. 2017;6:1140–5.
Lv Q, Cheng L, Lu Y, Zhang X, Wang Y, Deng J, et al. Thermosensitive exosome–liposome hybrid nanoparticle-mediated chemoimmunotherapy for improved treatment of metastatic peritoneal cancer. Adv Sci. 2020;7:2000515.
Morales-Kastresana A, Telford B, Musich TA, McKinnon K, Clayborn C, Braig Z, et al. Labeling extracellular vesicles for nanoscale flow cytometry. Sci Rep. 2017;7:1878.
Boott CE, Laine RF, Mahou P, Finnegan JR, Leitao EM, Webb SED, et al. In situ visualization of block copolymer self-assembly in organic media by super-resolution fluorescence microscopy. Chem - A Eur J. 2015;21(51):18539–42.
Wöll D, Flors C. Super-resolution fluorescence imaging for materials science. Small Methods. 2017;1:1700191.
Abouelmagd SA, Sun B, Chang AC, Ku YJ, Yeo Y. Release kinetics study of poorly water-soluble drugs from nanoparticles: are we doing it right ? Mol Pharamaceutics. 2015;12:997–1003.
Mathiasen S, Christensen SM, Fung JJ, Rasmussen SGF, Fay JF, Jorgensen SK, et al. Nanoscale high-content analysis using compositional heterogeneities of single proteoliposomes. Nat Methods. 2014;11:931–4.
Ural MS, Dartois E, Mathurin J, Desmaële D, Collery P, Dazzi A, et al. Quantification of drug loading in polymeric nanoparticles using AFM-IR technique: a novel method to map and evaluate drug distribution in drug nanocarriers. Analyst. 2022;147:5564–78.
Rabanel J-M, Adibnia V, Tehrani SF, Sanche S, Hildgen P, Banquy X, et al. Nanoparticle heterogeneity: an emerging structural parameter influencing particle fate in biological media? Nanoscale. 2019;11:383–406.
Wojnilowicz M, Glab A, Bertucci A, Caruso F, Cavalieri F. Super-resolution imaging of proton sponge-triggered rupture of endosomes and cytosolic release of small interfering RNA. ACS Nano. 2019;13:187–202.
Riera R, Feiner-Gracia N, Fornaguera C, Cascante A, Borrós S, Albertazzi L. Tracking the DNA complexation state of pBAE polyplexes in cells with super resolution. Nanoscale. 2019;11:17869–77.
Schaefer JJ, Crawford AC, Porter MD, Harris M. Confocal Raman microscopy for investigating synthesis and characterization of individual optically trapped vinyl- polymerized surfactant particles. 2014;68(6):633–41
Fox CB, Horton RA, Harris JM. Detection of drug-membrane interactions in individual phospholipid vesicles by confocal Raman microscopy. Anal Chem. 2006;78:4918–24.
Friedl JD, Nele V, De Rosa G, Bernkop-Schnürch A. Bioinert, stealth or interactive: how surface chemistry of nanocarriers determines their fate in vivo. Adv Funct Mater. 2021;31(34):2103347.
Blanco E, Shen H, Ferrari M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat Biotechnol. 2015;33(9):941–51.
Wang J, Chen G, Liu N, Han X, Zhao F, Zhang L, et al. Strategies for improving the safety and RNAi efficacy of noncovalent peptide/siRNA nanocomplexes. Adv Colloid Interface Sci. 2022;302: 102638.
Adjei IM, Peetla C, Labhastewar V. Heterogeneity in nanoparticles influences biodistribution and targeting. Nanomedicine (Lond). 2014;9(2):267–78.
Marques AC, Costa PC, Velho S, Amaral MH. Lipid nanoparticles functionalized with antibodies for anticancer drug therapy. Pharmaceutics. 2023;15:216.
Sharma R, Borah SJ, Bhawna, Kumar S, Gupta A, Singh P, et al. Functionalized peptide-based nanoparticles for targeted cancer nanotherapeutics: a state-of-the-art review. ACS Omega. 2022;7:36092–107.
Ståhl S, Gräslund T, Eriksson A, Frejd FY, Nygren P, Löfblom J. Affibody molecules in biotechnological and medical applications. Trends Biotechnol. 2017;35(8):691–712.
Jo H, Ban C. Aptamer–nanoparticle complexes as powerful diagnostic and therapeutic tools. Exp Mol Med. 2016;48: e230.
Yang A, Sun Z, Liu R, Liu X, Zhang Y, Zhou Y, et al. Transferrin-conjugated erianin- loaded liposomes suppress the growth of liver cancer by modulating oxidative stress. Front Oncol. 2021;11: 727605.
Narmani A, Rezvani M, Farhood B, Darkhor P, Mohammadnejad J, Amini B, et al. Folic acid functionalized nanoparticles as pharmaceutical carriers in drug delivery systems. Drug Dev Res. 2019;80:404–24.
Shen J, Hu Y, Putt KS, Singhal S, Han H, Visscher DW, et al. Assessment of folate receptor alpha and beta expression in selection of lung and pancreatic cancer patients for receptor targeted therapies. Oncotarget. 2018;9(4):4485–95.
Gonzalez-Carter D, Liu X, Tockary TA, Dirisala A, Toh K, Anraku Y, et al. Targeting nanoparticles to the brain by exploiting the blood-brain barrier impermeability to selectively label the brain endothelium. Proc Natl Acad Sci U S A. 2020;117(32):19141–50.
Bros M, Nuhn L, Simon J, Moll L, Mailänder V, Landfester K, et al. The protein corona as a confounding variable of nanoparticle-mediated targeted vaccine delivery. Front Immunol. 2018;9:1760.
Stater EP, Sonay AY, Hart C, Grimm J. The ancillary effects of nanoparticles and their implications for nanomedicine. Nat Nanotechnol. 2021;16:1180–94.
Wang Y, Soto Rodriguez PED, Woythe L, Sánchez S, Samitier J, Zijlstra P, et al. Multicolor super-resolution microscopy of protein corona on single nanoparticles. ACS Appl Mater Interfaces. 2022;14(33):37345–55.
Acknowledgements
We kindly acknowledge A. Najer for thorough reading of and critical suggestions for the text. For the purpose of open access, the authors have applied a CC BY public copyright license to any Author Accepted Manuscript version arising from this submission.
Funding
C.S. acknowledges funding from EPSRC Centre for Doctoral Training in the Advanced Characterisation of Materials (EP/S023259/1). M.M.S. acknowledges support from the Royal Academy of Engineering Chair in Emerging Technologies award (CiET2021\94).
Author information
Authors and Affiliations
Contributions
All authors have contributed to the conceptualisation of the work. C.S. and C.A.dV. have each written and edited individual sections. C.S. has provided synthesis and curation. M.M.S. has provided review, scientific editing, and supervision.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare the following competing financial interest(s): M.M.S. has filed a patent application (1810010.7) and has a registered trademark (US Reg. No. 6088213) covering the name SPARTA® and the techniques described in the manuscript by Penders et al. https://doi.org/10.1038/s41467-018-06397-6). M.M.S. is a founder of Sparta Biodiscovery Ltd.
Additional information
Communicated by Aliasger Salem
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Saunders, C., de Villiers, C.A. & Stevens, M.M. Single Particle Chemical Characterisation of Nanoformulations for Cargo Delivery. AAPS J 25, 94 (2023). https://doi.org/10.1208/s12248-023-00855-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1208/s12248-023-00855-w