Abstract
The mechanistic neuropharmacokinetic (neuroPK) model was established to predict unbound brain-to-plasma partitioning (Kp,uu,brain) by considering in vitro efflux activities of multiple drug resistance 1 (MDR1) and breast cancer resistance protein (BCRP). Herein, we directly compare this model to a computational machine learning approach utilizing physicochemical descriptors and efflux ratios of MDR1 and BCRP-expressing cells for predicting Kp,uu,brain in rats. Two different types of machine learning techniques, Gaussian processes (GP) and random forest regression (RF), were assessed by the time and cluster-split validation methods using 640 internal compounds. The predictivity of machine learning models based on only molecular descriptors in the time-split dataset performed worse than the cluster-split dataset, whereas the models incorporating MDR1 and BCRP efflux ratios showed similar predictivity between time and cluster-split datasets. The GP incorporating MDR1 and BCRP in the time-split dataset achieved the highest correlation (R2 = 0.602). These results suggested that incorporation of MDR1 and BCRP in machine learning is beneficial for robust and accurate prediction. Kp,uu,brain prediction utilizing the neuroPK model was significantly worse compared to machine learning approaches for the same dataset. We also investigated the predictivity of Kp,uu,brain using an external independent test set of 34 marketed drugs. Compared to machine learning models, the neuroPK model showed better predictive performance with R2 of 0.577. This work demonstrates that the machine learning model for Kp,uu,brain achieves maximum predictive performance within the chemical applicability domain, whereas the neuroPK model is applicable more widely beyond the chemical space covered in the training dataset.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
One of the biggest impediments in drug discovery to accomplish favorable pharmacological effects for central nervous system (CNS) diseases is to design drugs with high blood-brain barrier (BBB) permeability (1, 2). Two active efflux transporters, multiple drug resistance 1 (MDR1) and breast cancer resistance protein (BCRP), expressed at the BBB, are mainly responsible for decreased brain concentration of drugs (3,4,5,6,7). Since higher unbound brain-to-plasma partitioning (Kp,uu,brain) values are usually favorable to yield a lower dose and systemic exposure (1, 8), assessments of Kp,uu,brain are a key process in CNS drug discovery (9). The Kp,uu,brain is derived from not only in vitro binding in the plasma and brain homogenate, but also the in vivo brain to plasma concentration ratio derived in animal studies. These in vivo studies require significant resource, and also raise ethical issues. Therefore, a quantitative Kp,uu,brain prediction based on in silico and in vitro data consisting of in vitro cell line assessments is beneficial in reducing chemistry cycle times for chemical optimization.
Several machine learning approaches based on in silico data have been used for Kp,uu,brain prediction (10,11,12,13,14,15), and the predictivity was improved by incorporating the efflux activity of MDR1 as an explanatory variable (16). However, in a particular example by Dolgikh and colleagues using MDR1 expressing cells, which did not include BCRP efflux activity in the prediction, an influential aspect of their work is the source of the MDR1 cell line, where the authors used a cell line supplied by the Netherlands Cancer Institute. The cell line appears to be less sensitive compared to those established at National Institutes of Health (NIH), which would lessen the ability to derive accurate predictions (9, 17, 18). Hence, it is thought that there is room for further improvement in the predictivity of machine learning approaches by incorporating sensitive efflux activities of MDR1 cells from NIH and the additional activity of BCRP cell lines.
In addition to machine learning models, mechanistic neuroPK models considering efflux activities of MDR1 and/or BCRP are also powerful tools to predict Kp,uu,brain (17, 19,20,21). Mechanistic neuroPK models enable the translation across species through the use of relative activity factors (RAF) and relative expression factors (REF). These scaling factors bridge in vitro cell lines over-expressing transporters to tissue/cellular fractions by incorporating proteomic quantitation of the transporters and associating to activity (22). Both of these models have been useful for predicting human Kp,uu,brain; however, the predictive ability of the neuroPK model has not been directly compared to machine learning approaches using the same dataset; therefore, it is still unclear which approach is more predictive, and better suited in particular situations.
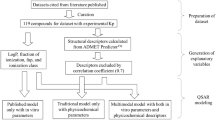
This study directly compares the predictability of Kp,uu,brain predictions using both the neuroPK model and machine learning approach, which incorporates both the efflux transporter activities of MDR1 and BCRP across the same dataset.
MATERIALS AND METHODS
Materials
A total of 640 internal compounds were obtained from Takeda Pharmaceutical Company (Fujisawa, Japan). All other reagents and solvents were of analytical grade or better and were commercially available.
Animals
All the experimental protocols and procedures were approved by the Institutional Animal Care and Use Committee of the Shonan Health Innovation Park, Takeda Pharmaceutical Company Ltd., and all the animal experiments were performed at an animal research facility in Shonan Health Innovation Park accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International. Male Sprague-Dawley rats (7–9 weeks old) were purchased from Charles River Laboratories (Wilmington, MA).
In Vivo Compound Administration and Sampling of Plasma and Brain
Plasma and brain concentrations of proprietary compounds after intravenous, subcutaneous, intraperitoneal, and oral administration were determined. At two time points after the administration (Supporting Information, Table S5), whole blood and whole brain were collected. The blood samples were collected in a heparinized tube from the abdominal aorta and immediately centrifuged to obtain plasma. Brain samples were homogenized immediately after collection in 4 times the volume of the brain using saline under ice-cold conditions. All samples were stored at − 80°C until analysis by liquid chromatography-tandem mass spectrometry (LC-MS/MS, Applied Biosystems, Foster City, CA, USA). Before conducting this analysis, samples were thawed on ice and mixed with three volumes of acetonitrile. The supernatants were diluted and injected into the LC-MS/MS system to quantify compound concentrations using a calibration standard curve.
In Vitro Permeability MDR1 and BCRP-Expressing Cells
Test compound solubilized in dimethyl sulfoxide (DMSO) were added to transport buffer (Hanks’ balanced salt solution with 10 mM HEPES, pH 7.4) at a final concentration of 2 μM (DMSO < 1%), on either the apical or basolateral side of the transwell chamber with the Madin-Darby canine kidney (MDCK)-MDR1 from NIH and MDCK-BCRP from Solvo Biotechnology (Szeged, Hungary). The confluent cell monolayers on the transwell were incubated for 1 h at 37°C with 5% CO2. Test compounds were quantified by LC/MS/MS analysis. Permeation of the test compounds from apical to basolateral (A to B) direction or B to A direction and the efflux ratio were determined. The apparent permeability coefficient Papp (cm/s) was calculated by using the following equation:
where dCr/dt is the cumulative concentration of the compound in the receiver chamber as a function of time (μM/s); Vr is the volume of the solution in the receiver chamber (0.075 mL on the apical side, 0.25 mL on the basolateral side); A is the surface area for transport, i.e., 0.0804 cm2 for the area of the monolayer; and C0 is the initial concentration in the donor chamber (μM).
The efflux ratio (ER) was calculated using the following equation:
Unbound Fractions in Brain and Plasma of Rats
The unbound fraction in rat plasma and brains for each compound was evaluated using the equilibrium dialysis method, details of which have been reported previously (17). Briefly, test compound solutions in DMSO were diluted in rat plasma or 20% (w/v) rat brain homogenate in 100 mM sodium phosphate buffer (pH 7.4) to a concentration of 1 μM. Dialysis was conducted against an equal volume of 10 mM phosphate buffer (plasma) and 100 mM sodium phosphate buffer (brain homogenate) at 37°C for 16–20 h with 8% CO2 (plasma) and without CO2 (brain homogenate). Both the plasma and the brain homogenate obtained from the apparatus were added to equal volumes of control buffer, and then mixed with three volumes of acetonitrile. After centrifugation, the supernatants were analyzed by LC-MS/MS.
The unbound fraction in the incubation mixture (fu,plasma) was calculated using the following equation:
The unbound fractions in the brain (fu,brain) were calculated using the following equation:
where D and fu,brain′ represent the dilution factor for the brain homogenate and unbound fraction determined in the 20% (w/v) brain homogenate, respectively.
Determination of K p,uu,brain
After determination of concentration in plasma (Cplasma) and brain (Cbrain), Kp,uu,brain was calculated by using the following equation:
Prediction of K p,uu,brain in Rats Based on NeuroPK Model
The neuroPK model used in this study was developed in a previous study (17). The major assumptions of this model are that the drug penetration is at steady state, active transport is governed only by two efflux transporters (whose flux is measured in vitro as ERMDR1 and ERBCRP), drug flux from bulk flow is minimal in vivo, and paracellular diffusion is absent in vitro. Based on these assumptions, Kp,uu,brain can be described using the following equation:
where α represents a scaling factor of the in vitro efflux activity of MDR1 in MDCK cells against that of MDR1 in vivo and β represents a scaling factor of the in vitro efflux activity of BCRP in MDCK cells against that of BCRP in vivo. The detailed deviation method leading to Eq. 6 is discussed in two reported literature references (17, 20).
The parameters α and β in Eq. 6 were fitted simultaneously to the observed Kp,uu,brain values in rats using a nonlinear least-square methods provided by Curve Fitting Toolbox 3.5.7 equipped with Matlab R2018a (Mathworks Inc.).
Model Building Using Machine Learning
Each prediction model was generated with StarDrop (StarDrop v6.5.0, Optibrium Ltd, Cambridge, UK) according to the previously described method (23, 24). StarDrop uses 2D SMARTS-based descriptors, which are counts of atom types and functionalities, along with whole molecule properties such as molecular weight (M.W.), topological polar surface area (TPSA), and logP (for a total of 330 descriptors). The descriptors were scaled to unit variance and mean-centered on zero. The rules for descriptor exclusion were as follows:
-
Descriptors with a standard deviation less than 0.0005,
-
Descriptors represented by less than 4% of compounds, and
-
If the pair-wise correlation between any two descriptors exceeds 0.95, then the descriptor of the pair with the lowest correlation with the Y column is excluded.
All in silico models were built using random forest regression (RF) and Gaussian process (GP) model. RF is a flexible, easy to use machine learning algorithm that produces a great result frequently, even without hyper-parameter tuning (25, 26). We used 400 random forest trees for each model. The concept and detailed implementation of the GP method for regression problems were described in Obrezanova et al. (27). Briefly, 4 hyperparameters, θ1, θ2, θ3, and γi (i = 1…K), were optimized. The overall scale for the property values is given by θ1, and the γi are a set of length scale parameters, one for each descriptor. An overall constant shift in the function away from zero is given by θ2. The variance of the assumed noise in the data is described by hyperparameter θ3. A small value on the γi means that differences in the corresponding descriptor influence property values greatly. Hyperparameter tuning of γi was conducted by conjugate gradient optimization (GPOPT) (28). The descriptors ranked in the top 20 of feature importance used in GPOPT model for Kp,uu,brain prediction are summarized in Table S4 and Figure S4.
Model Evaluation
Predictive performance in each model was assessed based on the coefficient of determination (R2) values and residual mean squared error (RMSE) as the statistical indexes. R2 and RMSE were calculated by the following equations:
where n represents the size of the dataset and k represents kth data; the resulting RMSE depicts the magnitude of difference from the observed value. R2 and RMSE were calculated using log(Kp,uu,brain) and log(ER). The percentage of correct answers was evaluated using the number of predicted values against observed values within 2-fold variabilities. Paired t tests of squared errors were used to assess statistical differences between machine learning and the neuroPK models.
Three validation approaches were applied to investigate the model performance. In the first method, the data set is clustered using fingerprints of the molecule structures with a specified Tanimoto coefficient (29), and then split into training and test set with a ratio of 80:20 depending on the property value. This approach enables us to estimate the predictivity for the case that compounds with similar scaffold were included in training and test data set. In the second method, time-split validation was applied to estimate the predictivity for new chemical entities that chemists are likely to investigate in the future (30). The total data set was split by date of assay, i.e., 80% of the total data set before a certain date were assigned as the time-split training set while the remaining 20% of data after that date were assigned as the time-split test set. In the third method, 34 commercially available compounds were used to assess the model performance as an external dataset.
RESULTS
Dataset Analysis
In vivo Kp, in vitro brain tissue and plasma protein binding, and efflux activities of MDR1 and BCRP were experimentally determined for 640 proprietary compounds. This study covered a broad chemical diversity representing a wide range of physicochemical properties, ER and Kp,uu,brain (Supporting Information, Figure S1 and S2, M.W., 187 to 555; clogP, 0.291 to 5.20; TPSA, 12.0 to 157; ER in MDR1 cells, 0.563 to 151; ER in BCRP cells, 0.522 to 100; Kp,uu,brain, 0.00354 to 2.82). Data of both Kp,uu,brain and ER in MDCK-MDR1 from NIH cells and MDCK-BCRP cells did not follow the normal distribution and were log transformed to reduce unequal error variances (Fig. 1). The cluster-split training and test sets showed similar distributions of log(Kp,uu,brain) (Supporting Information, Figure S1), indicating that the splitting method adopted was reasonable. However, M.W., clogP, and TPSA in the time-split test sets tended to have higher values than those in the time-split training sets (Supporting Information, Figure S2). The Kp,uu,brain at the earlier time point was within 2-fold of that at the later point for 95.0% of the compounds (Supporting Information, Figure S3), and therefore, Kp,uu,brain was assumed to be determined under steady-state conditions. Kp,uu,brain values at the later time point were used for model building.
Prediction of Efflux Activities of MDR1 and BCRP Using Machine Learning Approach
Transcellular transport activities characterized using MDCK-MDR1 cells from NIH and MDCK-BCRP cells were used to develop the machine learning model. Among tested compounds, 84.4% of compounds had efflux ratios > 2 in MDR1 assay and 56.1% for BCRP. The machine learning models for MDR1 and BCRP developed by GPOPT showed R2 of 0.581 and 0.499, respectively, in the cluster-split test set (Table I). The prediction of efflux activities in the cluster-split dataset also indicated that 71.1% and 75.0% of those compounds in MDR1 and BCRP models, respectively, were predicted within 2-fold of the observed values (Fig. 2). The predictivity of the machine learning models in the time-split dataset was less than that of the cluster-split dataset, suggesting that the prediction models for efflux activities may not be applied to chemotypes different from compounds included in the training set. In particular, the model for BCRP showed poor prediction. Therefore, Kp,uu,brain prediction using predicted efflux ratio for MDR1 and BCRP was conducted only in the cluster-split test set (Supporting Information, Table S1).
Prediction of K p,uu,brain Using Machine Learning Approaches
The log(Kp,uu,brain) values were predicted by RF and GPOPT. For the determination of the percentage of compounds falling within the 2-fold variability, Kp,uu,brain was transformed from predicted log(Kp,uu,brain) to compare with the neuroPK model that directly predicted Kp,uu,brain based on Eq. 6 (Fig. 3 and Table II). The predictivity based on R2 and RMSE was improved by the incorporation of MDR1 and BCRP efflux activities in both cluster and test-split datasets. Overall, MDR1 showed the greatest impact on the predictive performance of Kp,uu,brain compared to the BCRP addition. The highest R2 of 0.602 was obtained by using GPOPT and both MDR1 and BCRP activities were taken into account pertaining to the time-split dataset with 73.4% of the compounds being predicted within 2-fold of the observed value (Fig. 3b). When leaving the MDR1 and BCRP activities out of the prediction, the cluster-split approach resulted in superior performance compared to the time-split approach (Table II). Meanwhile, the time-split approach incorporating both the MDR1 and the BCRP activities performed equally to the cluster-split approach. The top 20 features obtaining the lowest length scale were extracted in RF and GPOPT models incorporating both MDR1 and BCRP activities (Supporting Information, Figure S4 and Table S4). Efflux ratios of MDR1 and BCRP were ranked in the top 20 of feature importance.
Prediction of K p,uu,brain Using Translational NeuroPK Model
By using the neuroPK model, scaling factors for MDR1 (α) and BRCP (β) were estimated by fitting against Kp,uu,brain (Table III). Using the model incorporating both efflux transporter activities, 58.6% of compounds were predicted within 2-fold of the observed Kp,uu,brain value with a trending R2 value of 0.386 in cluster test sets (Table III and Fig. 4). Meanwhile, the time-splitting approach resulted in a lower R2 value of 0.265. Kp,uu,brain prediction utilizing the neuroPK model was significantly worse compared to machine learning approaches for the same dataset (p < 0.01). The models incorporating in silico MDR1 and BCRP showed an even lower predictive performance than that using experimental values with an R2 value of 0.268 in the cluster test set (Supporting Information, Table S2).
Comparison of the observed Kp,uu,brain and values predicted by the neuroPK model in a cluster-split and b time-split test set. Each figure represents results of 128 compounds in the test set. Fitting was performed to estimate scaling factors α and β against observed values of Kp,uu,brain in rats. Solid line is the line of unity. Dashed lines indicate 2-fold deviation
Model Comparison Using External Dataset
The MDR1 and BCRP activities were determined for 34 commercially available compounds in which Kp,uu,brain values were reported by Friden et al. (11), Summerfield et al. (31), Kodaira et al. (32), and our previous research (17). In this external test set for the established models, 18 and 10 of 34 compounds were substrates of MDR1 and BCRP based on in vitro assessment, respectively, with efflux ratios > 2 (Supporting Information, Table S3). We then applied the machine learning models and the neuroPK model established by the 640 compound internal data to the external test set. The R2 in neuroPK model was 0.577, which was higher than those of 0.422 and 0.363 in GP and RF, respectively (Table IV).
DISCUSSION
It has been reported that MDCK-MDR1 cells obtained from NIH provide a more sensitive in vitro tool for determining MDR1-mediated efflux liability than MDCK-MDR1 from the Netherlands Cancer Institute, or MDR1-overexpressing Lilly Laboratories cell porcine kidney 1 cells (LLC-PK1-MDR1) (9, 18). Previously, we also reported that the NIH cell line MDCK-MDR1 was superior as an MDR1-overexpressing cell line to LLC-PK1-MDR1 for quantitatively predicting brain disposition (17). This work is the first report to establish machine learning models using efflux activity data of MDCK-MDR1 from the NIH-supplied cell line. In addition, there are few reports about quantitative prediction of the efflux transporter BCRP by in silico modeling methods, although several qualitative classification models have been reported (33,34,35,36,37,38,39). The in vitro prediction results of the MDR1 and the BCRP efflux assays indicate that the predictivity of MDR1 efflux activity was higher than the predictivity of BCRP efflux activity (Table I). Based on the results, this difference in predictivity appears to be caused by the differences of distribution patterns, which log(ER) in MDR1 showed relatively higher than that in BCRP cells (Fig. 1d and f). The predictivity in the time-split dataset was worse than that in the cluster-split dataset, suggesting that in silico MDR1 and BCRP models are more applicable to compounds with similar structure, or with timely updating of the model for practical use.
The efflux activities predicted by in silico MDR1 and BCRP models were used for the Kp,uu,brain prediction as additional explanatory variables (Supporting Information, Table S1). In silico predictions of MDR1 and BCRP efflux resulted in a limited predictivity of the machine learning model for Kp,uu,brain, suggesting that current dataset scale and predictivity for MDR1 and BCRP models are not sufficient to apply the Kp,uu,brain prediction. Meanwhile, in silico approaches, underpinned by in vitro assessments, can utilize larger datasets than that by in vivo studies in early discovery, and therefore, the predictivity will be improved when larger training sets are employed. Indeed, Dolgikn et al. succeeded in improving the predictive performance of their model for Kp,uu,brain by using an internal MDR1 model trained with a significantly larger dataset (16).
Although several in silico Kp,uu,brain prediction models have been reported (10,11,12,13,14,15,16), there are few Kp,uu,brain prediction models considering experimentally determined MDR1 and/or BCRP activities. This study revealed that the predictive performance of Kp,uu,brain tended to be improved by considering BCRP activities in both GPOPT and RF models (Table II), suggesting that the BCRP activity also contributes in part to the predictivity of Kp,uu,brain. However, the predictive performance using only MDR1 activity was better than that of using the BCRP activity alone in both the cluster and time-splinting datasets. Considering that the expression level of Mdr1a has been shown to be higher than Bcrp in brain capillaries of the rat (40), it therefore seems reasonable that the substrates for MDR1 would have a larger impact on the Kp,uu,brain assessments compared to BCRP.
In this study, the machine learning models were based on rational selection using the clustering approach, which often provided an optimistic prediction result compared to the time-split approach. The authors believe this is due in part to the addition of new chemical scaffolds being added over time (30, 41,42,43). Indeed, the models developed by only in silico descriptors showed higher predictivity in the cluster-split dataset than that in the time-split dataset (Table II), suggesting that practical operation of a machine learning model warrants timely model updates to cover new chemical space. Interestingly, the machine learning models incorporating MDR1 and BCRP activities showed similar predictivity between the time-split and the cluster-split datasets. This result indicated that the drawback of the machine learning model could be resolved by considering MDR1 and BCRP activities as an explanatory variable.
In the neuroPK model, the α and β in this study were lower than those in previous studies using Kp,brain ratio obtained from Kp,brain in wild-type and Mdr1a (−/−)/Bcrp(−/−) (dual KO) rats even though the same MDR1 and BCRP cell lines were used. Although the Kp,brain ratio is reported to be a direct index of Kp,uu,brain based on the effect of the in vivo MDR1 and BCRP expressed at the BBB (21), actual Kp,brain ratios tend to be higher than Kp,uu,brain (17). Thus, the difference between Kp,brain ratio and Kp,uu,brain would lead to different α and β correction factors. When setting α and β in the neuroPK model, consideration of both efflux activities in cell lines and predictive endpoints are necessary for accurate prediction.
While the time-split approach using in-house data is most useful in estimating the predictive performance of ongoing internal drug discovery programs, the authors also investigated whether the established model was applicable to external compounds (Table IV). Out of 34 external compounds, 44.1% compounds were predicted to within 2-fold of the observed values in the GPOPT and the neuroPK models. Meanwhile, based on R2 and RMSE, the neuroPK models showed better prediction performance compared to machine learning models. The distribution of M.W., clogP, and TPSA in the external test set is less consistent with the external test set (Supporting Information, Figure S5), indicating that the coverage of a chemical space is different between internal and external datasets. These results suggested that the neuroPK model is widely applicable over structurally diverse datasets, while machine learning would achieve maximum performance for predicting compounds with similar chemical properties to those used in the compound training set.
Another advantage of the neuroPK model is the utilization of MDR1 and BCRP variables to establish in vitro to in vivo correlations (IVIVE) to be used in translation (22). These are presented by α and β, in the aforementioned equations. RAF approach corresponding to the ratio of in vitro to in vivo efflux activities for MDR1 and BCRP and REF approach using the ratio of transporter protein levels between in vitro cell lines and in vivo BBB are applicable to estimate these variables across the species. In comparison, the machine learning model suggests that large datasets are required to build accurate models. Therefore, building an inclusive model is limited by the lack of species-specific data, such as non-human primate and human. In order to estimate the minimum number of data required for the machine learning model, the model performance was investigated by changing the number of compounds used for training via the cluster-split dataset (Supporting Information, Figure S6). This optimistic assessment suggested that more than 50 data points were required to obtain equal to or greater predictivity compared to the neuroPK model. Another way to extend the Kp,uu,brain prediction model from rat to other species is that efflux activities in the rat model can be corrected by the ratio of in vivo transporter expression levels between rat and the other species. This hybrid approach could be applicable for both neuroPK and machine learnings models; however, additional validation is needed.
CONCLUSIONS
This work has clarified the characterization of the neuroPK model and machine learning approaches for Kp,uu,brain prediction. Incorporating in vitro MDR1 and BCRP activities is useful to improve the predictivity and coverage of application by machine learning approaches for Kp,uu,brain prediction. Machine learning models have advantages for a homologous series of compounds in internal Kpuu,brain prediction where sufficient data can be secured. Since the machine leaning approach requires a large dataset for model building, the neuroPK model is preferred in translation of Kp,uu,brain from rodent to monkey and human by considering RAF and REF approaches at this time. Additionally, the neuroPK model provides better predictivity of Kp,uu,brain for external compounds which are outside the chemical space in which the model was derived.
Abbreviations
- MDR1:
-
multiple drug resistance 1
- BCRP:
-
breast cancer resistance protein
- GP:
-
Gaussian processes
- RF:
-
random forest regression
- CNS:
-
central nervous system
- BBB:
-
blood-brain barrier
- LLC-PK1:
-
Lilly Laboratories cell porcine kidney 1
- NIH:
-
National Institutes of Health
- M.W.:
-
molecular weight
- TPSA:
-
topological polar surface area
References
Hammarlund-Udenaes M, Bredberg U, Friden M. Methodologies to assess brain drug delivery in lead optimization. Curr Top Med Chem. 2009;9(2):148–62. https://doi.org/10.2174/156802609787521607.
Rankovic Z. CNS drug design: balancing physicochemical properties for optimal brain exposure. J Med Chem. 2015;58(6):2584–608. https://doi.org/10.1021/jm501535r.
Breedveld P, Pluim D, Cipriani G, Wielinga P, van Tellingen O, Schinkel AH, et al. The effect of Bcrp1 (Abcg2) on the in vivo pharmacokinetics and brain penetration of imatinib mesylate (Gleevec): implications for the use of breast cancer resistance protein and P-glycoprotein inhibitors to enable the brain penetration of imatinib in patients. Cancer Res. 2005;65(7):2577–82. https://doi.org/10.1158/0008-5472.CAN-04-2416.
Enokizono J, Kusuhara H, Ose A, Schinkel AH, Sugiyama Y. Quantitative investigation of the role of breast cancer resistance protein (Bcrp/Abcg2) in limiting brain and testis penetration of xenobiotic compounds. Drug Metab Dispos. 2008;36(6):995–1002. https://doi.org/10.1124/dmd.107.019257.
Kodaira H, Kusuhara H, Ushiki J, Fuse E, Sugiyama Y. Kinetic analysis of the cooperation of P-glycoprotein (P-gp/Abcb1) and breast cancer resistance protein (Bcrp/Abcg2) in limiting the brain and testis penetration of erlotinib, flavopiridol, and mitoxantrone. J Pharmacol Exp Ther. 2010;333(3):788-796. doi: https://doi.org/10.1124/jpet.109.162321.
Sakata A, Tamai I, Kawazu K, Deguchi Y, Ohnishi T, Saheki A, et al. In vivo evidence for ATP-dependent and P-glycoprotein-mediated transport of cyclosporin A at the blood-brain barrier. Biochem Pharmacol. 1994;48(10):1989–92. https://doi.org/10.1016/0006-2952(94)90601-7.
Schinkel AH, Smit JJ, van Tellingen O, Beijnen JH, Wagenaar E, van Deemter L, et al. Disruption of the mouse mdr1a P-glycoprotein gene leads to a deficiency in the blood-brain barrier and to increased sensitivity to drugs. Cell. 1994;77(4):491–502. https://doi.org/10.1016/0092-8674(94)90212-7.
Hammarlund-Udenaes M, Friden M, Syvanen S, Gupta A. On the rate and extent of drug delivery to the brain. Pharm Res. 2008;25(8):1737–50. https://doi.org/10.1007/s11095-007-9502-2.
Liu H, Dong K, Zhang W, Summerfield SG, Terstappen GC. Prediction of brain:blood unbound concentration ratios in CNS drug discovery employing in silico and in vitro model systems. Drug Discov Today. 2018;23(7):1357–72. https://doi.org/10.1016/j.drudis.2018.03.002.
Chen H, Winiwarter S, Friden M, Antonsson M, Engkvist O. In silico prediction of unbound brain-to-plasma concentration ratio using machine learning algorithms. J Mol Graph Model. 2011;29(8):985–95. https://doi.org/10.1016/j.jmgm.2011.04.004.
Friden M, Winiwarter S, Jerndal G, Bengtsson O, Wan H, Bredberg U, et al. Structure-brain exposure relationships in rat and human using a novel data set of unbound drug concentrations in brain interstitial and cerebrospinal fluids. J Med Chem. 2009;52(20):6233–43. https://doi.org/10.1021/jm901036q.
Loryan I, Sinha V, Mackie C, Van Peer A, Drinkenburg WH, Vermeulen A, et al. Molecular properties determining unbound intracellular and extracellular brain exposure of CNS drug candidates. Mol Pharm. 2015;12(2):520–32. https://doi.org/10.1021/mp5005965.
Spreafico M, Jacobson MP. In silico prediction of brain exposure: drug free fraction, unbound brain to plasma concentration ratio and equilibrium half-life. Curr Top Med Chem. 2013;13(7):813–20. https://doi.org/10.2174/1568026611313070004.
Varadharajan S, Winiwarter S, Carlsson L, Engkvist O, Anantha A, Kogej T, et al. Exploring in silico prediction of the unbound brain-to-plasma drug concentration ratio: model validation, renewal, and interpretation. J Pharm Sci. 2015;104(3):1197–206. https://doi.org/10.1002/jps.24301.
Zhang YY, Liu H, Summerfield SG, Luscombe CN, Sahi J. Integrating in silico and in vitro approaches to predict drug accessibility to the central nervous system. Mol Pharm. 2016;13(5):1540–50. https://doi.org/10.1021/acs.molpharmaceut.6b00031.
Dolgikh E, Watson IA, Desai PV, Sawada GA, Morton S, Jones TM, et al. QSAR model of unbound brain-to-plasma partition coefficient, Kp,uu,brain: incorporating P-glycoprotein efflux as a variable. J Chem Inf Model. 2016;56(11):2225–33. https://doi.org/10.1021/acs.jcim.6b00229.
Sato S, Tohyama K, Kosugi Y. Investigation of MDR1-overexpressing cell lines to derive a quantitative prediction approach for brain disposition using in vitro efflux activities. Eur J Pharm Sci. 2020;142:105119. https://doi.org/10.1016/j.ejps.2019.105119.
Zheng Y, Chen X, Benet LZ. Reliability of in vitro and in vivo methods for predicting the effect of P-glycoprotein on the delivery of antidepressants to the brain. Clin Pharmacokinet. 2016;55(2):143–67. https://doi.org/10.1007/s40262-015-0310-2.
Adachi Y, Suzuki H, Sugiyama Y. Comparative studies on in vitro methods for evaluating in vivo function of MDR1 P-glycoprotein. Pharm Res. 2001;18(12):1660–8. https://doi.org/10.1023/a:1013358126640.
Trapa PE, Belova E, Liras JL, Scott DO, Steyn SJ. Insights from an integrated physiologically based pharmacokinetic model for brain penetration. J Pharm Sci. 2016;105(2):965–71. https://doi.org/10.1016/j.xphs.2015.12.005.
Uchida Y, Ohtsuki S, Kamiie J, Terasaki T. Blood-brain barrier (BBB) pharmacoproteomics: reconstruction of in vivo brain distribution of 11 P-glycoprotein substrates based on the BBB transporter protein concentration, in vitro intrinsic transport activity, and unbound fraction in plasma and brain in mice. J Pharmacol Exp Ther. 2011;339(2):579–88. https://doi.org/10.1124/jpet.111.184200.
Trapa PE, Troutman MD, Lau TY, Wager TT, Maurer TS, Patel NC, et al. In vitro-in vivo extrapolation of key transporter activity at the blood-brain barrier. Drug Metab Dispos. 2019;47(4):405–11. https://doi.org/10.1124/dmd.118.083279.
Kosugi Y, Hosea N. Prediction of oral pharmacokinetics using a combination of in silico descriptors and in vitro ADME properties. Mol Pharm. 2021;18:1071–9. https://doi.org/10.1021/acs.molpharmaceut.0c01009.
Kosugi Y, Hosea N. Direct comparison of total clearance prediction: computational machine learning model versus bottom-up approach using in vitro assay. Mol Pharm. 2020;17(7):2299–309. https://doi.org/10.1021/acs.molpharmaceut.9b01294.
Breiman L. Random Forests. Machine Learning. 2001;45(1):5–32. https://doi.org/10.1023/A:1010933404324.
Svetnik V, Liaw A, Tong C, Culberson JC, Sheridan RP, Feuston BP. Random forest: a classification and regression tool for compound classification and QSAR modeling. J Chem Inf Comput Sci. 2003;43(6):1947–58. https://doi.org/10.1021/ci034160g.
Obrezanova O, Csanyi G, Gola JM, Segall MD. Gaussian processes: a method for automatic QSAR modeling of ADME properties. J Chem Inf Model. 2007;47(5):1847–57. https://doi.org/10.1021/ci7000633.
Obrezanova O, Gola JM, Champness EJ, Segall MD. Automatic QSAR modeling of ADME properties: blood-brain barrier penetration and aqueous solubility. J Comput Aided Mol Des. 2008;22(6-7):431–40. https://doi.org/10.1007/s10822-008-9193-8.
Butina D. Unsupervised data base clustering based on daylight’s fingerprint and tanimoto similarity: a fast and automated way to cluster small and large data sets. J Chem Inf Comput Sci. 1999;39(4):747–50.
Sheridan RP. Time-split cross-validation as a method for estimating the goodness of prospective prediction. J Chem Inf Model. 2013;53(4):783–90. https://doi.org/10.1021/ci400084k.
Summerfield SG, Lucas AJ, Porter RA, Jeffrey P, Gunn RN, Read KR, et al. Toward an improved prediction of human in vivo brain penetration. Xenobiotica. 2008;38(12):1518–35. https://doi.org/10.1080/00498250802499459.
Kodaira H, Kusuhara H, Fujita T, Ushiki J, Fuse E, Sugiyama Y. Quantitative evaluation of the impact of active efflux by p-glycoprotein and breast cancer resistance protein at the blood-brain barrier on the predictability of the unbound concentrations of drugs in the brain using cerebrospinal fluid concentration as a surrogate. J Pharmacol Exp Ther. 2011;339(3):935–44. https://doi.org/10.1124/jpet.111.180398.
Hazai E, Hazai I, Ragueneau-Majlessi I, Chung SP, Bikadi Z, Mao Q. Predicting substrates of the human breast cancer resistance protein using a support vector machine method. BMC Bioinformatics. 2013;14:130. https://doi.org/10.1186/1471-2105-14-130.
Zhong L, Ma CY, Zhang H, Yang LJ, Wan HL, Xie QQ, et al. A prediction model of substrates and non-substrates of breast cancer resistance protein (BCRP) developed by GA-CG-SVM method. Comput Biol Med. 2011;41(11):1006–13. https://doi.org/10.1016/j.compbiomed.2011.08.009.
Gantner ME, Peroni RN, Morales JF, Villalba ML, Ruiz ME, Talevi A. Development and validation of a computational model ensemble for the early detection of BCRP/ABCG2 substrates during the drug design stage. J Chem Inf Model. 2017;57(8):1868–80. https://doi.org/10.1021/acs.jcim.7b00016.
Sedykh A, Fourches D, Duan J, Hucke O, Garneau M, Zhu H, et al. Human intestinal transporter database: QSAR modeling and virtual profiling of drug uptake, efflux and interactions. Pharm Res. 2013;30(4):996–1007. https://doi.org/10.1007/s11095-012-0935-x.
Garg P, Dhakne R, Belekar V. Role of breast cancer resistance protein (BCRP) as active efflux transporter on blood-brain barrier (BBB) permeability. Mol Divers. 2015;19(1):163–72. https://doi.org/10.1007/s11030-014-9562-2.
Eric S, Kalinic M, Ilic K, Zloh M. Computational classification models for predicting the interaction of drugs with P-glycoprotein and breast cancer resistance protein. SAR QSAR Environ Res. 2014;25(12):939–66. https://doi.org/10.1080/1062936X.2014.976265.
Ose A, Toshimoto K, Ikeda K, Maeda K, Yoshida S, Yamashita F, et al. Development of a support vector machine-based system to predict whether a compound is a substrate of a given drug transporter using its chemical structure. J Pharm Sci. 2016;105(7):2222–30. https://doi.org/10.1016/j.xphs.2016.04.023.
Hoshi Y, Uchida Y, Tachikawa M, Inoue T, Ohtsuki S, Terasaki T. Quantitative atlas of blood-brain barrier transporters, receptors, and tight junction proteins in rats and common marmoset. J Pharm Sci. 2013;102(9):3343–55. https://doi.org/10.1002/jps.23575.
Lombardo F, Desai PV, Arimoto R, Desino KE, Fischer H, Keefer CE, et al. In silico absorption, distribution, metabolism, excretion, and pharmacokinetics (ADME-PK): utility and best practices. An Industry Perspective from the International Consortium for Innovation through Quality in Pharmaceutical Development. J Med Chem. 2017;60(22):9097–113. https://doi.org/10.1021/acs.jmedchem.7b00487.
Martin TM, Harten P, Young DM, Muratov EN, Golbraikh A, Zhu H, et al. Does rational selection of training and test sets improve the outcome of QSAR modeling? J Chem Inf Model. 2012;52(10):2570–8. https://doi.org/10.1021/ci300338w.
Mayr A, Klambauer G, Unterthiner T, Steijaert M, Wegner JK, Ceulemans H, et al. Large-scale comparison of machine learning methods for drug target prediction on ChEMBL. Chem Sci. 2018;9(24):5441–51. https://doi.org/10.1039/c8sc00148k.
Acknowledgements
We acknowledge experimental and technical contribution from members of Research in Takeda Pharmaceutical Company Ltd. We also acknowledge members of Global DMPK for useful exchange of ideas and discussions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 657 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kosugi, Y., Mizuno, K., Santos, C. et al. Direct Comparison of the Prediction of the Unbound Brain-to-Plasma Partitioning Utilizing Machine Learning Approach and Mechanistic Neuropharmacokinetic Model. AAPS J 23, 72 (2021). https://doi.org/10.1208/s12248-021-00604-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1208/s12248-021-00604-x