Abstract
Trastuzumab emtansine (T-DM1) is an antibody–drug conjugate (ADC) therapeutic for treatment of human epidermal growth factor receptor 2 (HER2)-positive cancers. The T-DM1 dose product contains a mixture of drug-to-antibody ratio (DAR) moieties whereby the small molecule DM1 is chemically conjugated to trastuzumab antibody. The pharmacokinetics (PK) underlying this system and other ADCs are complex and have not been elucidated. Accordingly, we have developed two PK modeling approaches from preclinical data to conceptualize and understand T-DM1 PK, to quantify rates of DM1 deconjugation, and to elucidate the link between trastuzumab, T-DM1, and DAR measurements. Preclinical data included PK studies in rats (n = 34) and cynomolgus monkeys (n = 18) at doses ranging from 0.3 to 30 mg/kg and in vitro plasma stability. T-DM1 and total trastuzumab (TT) plasma concentrations were measured by enzyme-linked immunosorbent assay. Individual DAR moieties were measured by affinity capture liquid chromatography-mass spectrophotometry. Two PK modeling approaches were developed for T-DM1 using NONMEM 7.2 software: a mechanistic model fit simultaneously to TT and DAR concentrations and a reduced model fit simultaneously to TT and T-DM1 concentrations. DAR moieties were well described with a three-compartmental model and DM1 deconjugation in the central compartment. DM1 deconjugated fastest from the more highly loaded trastuzumab molecules (i.e., DAR moieties that are ≥3 DM1 per trastuzumab). T-DM1 clearance (CL) was 2-fold faster than TT CL due to deconjugation. The two modeling approaches provide flexibility based on available analytical measurements for T-DM1 and a framework for designing ADC studies and PK–pharmacodynamic modeling of ADC efficacy- and toxicity-related endpoints.






Similar content being viewed by others
REFERENCES
Chen J, Jaracz S, Zhao X, Chen S, Ojima I. Antibody–cytotoxic agent conjugates for cancer therapy. Expert Opin Drug Deliv. 2005;2(5):873–90.
Wu AM, Senter PD. Arming antibodies: prospects and challenges for immunoconjugates. Nat Biotechnol. 2005;23(9):1137–46.
Lopus M. Antibody–DM1 conjugates as cancer therapeutics. Cancer Lett. 2011;307(2):113–8.
Chari RV, Martell BA, Gross JL, Cook SB, Shah SA, Blattler WA, et al. Immunoconjugates containing novel maytansinoids: promising anticancer drugs. Cancer Res. 1992;52(1):127–31.
Lewis Phillips GD, Li G, Dugger DL, Crocker LM, Parsons KL, Mai E, et al. Targeting HER2-positive breast cancer with trastuzumab emtansine, an antibody–cytotoxic drug conjugate. Cancer Res. 2008;68(22):9280–90.
Burris 3rd HA, Tibbitts J, Holden SN, Sliwkowski MX, Lewis Phillips GD. Trastuzumab emtansine (T-DM1): a novel agent for targeting HER2+ breast cancer. Clin Breast Cancer. 2011;11(5):275–82.
Shen B-Q, Bumbaca D, Saad O, Yue Q, Pastuskovas CV, Khojasteh SC, et al. Catabolic fate and pharmacokinetic characterization of trastuzumab emtansine (T-DM1): an emphasis on preclinical and clinical catabolism. Curr Drug Metab. 2012;13:901–10.
Erickson HK, Phillips GL, Leipold DD, Provenzano CA, Mai E, et al. The effect of different linkers on target cell catabolism and pharmacokinetics/pharmacodynamics of trastuzumab maytansinoid conjugates. Mol Cancer Ther. 2012;11(5):1133–42.
Junttila TT, Guangmin L, Parsons K, Phillips GL, Sliwkowski MX. Trastuzumab-DM1 (T-DM1) retains all the mechanisms of action of trastuzumab and efficiently inhibits growth of lapatinib insensitive breast cancer. Breast Cancer Res Treat. 2011;128(2):347–56.
Slikowski MX, Lofgre J, Lewis GD, Hotalin TE, Fendly BM, Fox JA. Nonclinical studies addressing the mechanism of action of Herceptin® (Trastuzumab). Semin Oncol. 1999;26(4 Suppl 12):60–70.
Lin K, Tibbitts J. Pharmacokinetic considerations for antibody drug conjugates. Pharm Res. 2012;29(9):2354–66.
Xu K, Liu L, Saad OM, Baudys J, Williams L, Leipold D, et al. Characterization of intact antibody–drug conjugates from plasma/plasma in vivo by affinity capture capillary liquid chromatography-mass spectrometry. Anal Biochem. 2011;412:56–66.
Xu K, Liu L, Dere R, Mai E, Erickson R, Hendricks A, et al. Characterization of the drug-to-antibody ratio distribution for antibody–drug conjugates in plasma/serum. Bioanalysis. 2013;5(9):1057–71.
Gibiansky L, Gibiansky E. Target-mediated drug disposition model and its approximations for antibody–drug conjugates. J Pharmacokinet Pharmacodyn. 2014;41:35–47.
Beal SL, Boeckman AJ, Sheiner LB, editors. NONMEM User’s guide, part IV. San Francisco: University of California at San Francisco; 1988–1992.
Linbolm L, Pihlgren P, Jonsson EN. Psn-Toolkit: a collection of computer intensive statistical methods for non-linear mixed effect modeling using NONMEM. Comput Methods Programs Biomed. 2005;79(3):241–57.
Deng R, Iyer S, Theil FP, Mortensen DL, Fielder PJ, Prabhu S. Projecting human pharmacokinetics of therapeutic antibodies from nonclinical data. mAbs. 2011;3(1):61–6.
Davies B, Morris T. Physiological parameters in laboratory animals and humans. Pharm Res. 1993;10(7):1093–5.
Mager DE, Jusko WJ. General pharmacokinetic model for drugs exhibiting target-mediated drug disposition. J Pharmacokin Pharmacodyn. 2001;28:507–32.
Shen B-Q, Xu K, Liu K, Raab H, Bhakta S, et al. Conjugation site modulated the in vivo stability and therapeutic activity of antibody–drug conjugates. Nat Biotechnol. 2012;30(2):184–9.
Erickson HK, Lambert JM. ADME of antibody–maytansinoid conjugates. AAPS J. 2012;14(4):799–805.
Xie H, Audette C, Hoffee M, Lambert JM, Blattler WA. Pharmacokinetics and biodistribution of the antitumor immunoconjugate, cantuzumab mertansine (huc242-dm1), and its two components in mice. J Pharmacol Exp Ther. 2004;308(3):1073–82.
Jumbe NL, Xim Y, Leipold D, Crocker L, Dugger D, Mai E, et al. Modeling the efficacy of trastuzumab-DM1, and antibody drug conjugate, in mice. J Pharmacokinet Pharmacodyn. 2010;37(3):221–42.
Bender BC, Schaedeli-Stark F, Koch R, Joshi A, Chu YW, Rugo H, et al. A population pharmacokinetic/pharmacodynamic model of thrombocytopenia characterizing the effect of trastuzumab emtansine (T-DM1) on platelet counts in patients with HER2-positive metatstatic breast cancer. Cancer Chemother Pharmacol. 2012;70(4):591–601.
Hamblett KJ, Senter PD, Chace DF, Sun MM, Lenox J, Cerveny CG, et al. Effects of drug loading on the antitumor activity of a monoclonal antibody drug conjugate. Clin Cancer Res. 2004;10(20):7063–70.
Girish S, Gupta M, Wang B, Lu D, Krop IE, Vogel CL, et al. Clinical pharmacology of trastuzumab emtansine (T-DM1): an antibody–drug conjugate in development for the treatment of HER2-postive cancer. Clin Chemother Pharmacol. 2012;69(5):1229–40.
Gupta M, LoRusso PM, Wang B, Yi JH, Burris 3rd HA, Beeram M, et al. Clinical implications of pathophysiological and demographic covariates on the population pharmacokinetics of trastuzumab emtansine, a HER2-targeted antibody–drug conjugate, in patients with HER2-positive metastatic breast cancer. J Clin Pharmacol. 2012;52(5):691–703.
Bruno R, Washington CB, Lu JF, Lieberman G, Banken L, Klein P. Population pharmacokinetics of trastuzumab in patients with HER2+ metastatic breast cancer. Cancer Chemother Pharmacol. 2005;56(4):361–9.
Lu D, Joshi A, Wang B, Olsen S, Yi J-H, Krop IE, et al. An integrated multiple-analyte pharmacokinetic model to characterize trastuzumab emtansine (T-DM1) clearance pathways and to evaluate reduced pharmacokinetic sampling in patients with HER2-positive metastatic breast cancer. Clin Pharmacokinet. 2013;52(8):657–72.
Junutula JR, Raab H, Clark S, Bhakta S, Leipold DD, et al. Site-specific conjugation of a cytotoxic drug to an antibody improves the therapeutic index. Nat Biotechnol. 2008;26(8):925–32.
Panowksi S, Bhakta S, Raab H, Polakis P, Junutula JR. Site-specific antibody drug conjugates for cancer therapy. MAbs. 2014;6(1):34–45.
Valliere-Douglass JF, McFee WA, Salas-Solano O. Native intact mass determination of antibodies conjugated with monomethyl auristatin E and F at interchain cysteine residues. Anal Chem. 2012;84(6):2843–9.
Hengel SM, Sanderson RJ, Valliere-Douglass JF, Nicholas N, Leiske C, et al. Measurement of in vivo drug load distribution of cysteine-linked antibody–drug conjugates using microscale liquid chromatography mass spectrometry. Anal Chem. 2014. doi:10.1021/ac403860c.
Sievers EL, Senter PD. Antibody–drug conjugates in cancer therapy. Annu Rev Med. 2013;64:15–29.
Author information
Authors and Affiliations
Corresponding author
Additional information
Jay Tibbitts and Lena E. Friberg contributed equally to this work.
Appendices
Appendix 1: Calculation of exposure (AUC) for T-DM1 and its individual DAR moieties
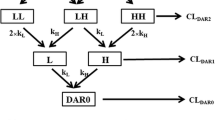
As illustrated in Fig. 1a, each DAR moiety is eliminated from its respective compartment via two rates: (1) deconjugation to the next DAR moiety, and (2) antibody clearance mechanisms. In order to derive the equations for the individual DAR exposures (i.e., AUCDAR0–AUCDAR7) and T-DM1 exposure (i.e., the sum of AUCDAR1–AUCDAR7), we define the following:
-
1.
Dose = T-DM1 dose (mg/kg) × weight (kg)
-
2.
fnn = fraction of DAR moiety (n) in the T-DM1DAR 3.1 dose product; e.g., fn7 = 0.02 (See Methods: Preparation of T-DM1DAR 3.1 and T-DM1DAR 1.5 Dose Products)
-
3.
k TT = rate constant for antibody clearance mechanisms = (CLTT/V1); e.g., k TT = 17.4/148 = 0.118 day−1; Table II, cynomolgus monkeys
-
4.
k n,TOT = total elimination rate for each DAR moiety (n) = (k n → n − 1 + kTT); e.g., k 7,TOT = k 7 → 6 + k TT = 0.341 + 0.118 = 0.459 day−1; Table II, cynomolgus monkeys
The system of T-DM1 deconjugation is a catenary chain containing the moieties DAR0–DAR7. Not only does the exposure of each moiety derive from its initial percentage in the dose product but also from input from higher DAR species. The fraction of each DAR moiety serving as input into the other compartments is calculated as follows:
-
F n → n − 1 = fraction of DAR moiety (n) as input to next DAR (n − 1) compartment = (k n → n − 1/(k n → n − 1 + k TT)); e.g., F 7 → 6 = (k 7 → 6/(k 7 → 6 + k TT)) = (0.341/(0.341 + 0.118)) = 0.742; Table II, cynomolgus monkeys
-
The fraction of DAR moiety as input to all DAR compartments is as follows, using DAR7 as an example:
-
1.
F 7 → 6 = the fraction of DAR7 as input to DAR6
-
2.
F 7 → 5 = F 7 → 6 × F 6 → 5; the fraction of DAR7 as input to DAR5
-
3.
F 7 → 4 = F 7 → 6 × F 6 → 5 × F 5 → 4; the fraction of DAR7 as input to DAR4
-
4.
F 7 → 3 = F 7 → 6 × F 6 → 5 × F 5 → 4 × F 4 → 3; the fraction of DAR7 as input to DAR3
-
5.
F 7 → 2 = F 7 → 6 × F 6 → 5 × F 5 → 4 × F 4 → 3 × F 3 → 2; the fraction of DAR7 as input to DAR2
-
6.
F 7 → 1 = F 7 → 6 × F 6 → 5 × F 5 → 4 × F 4 → 3 × F 3 → 2 × F 2 → 1; the fraction of DAR7 as input to DAR1
-
7.
F 7 → 0 = F 7 → 6 × F 6 → 5 × F 5 → 4 × F 4 → 3 × F 3 → 2 × F 2 → 1 × F 1 → 0; the fraction of DAR7 as input to DAR0
-
1.
Equations for the AUC of the individual DAR0–DAR7 moieties are shown below. Here, the total amount of each DAR, deriving from its percentage in the dose product and from input from the higher DAR moieties, is divided by its respective clearance to yield AUC.
-
1.
AUCDAR7 = Dose × fn7/(V 1 × k 7,TOT)
-
2.
AUCDAR6 = dose × (F 7 → 6 × fn7 + fn6)/(V 1 × k6,TOT)
-
3.
AUCDAR5 = Dose × (F7 → 5 × fn7 + F6 → 5 × fn6 + fn5)/(V 1 × k 5,TOT)
-
4.
AUCDAR4 = dose × (F 7 → 4 × fn7 + F 6 → 4 × fn6 + F 5 → 4 × fn5 + fn4)/(V 1 × k 4,TOT)
-
5.
AUCDAR3 = dose × (F 7 → 3 × fn7 + F 6 → 3 × fn6 + F 5 → 3 × fn5 + F 4 → 3 × fn4 + fn3)/(V 1 × k 3,TOT)
-
6.
AUCDAR2 = dose × (F 7 → 2 × fn7 + F 6 → 2 × fn6 + F 5 → 2 × fn5 + F 4 → 2 × fn4 + F 3 → 2 × fn3 + fn2)/(V 1 × k 2,TOT
-
7.
AUCDAR1 = dose × (F 7 → 1 × fn7 + F 6 → 1 × fn6 + F 5 → 1 × fn5 + F 4 → 1 × fn4 + F 3 → 1 × fn3 + F 2 → 1 × fn2 + fn1)/(V 1 × k 1,TOT)
-
8.
AUCDAR0 = dose × (F 7 → 0 × fn7 + F 6 → 0 × fn6 + F 5 → 0 × fn5 + F 4 → 0 × fn4 + F 3 → 0 × fn3 + F 2 → 0 × fn2 + F 1 → 0 × fn1 + fn0)/(V 1 × k TT)
The overall T-DM1 exposure is the sum of the individual DAR1–DAR7 AUCs.
-
1.
AUCTDM1 = AUC DAR7 + AUCDAR6 + AUCDAR5 + AUCDAR3 + AUC DAR2 + AUCDAR1, thus
AUCTDM1 = dose × fn7/(V 1 × k 7,TOT) + dose × (F 7 → 6 × fn7 + fn6)/(V 1 × k 6,TOT) + …etc.
-
2.
Substitution, in the above equation, with the Table II parameter values for cynomolgus monkeys and the fn1–fn7 values for the T-DM1DAR 3.1 dose product (see “Preparation of T-DM1DAR 3.1 and T-DM1DAR 1.5 Dose Products”) yields:
-
T-DM1DAR 3.1 dose product at 3.6 mg/kg has an AUCTDM1 = 538 μg/mL × day−1
-
Appendix 2: Dose calculations (mg/kg) for T-DM1 dose products targeting a specified T-DM1 AUC
Appendix 1 derived the mathematical equations for T-DM1 AUC and its individual DAR AUCs. From these equations, the dose (mg/kg) of the T-DM1DAR 1.5 dose product equivalent to 3.6 mg/kg T-DM1DAR 3.1 by AUC is calculated here.
-
1.
From Appendix 1:
-
T-DM1DAR 3.1 dose product at 3.6 mg/kg has an AUCTDM1 = 538 μg/mL × day−1
-
AUCTDM1 = dose × fn7/(V 1 × k 7,TOT) + dose × (F 7 → 6 × fn7 + fn6)/(V 1 × k 6,TOT) + … etc.
-
-
2.
Rearrangement yields:
-
Dose = AUCTDM1/(fn7/(V 1 × k 7,TOT) + (F 7 → 6 × fn7 + fn6)/(V 1 × k 6,TOT) + … etc.)
-
-
3.
Substitution, in the above equation with the Table II parameter values for cynomolgus monkeys, and the appropriate fn1–fn7 values for the T-DM1DAR 1.5 dose product (see “Preparation of T-DM1DAR 3.1 and T-DM1DAR 1.5 Dose Products”) yields:
-
T-DM1DAR 1.5 dose product at 5.16 mg/kg has an AUCTDM1 = 538 μg/mL × day−1
-
Rights and permissions
About this article
Cite this article
Bender, B., Leipold, D.D., Xu, K. et al. A Mechanistic Pharmacokinetic Model Elucidating the Disposition of Trastuzumab Emtansine (T-DM1), an Antibody–Drug Conjugate (ADC) for Treatment of Metastatic Breast Cancer. AAPS J 16, 994–1008 (2014). https://doi.org/10.1208/s12248-014-9618-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1208/s12248-014-9618-3