Abstract
Background
Since the beginning of coronavirus disease 2019 (COVID-19), the development of predictive models has sparked relevant interest due to the initial lack of knowledge about diagnosis, treatment, and prognosis. The present study aimed at developing a model, through a machine learning approach, to predict intensive care unit (ICU) mortality in COVID-19 patients based on predefined clinical parameters.
Results
Observational multicenter cohort study. All COVID-19 adult patients admitted to 25 ICUs belonging to the VENETO ICU network (February 28th 2020-april 4th 2021) were enrolled. Patients admitted to the ICUs before 4th March 2021 were used for model training (“training set”), while patients admitted after the 5th of March 2021 were used for external validation (“test set 1”). A further group of patients (“test set 2”), admitted to the ICU of IRCCS Ca’ Granda Ospedale Maggiore Policlinico of Milan, was used for external validation. A SuperLearner machine learning algorithm was applied for model development, and both internal and external validation was performed. Clinical variables available for the model were (i) age, gender, sequential organ failure assessment score, Charlson Comorbidity Index score (not adjusted for age), Palliative Performance Score; (ii) need of invasive mechanical ventilation, non-invasive mechanical ventilation, O2 therapy, vasoactive agents, extracorporeal membrane oxygenation, continuous venous-venous hemofiltration, tracheostomy, re-intubation, prone position during ICU stay; and (iii) re-admission in ICU.
One thousand two hundred ninety-three (80%) patients were included in the “training set”, while 124 (8%) and 199 (12%) patients were included in the “test set 1” and “test set 2,” respectively. Three different predictive models were developed. Each model included different sets of clinical variables. The three models showed similar predictive performances, with a training balanced accuracy that ranged between 0.72 and 0.90, while the cross-validation performance ranged from 0.75 to 0.85. Age was the leading predictor for all the considered models.
Conclusions
Our study provides a useful and reliable tool, through a machine learning approach, for predicting ICU mortality in COVID-19 patients. In all the estimated models, age was the variable showing the most important impact on mortality.
Similar content being viewed by others
Background
Predictive modeling has been a hot topic of coronavirus disease 2019 (COVID-19) research [1]. Since the very beginning of the epidemic, there was a significant push towards developing predictive models for COVID-19 diagnosis and prognosis. The interest in predictive models' development was associated with the initial lack of knowledge about COVID-19 diagnosis/treatment/prognosis and the unexpected and dramatic pressure on the healthcare system, especially on intensive care units (ICU) [2]. Such predictive models were aimed at helping physicians stratify patients’ risk of developing the outcome of interest, e.g., need of hospitalization and mechanical ventilation.
A systematic review of the literature by Wynants et al. identified more than sixty predictive models already published at the beginning of the pandemic, i.e., April 2020. The update of this systematic review recorded more than two hundred models [1]. Initially, most models focused on COVID-19 diagnosis, while the update of the revision showed that much more published models focused on patients' prognosis and particularly on predicting death risk.
The idea behind such algorithms was to characterize patients at higher risk of death from severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection to help physicians identify the best treatment for each patient according to his/her characteristics. The final aim was to guarantee an efficient allocation of the healthcare resources given the dramatic shortage resulting from the outbreak.
Italy was the first European country hit by the COVID-19 outbreak. Lombardy and Veneto were the two Italian regions where COVID-19 spread first. In a short time, healthcare authorities tried to activate emergency measures to contain the virus spread at the population level and organize the healthcare system response to face the sudden and unexpected increased demand for healthcare assistance [2,3,4,5]. In the Veneto Region, the “COVID-19 VENETO ICU Network” was established [5]. It is an official task force aimed at optimizing ICU resources management through the identification of dedicated COVID-19 pathways and the increase of ICU beds capacity. Furthermore, the network aims to share experience on COVID-19 patients’ treatment among intensive care medicine specialists to standardize patient care. Finally, data on COVID-19 patients admitted to the COVID-19 ICUs of the network have been collected routinely, allowing the epidemiological surveillance of the phenomenon, e.g., to plan the activation of additional ICU beds and clinical research.
The aim of the present study was to develop and validate a predictive model through a machine learning approach for ICU mortality in COVID-19 patients using VENETO ICU Network data.
Methods
We prospectively screened the records of all adult patients with confirmed SARS-CoV-2 infection admitted to the ICUs of the COVID-19 VENETO ICU network, between 28th of February 2020 and 4th of April 2021 [5, 6]. COVID-19 diagnosis was made according to the World Health Organization interim guidance (http://www.who.int/docs/default-source/coronaviruse/clinical-management-of-novel-cov.pdf).
The study was approved by the Institutional Ethical Committee of each participating center (coordinator center approval reference number 4853AO20) and informed consent was obtained for each patient in compliance with national regulation and the recommendations of the Institutional Ethical Committee of Padova University Hospital.
The study cohort was divided into two groups, according to the time of ICU admission. The first group, i.e., “training set,” included patients admitted to the ICUs from 28th of February to 28th of April 2020 plus from 27th of November to 4th of March 2021, and was used for model training. The second group (named “test set 1”), composed by patients admitted to the ICUs from 5th of March to 4th of April 2021, was used for external validation of the model.
In addition to that, a third group (named “test set 2”), composed by patients admitted to the ICU of IRCCS Ca’ Granda Ospedale Maggiore Policlinico of Milan (Lombardy Region) in the same period of time, was also used for external validation.
At ICU admission, the physicians in charge of the patients prospectively collected a predefined set of clinical variables at ICU admission, as listed in Supplementary Materials (Table S1), and entered data into a predesigned data collection form implemented in a web-based system. Moreover, the physicians recorded the need of respiratory support, tracheostomy, re-intubation, prone positioning, extracorporeal membrane oxygenation, continuous venous-venous hemofiltration, vasoactive agents during ICU stay, or re-admission. Each investigator had a personal username and password. Patients’ privacy was protected by assigning a de-identified patient code. Prior to data analysis, two independent investigators and a statistician screened the database for errors against standardized ranges and contacted local investigators with any queries. Then, validated data were entered in the database for final analysis.
Models estimation
Three SuperLearner (SL) prediction tools were developed and validated on the ICU data (see Additional file 1, Table S1, for the complete list of variables included in each model)
-
1)
Model 1. The first model was tuned considering only the variables collected at ICU admission having less than 85% of missing data (see Additional file 1, Table S1, for the complete list of variables included in the model). The external validation was performed on the “test set 1 and 2.”
-
2)
Model 2. The second model was tuned considering all the variables collected at ICU admission, even though missing data were more than 85% (Additional file 1, Table S1). The external validation was performed on the “test set 1.”
-
3)
Model 3. The third model was tuned considering the variables collected at ICU admission and during ICU stay, even though the missing data were more than 85% (Additional file 1, Table S1). The external validation was performed on the “test set 1.”
SuperLearner approach
SuperLearner (SL) is an ensemble Machine Learning algorithm that combines multiple Machine Learning Techniques (MLTs), i.e., base learners, to achieve the best possible weighted performance of the base learners [7,8,9]. The detailed description of the algorithms is provided in the Additional file 1, Methods S2. Figure 1 presents the schematic representations of the base learners.
Performance measures
The sensitivity, specificity, F1 statistics, the balanced accuracy, and were computed. The training ROC plots were reported.
Internal cross-validation
The base models and the SuperLearner models underwent internal cross-validation performing a 5-fold cross-validation procedure.
Variable importance plot
The variable importance plots were reported. The importance measure was computed considering the mean decrease in the ROC measure resulting from the removal of the variable within the permutations, as recommended in the literature [10].
Descriptive statistics
Continuous data were reported I quartile/median/III quartile categorical data were reported as a percentage and absolute frequencies.
Shiny web application
A shiny web application was developed. The tool calculates the ICU death probability, according to the patients’ characteristics based on each one of the models estimated.
Results
Study population
The overall population included 1616 patients. The first 1293 (80%) patients admitted to the ICUs of the VENETO ICU Network were used for models training (“training set”), while the following 124 (8%) patients were used for external validation (“test set 1”). As well, a further cohort of 199 (12%) patients, admitted to the IRCCS Ca’ Granda Ospedale Maggiore Policlinico, was used as additional external validation (“test set 2”).
Table 1 presents the training and validation cohorts’ characteristics. The proportion of deaths was of 39% in the cohort of 1417 patients admitted to the ICUs of the VENETO ICU Network, and 28% in the cohort of 199 patients admitted to the IRCCS Ca’ Granda Ospedale Maggiore Policlinico.
Model 1 was trained on the overall ‘training set’ of patients (Table 1) because the model included a limited set of variables measured at ICU admission (Supplementary materials, Table S1) with less than 85% of missing data. Models 2 and 3 were trained on 656 out of 1293 patients because they also included variables with more than 85% of missing data (Table 1) (see Supplementary materials, Table S1, for the complete list of variables included in each model). The main difference between Model 2 and Model 3 is that Model 2 included only variables measured at ICU admission, while Model 3 included variables recorded at admission and also during the ICU stay.
Model 1 was validated on both the cohort of 124 patients belonging to the “test set 1” and on the cohort of 199 patients named “test set 2.” Models 2 and 3 were validated on the external cohort of 124 patients admitted to the ICUs of the COVID-19 VENETO ICU Network (“test set 1”).
Models’ performance
The three models showed similar performances in predicting ICU mortality (Table 2 and Additional file 1, Figure S3), with a training balanced accuracy that ranged between 0.72 and 0.90.
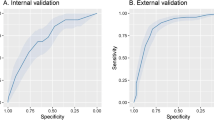
The cross-validation performance is in Fig. 2. The best performance was achieved by Model 3, with a ROC of 0.85, while both Models 1 and 2 presented a ROC value of 0.75.
With regards to the performance of the algorithms on which the SuperLearner was based, the RF was the one with the best performance on Model 3, as well as for Model 2, together with the GBM. For Model 1, the best performance was achieved by a Bayesian Machine Regression Trees (BartMachine) (Fig. 2).
Variables importance in relation to the outcome
Age was the leading predictor for all the considered models, followed by total SOFA score at ICU admission and the arterial partial pressure of oxygen to inspired oxygen fraction ratio used for SOFA calculation (SOFA PaO2/FiO2) in Model 1. The SOFA PaO2/FiO2 was a relevant predictor for Model 2, as well the Palliative Predictive Score (PPS) Activity variable. The PPS Activity was also in the top five parameters for Model 3, together with the need of O2 therapy, non-invasive or invasive ventilation (Fig. 3).
Variable importance plots. The ten most important predictors are reported in the plots. Abbreviations: PaO2/FiO2, arterial partial pressure of oxygen to inspired oxygen fraction ratio; SOFA, Sequential Organ Failure Assessment; CCI, Charlson Comorbidity Index; GCS, Glasgow Coma Scale; PPS, Palliative Performance Score.
The shiny app reporting the three ICU mortality prediction tools is available at https://r-ubesp.dctv.unipd.it/shiny/CoViD-19%20icupred/.
Discussion
The present study provides a tool for predicting ICU mortality in COVID-19 patients using data from a large cohort of patients admitted to the ICUs of the COVID-19 VENETO ICU Network. The three models, systematically built through a machine learning approach, showed good training and validation performances, yielding similar results to predict ICU mortality.
In particular, age was identified as the most important predictive parameter in every model investigated. Secondary, total SOFA score at ICU admission, the level of daily activity and the need of different types of respiratory supports were important parameters for Model 1, 2 and 3.
This finding is in line with current literature, describing a great impact of age on mortality in COVID-19 patients undergoing invasive and non-invasive ventilation [6, 11,12,13,14,15]. Karagiannidis C et al, in the widest cohort of hospitalized COVID-19 patients, showed that mortality has been high for patients receiving mechanical ventilation, particularly for patients aged 80 years or older and those requiring dialysis, and has been considerably lower for patients younger than 60 years [11].
Similar findings were reported by Boscolo et al. and Vaschetto et al. investigating in-hospital mortality of COVID-19 mechanically ventilated. In both studies, the cumulative incidence of mortality at 60 days was higher in the older ones [6, 12].
Worth noting, from the beginning of the pandemic, several tools have been proposed for mortality prediction of COVID-19 patients; however, it is difficult to compare their performance because each model was developed on patients with different characteristics, using different sets of variables, and using different techniques for model development. Indeed, Wynants and colleagues have shown that all published models have several limitations [1], including small sample size and lack of information and clarity on algorithm development reporting. For these reasons, it is difficult to compare models’ performance and to identify the most feasible model to be used in everyday clinical practice to assist physicians’ decisions. Our findings show that the SL is a feasible approach to be used with clinical data, providing good predictive performances and good generalizability. Although machine learning approaches are increasingly used in the clinical setting, also in COVID-19 research [7-10], more traditional techniques, i.e., traditional logistic regression for binary outcomes and survival regression models for time-to-event outcomes, are still widely used since they are much simpler to be implemented and interpreted. However, the use of machine learning approaches represents an added value to predictive modeling, as it allows the detection of complex relationships between the outcomes of interest and the covariates, overcoming the limits of traditional analysis, especially when a high number of predictors is evaluated in front of a low number of events.
The predictive tools described in the present paper have several strengths, including the fact that they have been developed on a large multicenter cohort of patients admitted to the ICUs of one of the Italian regions most severely affected by the COVID-19 pandemic, the use of both internal and external validation, and the use of a machine learning tool instead of more traditional techniques to build the predictive model.
However, our study has some limitations. First, clinical variables investigated in our study represent only a small number of parameters potentially relevant and able to affect critically ill patients’ outcomes. Second, several patients had incomplete records, which depended on the overwhelming workload for ICU physicians during the COVID-19 pandemic.
Conclusions
Our study provides a useful and reliable tool, through a machine learning approach using the SL algorithm, for predicting ICU mortality in COVID-19 patients. Age was the most predictive parameter in all the estimated Models.
Availability of data and materials
The data that support the findings of this study are available from the corresponding author, PN, upon request.
Abbreviations
- SARS-CoV-2:
-
Severe acute respiratory syndrome coronavirus 2
- COVID:
-
Coronavirus disease
- ICU:
-
Intensive care unit
- PaO2/FiO2 :
-
The ratio between arterial partial pressure of oxygen and inspired fraction of oxygen
- SL:
-
SuperLearner
- SOFA:
-
Sequential Organ Failure Assessment
- MLT:
-
Machine learning technique
- ROC:
-
Receiving operative characteristics
- CCI:
-
Charlson Comorbidity Index
- PPS:
-
Palliative Predictive Score
- IMV:
-
Invasive mechanical
- OR:
-
Odds ratio
- CI:
-
Confidence interval
- Sens:
-
Sensitivity
- Spec:
-
Specificity
References
Wynants L, Calster BV, Collins GS, Riley RD, Heinze G, Schuit E et al (2020) Prediction models for diagnosis and prognosis of covid-19: systematic review and critical appraisal. BMJ 369:m1328
Tonetti T, Grasselli G, Zanella A, Pizzilli G, Fumagalli R, Piva S et al (2020) Use of critical care resources during the first 2 weeks (February 24–March 8, 2020) of the Covid-19 outbreak in Italy. Ann Intensive Care 10:1–8
Lorenzoni G, Lanera C, Azzolina D, Berchialla P, Gregori D, Gregori D et al (2020) Is a more aggressive COVID-19 case detection approach mitigating the burden on ICUs? Some reflections from Italy. Crit Care 24:175
Cavarretta E, Biondi-Zoccai G, Frati G, Versaci F (2020) Veneto’s successful lesson for a world shocked by COVID-19: Think globally and act locally. J Cardiothor Vasc Anesth 34:2346–2348
Pasin L, Sella N, Correale C, Boscolo A, Rosi P, Saia M et al (2020) Regional COVID-19 network for coordination of SARS-CoV-2 outbreak in Veneto, Italy. J Cardiothor Vasc Anesth 34:2341–2345
Boscolo A, Sella N, Lorenzoni G, Pettenuzzo T, Pasin L, Pretto C et al (2021) Static compliance and driving pressure are associated with ICU mortality in intubated COVID-19 ARDS. Crit Care 2021;25(1):263
Van der Laan MJ, Polley EC, Hubbard AE (2007) Super Learner. Stat Appl Genet Mol Biol 6:25
Greenwell BM, Boehmke BC, McCarthy AJ (2018) A simple and effective model-based variable importance measure. arXiv:1805.04755[cs.stat] 2018.
Tezza F, Lorenzoni G, Azzolina D, Barbar S, Leone LAC, Gregori D (2021) Predicting in-hospital mortality of patients with COVID-19 using machine learning techniques. J Personal Med 11:343
Calabrese F, Pezzuto F, Fortarezza F, Boscolo A, Lunardi F et al (2021) Machine learning-based analysis of Alveolar and Vascular injury in SARS-CoV-2 acute respiratory failure. J Pathol 254:173–184
Karagiannidis C, Mostert C, Hentschker C, Voshaar T, Malzahn J, Schillinger G et al (2020) Case characteristics, resource use, and outcomes of 10 021 patients with COVID-19 admitted to 920 German hospitals: an observational study. Lancet Respir Med 8(9):853–862
Vaschetto R, Barone-Adesi F, Racca F, Pissaia C, Maestrone C, Colombo D et al (2021) Outcomes of COVID-19 patients treated with continuous positive airway pressure outside the intensive care unit. ERJ Open Res 7(1):00541–02020
Grasselli G, Zangrillo A, Zanella A, Antonelli M, Cabrini L, Castelli A et al (2020) Baseline characteristics and outcomes of 1591 patients infected with SARS-CoV-2 admitted to ICUs of the Lombardy Region, Italy. JAMA 323(16):1574–1581
Yang X, Yu Y, Xu J, Shu H, Xia J, Liu H et al (2020) Clinical course and outcomes of critically Ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med 8(5):475–481
Alhazzani W, Møller MH, Arabi YM, Loeb M, Gong MN, Fan E et al (2020) Surviving Sepsis Campaign: Guidelines on the Management of Critically Ill Adults with Coronavirus Disease 2019 (COVID-19). Crit Care Med 48(6):e440–e469
Acknowledgements
We are indebted with all ICUs personnel who made this work possible.
Members of COVID-19 VENETO ICU Network, to be searchable through their individual PubMed records, are listed below:
(i) U.O.C. Istituto di Anestesia e Rianimazione, Padova (PD) (Martina Tocco, MD; Chiara Pretto, MD; Enrico Tamburini, MD; Davide Fregolent, MD; Pier Francesco Pirelli, MD; Davide Marchesin, MD; Matteo Perona, MD; Nicola Franchetti, MD; Michele Della Paolera, MD; Caterina Simoni, MD; Tatiana Falcioni, MD; Alessandra Tresin, MD; Chiara Schiavolin, MD; Aldo Schiavi, MD; Sonila Vathi, MD; Daria Sartori, MD; Alice Sorgato, MD; Elisa Pistollato, MD; Federico Linassi, MD; Sara Gianoli MD; Silvia Gaspari MD; Francesco Gruppo MD; Alessandra Maggiolo MD; Elena Giurisato MD; Elisa Furlani MD; Alvise Calore MD); (ii) Azienda Ospedale-Università di Padova (PD) (Eugenio Serra, MD; Demetrio Pittarello, MD; Ivo Tiberio, MD; Ottavia Bond, MD; Elisa Michieletto, MD; Luisa Muraro, MD; Arianna Peralta, MD; Paolo Persona, MD; Enrico Petranzan, MD; Francesco Zarantonello, MD; Alessandro Graziano, MD; Eleonora Piasentini, MD); (iii) U.O.C. Anestesia e Rianimazione, Presidio Ospedaliero San Martino, Belluno (BL), AULSS 1 Dolomiti (Lorenzo Bernardi, MD; Roberto Pianon, MD, Davide Mazzon MD, Daniele Poole MD); (iv) U.O.C. Anestesia e Rianimazione, Ospedale di Vittorio Veneto, Vittorio Veneto (TV), AULSS 2 Marca Trevigiana (Flavio Badii, MD); (v) U.O.C. Anestesia e Rianimazione, Ospedale di Conegliano, Conegliano (TV), AULSS 2 Marca Trevigiana (Enrico Bosco, MD); (vi) U.O.C. Anestesia e Rianimazione, Ospedale di Montebelluna, Montebelluna (TV), AULSS 2 Marca Trevigiana (Moreno Agostini, MD); (vii) U.O.C. Anestesia e Rianimazione, Ospedale di Oderzo, Oderzo (TV), AULSS 2 Marca Trevigiana (Paride Trevisiol, MD); (viii) U.O.C. Anestesia e Rianimazione, Ospedale Ca’ Foncello, Treviso, AULSS 2 Marca Trevigiana (Antonio Farnia, MD); (ix) U.O.C. Anestesia, Rianimazione e Terapia Antalgica, Presidio Ospedaliero di Dolo, Dolo (VE), AULSS 3 Serenissima (Lorella Altafini, MD); (xi) U.O.C. Anestesia, Rianimazione e Terapia Antalgica, Presidio Ospedaliero di Mirano, Mirano (VE) AULSS 3 Serenissima (Mauro Antonio Calò, MD); (xii) U.O.C. Anestesia e Rianimazione, Ospedale SS. Giovanni e Paolo, Venezia, AULSS 3 Serenissima (Marco Meggiolaro, MD); (x) U.O.C. Anestesia e Rianimazione, Ospedale dell’Angelo, Mestre (VE), AULSS 3 Serenissima (Francesco Lazzari, MD; Ivan Martinello, MD; Francesco Papaccio, MD); (xiii) U.O.C. Anestesia e Rianimazione, Ospedali di San Donà di Piave e Jesolo, San Donà di Piave (VE), AULSS 4 Veneto Orientale; (xiv) U.O.C. Anestesia e Rianimazione, Ospedale di Cittadella, Cittadella (PD), AULSS 6 Euganea (Guido di Gregorio MD; Alfeo Bonato, MD; Camilla Sgarabotto, MD); (xv) U.O.C. Anestesia e Rianimazione, Ospedali Riuniti Padova Sud, Monselice (PD), AULSS 6 Euganea (Francesco Montacciani, MD; Parnigotto Alessandra, MD); (xvi) U.O.C. Anestesia e Rianimazione, Ospedali di Rovigo e Trecenta, Rovigo, AULSS 5 Polesana (Giuseppe Gagliardi, MD; Gioconda Ferraro, MD); (xvii) U.O.C. Anestesia e Rianimazione, Ospedale Alto Vicentino, Santorso (VI), AULSS 7 Pedemontana (Luigi Ongaro, MD); (xviii) U.O.C. Anestesia e Rianimazione, Ospedale San Bassiano, Bassano del Grappa (VI), AULSS 7 Pedemontana (Marco Baiocchi, MD); (xix) U.O.C Anestesia e Rianimazione, Ospedale di Vicenza, Vicenza (VI), AULSS 8 Berica (Vinicio Danzi, MD); (xx) U.O.C. Anestesia e Rianimazione A, Azienda Ospedaliera Universitaria Integrata Verona, Verona (VR) (Paolo Zanatta, MD); (xxi) U.O. Anestesia e Rianimazione B, Azienda Ospedaliera Universitaria Integrata Verona, Verona (VR) (Katia Donadello MD; Leonardo Gottin MD); (xxii) U.O.C Anestesia e Rianimazione, Ospedale Mater Salutis di Legnago, Legnago (VR), AULSS 9 Scaligera (Ezio Sinigaglia, MD; Alessandra da Ros, MD); (xxiii) U.O.C Anestesia e Rianimazione, Ospedale Magalini di Villafranca, Legnago (VR), AULSS 9 Scaligera (Simonetta Marchiotto, MD; Silvia Bassanini, MD); (xxiv) Dipartimento di Anestesia, Rianimazione e Terapia Antalgica, IRCCS Sacro Cuore-Don Calabria, Negrar (VR) (Massimo Zamperini, MD; Ivan Daroui, MD); (xxv) U.O. Terapia Intensiva, Ospedale P. Pederzoli – Casa di Cura Privata SpA, Peschiera sul Garda (VR) (Walter Mosaner, MD).
Funding
This study was funded by a grant provided by the Regional Government, Veneto, Italy. The regional database was commissioned to the Contract Research Organization Aleph srl (Milan, Italy).
Author information
Authors and Affiliations
Consortia
Contributions
Concept and design: DG, PN, PR, AP, GG, GL, EP, NS. Acquisition, analysis, or interpretation of data: GL, NS, BA, DA, PB, LP, TP, ADC, FB, FT, SDR, GF, MP, AZ. Drafting of the manuscript: GL, NS, AB, BA, DA, PB, LP. Critical revision of the manuscript for important intellectual content: PR, DG, PN, AP, GG; the COVID-19 VENETO ICU Network contributed to collect and interpret data. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This was a multicenter, observational study performed in twenty-five hospitals of Veneto Region, Northern Italy, listed on the Acknowledgements and in the ICU of IRCCS Ca’ Granda Ospedale Maggiore Policlinico of Milan. All the participating centers obtained Ethics Committee approval for the present research project, initially approved by the Institutional Ethical Committee of Padova University hospital (Ref: 4853AO20). Local investigators were responsible for ensuring data integrity and validity. The study was conducted in accordance with the Helsinki declaration and national regulation on study involving humans. Informed consent was obtained for each patient in compliance with national regulation and the recommendations of the Institutional Ethical Committee of Padova University Hospital.
Consent for publication
Informed consent was obtained for each patient in compliance with national regulation and the recommendations of the Institutional Ethical Committee of Padova University Hospital.
Competing interests
None related to the present work. PN received royalties from Intersurgical for Helmet Next invention and speaking fees from Philips, Resmed, MSD, Drager and Novartis. The other authors have no other competing interests to declare.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Fabio Baratto, Fabio Toffoletto, Giorgio Fullin, Mario Peta, Enrico Polati, Paolo Navalesi: Coordinamento delle Terapie Intensive della Regione Veneto.
Supplementary Information
Additional file 1: Table S1.
Variables collected and included in each predictive model. Methods S2. SuperLearner algorithm details. Figure S3. ROC Curves estimated on the test sample.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lorenzoni, G., Sella, N., Boscolo, A. et al. COVID-19 ICU mortality prediction: a machine learning approach using SuperLearner algorithm. J Anesth Analg Crit Care 1, 3 (2021). https://doi.org/10.1186/s44158-021-00002-x
Published:
DOI: https://doi.org/10.1186/s44158-021-00002-x