Abstract
Background
The current study was conducted to identify the genetic polymorphism of ghrelin (GHRL) gene of sheep and goats, as well as to determine whether these polymorphisms were associated with the evolutionary genetic differences in the involved species. This study was performed on 233 sheep and 91 goats. Two genetic loci of 113 bp and 262 bp partially spanning over exon 2/intron 2 and intron 4/exon 5 of GHRL gene respectively were amplified and genotyped using polymerase chain reaction–single-strand conformation polymorphism (PCR-SSCP) and DNA sequencing methods.
Results
The SSCP banding pattern of 262-bp locus indicated the presence of four diplotypes (BC, BB, AC, and AB) in Awassi sheep, three diplotypes (BC, BB, and AB) in Karadi sheep, and only two diplotypes (BC and BB) in all goats’ samples. The current study detected several novel SNPs in the ovine–caprine populations as well as two SNPs that are observed only in sheep, including intron4:119 C>A and intron4:123 T>G. The phylogenetic analysis revealed that the observed diplotypes resided within ovine sequences and were closely related to caprine counterparts. Computational analyses indicated the presence of various intronic RNA motifs. However, all these motifs were gathered in Awassi breed.
Conclusion
It is stated that the intron 4 is highly diverse amongst goats and sheep as well as within sheep with a particular emphasis on Awassi. This genetic peculiarity may in turn suggest a high polymorphic pattern of this breed in comparison with other related counterparts.
Similar content being viewed by others
Background
Ghrelin plays important roles in maintaining growth hormone release and energy homeostasis in vertebrates [1]. Ovine ghrelin is made up of 27 amino acids [2], which are encoded by GHRL gene that is located on the chromosome 19 and composed of 5 exons and 4 introns (NC_019476.2), while in goats, it is positioned in chromosome 22 and composed of only 4 exons and 3 introns (NC_030829.1). The GHRL gene polymorphisms have been associated with milk fat and protein synthesis in water buffaloes [3]; enhanced food intake, growth, and body conformation in cattle [4]; several growth traits in sheep [5]; and obesity in human [6]. Mutations in the GHRL gene could potentially cause a defective or inactive ghrelin hormone and alter growth hormone secretion and energy homeostasis [7]. In this regard, it can be emphasized that detection of the effect of GHRL genotype on the economically important phenotypic traits that it controls can only be possible through the identification of mutations that existed at the genome. On the other hand, it has been reported that the feeding regimes are significantly different amongst sheep and goats [8]. These variable regimes are maybe the consequences of the GHRL genetic variation status amongst them. Mutations can not only explore the gateway for searching molecular markers but also enable to break out the evolutionary relationship amongst animals of various species [9]. Keeping these facts in mind, the present study was undertaken to unravel the pattern of genomic diversity of both ovine and caprine GHRL gene through utilizing the SSCP technique that is characterized with its ability to identify the unknown mutations [10]. Besides, due to the lack of comparative knowledge about the genetic characterization and nucleotide sequence variations of GHRL gene amongst sheep and goat breeds, this study provides the first report on the detailed molecular characterization of the GHRL gene polymorphism in these highly worthwhile populations.
Methods
Blood sampling and DNA extraction
A total of randomly selected 324 animals that are maintained at different grazing herds in the middle Euphrates region of Iraq were included in this study. Regarding sheep (Ovis aries), both Awassi and Karadi breeds were included in the study. The studied native breeds of Awassi and Karadi constituted about 58% and 20% respectively of the national Iraqi sheep, which are characterized with their fat tail and carpet wool production and have some potential to produce milk [11]. Awassi and Karadi breeds are characterized by their ability to survive and reproduce under the condition of drought and extreme climate fluctuations [12]. Regarding goats (Capra hircus), both Shami and Native breeds were included in the present study. Blood samples were collected from 157 Awassi sheep, 76 Karadi sheep, 48 Shami goats, and 43 Iraqi Native goats. Genomic DNA was isolated using a salting out method [13]. The extracted genomic DNA was evaluated by 1% agarose gel electrophoresis in 1× TAE (40 mM Tris-acetate, 2 mM EDTA; pH 8.3) and quantified using a Nanodrop spectrophotometer (BioDrop μLITE; Biodrop, UK).
PCR primer sequences
Two pairs of specific PCR primers were utilized in this study. The first PCR pair, including the forward primer 5′-CCCTGCTCTGGATGGACTTGGC-3′ and the reverse primer 5′-GGCTTTGGGCATTTAGGACGC-3′, was made of 113 bp [5]. This specific fragment partially covered the coding sequences (CDS) of exon 2 and its intronic downstream region of the ovine GHRL sequences (NC_019476.2), whereas the second PCR pair, including the forward primer 5′-GCCAAACTGGATGGCAACAG-3′ and the reverse primer 5′-AACAGACAGGTGGTTGGTCC-3′, that was made of 262 bp was designed in the present study using the NCBI primer BLAST software (https://www.ncbi.nlm.nih.gov/tools/primer-blast). This 262-bp GHRL-specific fragment partially covered intron 4, all the CDS of exon 5, and a partial downstream region of exon 5 of the same ovine GHRL sequences. The referred PCR amplicons were used to amplify both ovine and caprine DNA sequences.
PCR
The PCR reaction was performed using the AccuPower PCR PreMix (Bioneer, South Korea). Each 20 μl of PCR premix contained 250 μM of dNTPs, 10 mM of Tris-HCl (pH 9.0), 30 mM of KCl, 1 U of Top DNA polymerase, and 1.5 mM of MgCl 2. The PCR reaction mixture was completed with 10 pmol of each primer and 30–50 ng of genomic DNA. The optimum annealing temperatures were determined empirically in our extracted genomic DNA template using a gradient PCR thermocycler (ver. Mastercycler-nexus; Eppendorf). The PCR program was as follows: initial denaturation at 94 °C for 5 min, followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 62.0 °C for 30 s, elongation at 72 °C for 30 s, and a final extension at 72 °C for 5 min. After performing the PCR thermocycling, the PCR products were verified by electrophoresis on a 1.5% (w/v) agarose gel in 1× TBE buffer (2 mM of EDTA, 89 mM of Tris-Borate, pH 8.3). All SSCP non-suitable PCR amplicon bands were eliminated.
SSCP
SSCP experiments were performed according to Al-Shuhaib et al. [14] protocol with some modifications. Several parameters were optimized to avoid false positive results as well as to enhance sensitivity. Briefly, 2.5 μl of each amplification product was mixed with an equal volume of SSCP denaturing loading buffer (95% formamide, 0.05% xylene cyanol, 20 mM EDTA pH 8, and 0.05% bromophenol blue). The PCR amplicons were heat-treated at 95 °C for 10 min and chilled on ice for at least 5 min. Then, PCR amplicons were separated in a vertical mini-wide gel format, gel size (216 × 110 mm) with gel thickness 1.0 mm (model JY-CZ-B, Junyi-Dongfang Electrophoresis Equipment, China). Denatured PCR products were loaded into the wells of 8% acrylamide/bisacrylamide (37.5:1), containing 7% glycerol, and 1× TBE buffer. The gel was run at constant conditions (250 V/125 mA/210 min for both PCR amplicons) at room temperature. Gels were stained by silver nitrate [15] and photographed.
DNA sequencing and sequencing analysis
Each of the varying patterns of the SSCP samples for polymorphic fragments was purified and sequenced from both directions (Macrogen Inc. Geumchen, Seoul, South Korea). Only evident chromatographs obtained from ABI sequence files were further analyzed, ensuring that the annotation and variations are not because of the PCR or sequencing artifacts. The sheep reference sequences of 262-bp locus were retrieved from NCBI websites (GenBank acc. no. NC_019476.2). Then, the sequenced diplotypes were edited, aligned, and compared with their referring sequences using the BioEdit Software, version 7.1 (DNASTAR; Madison, USA). The representative sequences of BC, AC, BB, and AB diplotypes were deposited into the NCBI GenBank database with the accession numbers MG387135–MG387138 respectively.
Bio-computational analysis
The observed variations were analyzed by the FSPLICE, which is a web server that can provide the opportunity to search for both donor and acceptor splicing sites and to define their thresholds (http://www.softberry.com). The intronic regulatory motifs for referring sequences as well as the observed diplotypes were analyzed using RegRNA (a regulatory RNA motifs and elements finder) web server (http://regrna.mbc.nctu.edu.tw/html/prediction.html).
Statistical analysis
The bio-statistical genetic diversity calculations were performed for the polymorphic 262-bp genetic fragment to estimate haplotype and diplotype frequencies, as well as Nei’s heterozygosity. These criteria were calculated with PopGene32 software, version 1.31 [16]. A χ2 test was determined to verify possible deviations from Hardy–Weinberg equilibrium (HW) expectations for the distribution of diplotypes. Average heterozygosity was employed to estimate genetic diversity within the population.
Phylogenetic analysis
A comprehensive phylogenetic tree was constructed, in which the observed GHRL variants were compared with each other and with their GHRL-based DNA sequences of other livestock animals using NCBI-blastn server [17]. An inclusive tree was made using the neighbor joining tree option of Clustal Omega server (https://www.ebi.ac.uk/Tools/msa/clustalo/). Subsequently, a radial tree layout was made and visualized by Figtree software (http://tree.bio.ed.ac.uk/software/figtree/). The accession numbers of the tree-involved organisms were annotated and colored appropriately.
Results
SSCP banding patterns
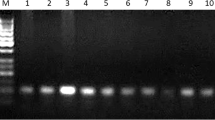
SSCP results showed the absence of any noticeable polymorphism in 113-bp fragment in all studied animals. Only two SSCP single-stranded DNA bands were revealed without any polymorphism in all 324 genomic DNA samples (Fig. 1a). In contrary to the 113-bp fragment, the 262-bp fragment showed four distinctly observed SSCP diplotypes, including BC, BB, AC, and AB, with three haplotypes, including A, B, and C. The SSCP results shown that the observed haplotypes were shared for both ovine and caprine fragments, while the distribution of these haplotypes was found to be different. The present study revealed two different SSCP diplotype distributions between sheep and goats. All four diplotypes were observed in Awassi sheep, including BC, BB, AC, and AB. In Karadi sheep, only three diplotypes were observed, including BC, BB, and AC with the absence of AB diplotype, whereas both BB and AB diplotypes were entirely absent in goats (Fig. 1).
PCR-SSCP sequencing patterns of GHRL gene in Awassi, Karadi, Shami, and Iraqi Native breeds. a The monomorphic nature of the 113 bp spanning over exon2/intron 2. b The polymorphic nature of 262 bp spanning over intron 4/exon 5. c The in silico consequences of several RNA motifs predicted in the referring sequences as well as each observed diplotype of the 262-bp fragment
Sequencing interpretation
Sequencing results confirmed the observed four electrophoretic SSCP patterns as several unique SNPs were detected amongst three resolved SSCP banding patterns, including BB, AC, and AB. Sequencing of AC diplotype revealed that it was characterized by the presence of intron4:123 T>G variant, whereas intron4:119 C>A variant was found to be shared with AB diplotype. The latter diplotype also had two other variations that were shared in turn with BB diplotype, namely intron4:29 T>G and intron4:36 A>C. In contrary to other SSCP resolved banding patterns, sequencing experiments shown that the most abundant BC diplotype had no unique variation(s). However, six in common variations were observed in the intron 4 of all identified diplotypes including g.7 C>A, g.61 C>T, g.71 C>G, g.93 C>T, g.129 C>T, and g.149 C>T (Table 1). The novelty of all observed variations was confirmed by reviewing the corresponding GHRL referring sequences using the ensemble genome browser 95 (https://asia.ensembl.org/index.html).
Bio-computational prediction
The FSPLICE prediction tool identified a noticeable alteration in the first position of the acceptor splicing site (1P) in all SSCP banding patterns. Various numbers in the threshold values for BC, AB, and BB diplotypes in comparison with the referring sequences were observed for the 1P, whereas this 1P position was entirely absent in AC diplotype (Table 2).
To add another layer of confirmation, the regulatory intronic RNA motifs of all diplotypes were analyzed using RegRNA. RegRNA is an integrated in silico web server for identifying the homologs of regulatory RNA motifs and elements to determine their roles in the transcriptional and post-transcriptional regulation of gene expression [18]. We indicated the presence of variable intronic RNA regulatory motifs in the intronic sequences of all observed diplotypes (Fig. 1c).
Statistical analysis
The most abundant SSCP diplotype was BC, which was observed in all studied population with a total frequency of 0.65 (n = 210). The highly dominant BC diplotype was followed by BB, which was found only in Awassi and Karadi sheep in a total frequency of 0.2 (n = 64). Then, AC diplotype, which was observed in all studied population, was recognized with only a small total frequency of 0.09 (n = 30), whereas it was found that AB diplotype that occupied the smallest total frequency of 0.06 (n = 20) was available only in Awassi sheep. The analysis of the GHRL 262-bp locus showed a prevalence of B haplotype, which is followed by C haplotype, while A haplotype was identified at a very low frequency in sheep in comparison to goats. The details of the diplotype distribution and haplotypes’ frequencies at the GHRL 262-bp locus are reported in Table 3.
The GHRL locus was tested for the Hardy–Weinberg equilibrium and characterized by observed (obs-Het) and expected (exp-Het) heterozygosity in all included four populations. All studied populations did not follow the Hardy–Weinberg equilibrium at the GHRL locus and showed medium heterozygosity as obs-Het equals 0.7834 in Awassi, 0.6 in Karadi, 1 in Shami, and 1 in Native and exp-Het equals 0.5497 in Awassi, 0.4676 in Karadi, 0.6015 in Shami, and 0.5496 in Native breeds respectively. The values of the obs-Het for GHRL 262-bp diplotypes were higher than their expected values. This refers to the high level of genetic variability in the studied populations.
Phylogenetic analysis
The phylogenetic analysis of the observed four diplotypes was conducted to explore the molecular taxonomic variations of GHRL variants. The phylogenetic tree provided a more detailed view of the specific taxonomic affinities shared by the investigated ovine–caprine sequences, reflecting their close evolutionary relationships. The total number of the aligned nucleic acid sequences, in addition to detected diplotypes, was 31, comprising the main livestock animals. The constructed phylogenetic tree revealed that all four GHRL diplotypes encompassed extremely close phylogenetic species positions (Fig. 2). These positions were positioned within ovine sequences, while caprine sequences occupied the nearest position in relation to their ovine counterparts. Unfortunately, the deposited caprine sequences were only one (AH0137212) and there were no other GHRL-based accession numbers regarding these sequences in caprine breeds, which may reduce the ability to exactly discriminate the observed diplotypes with the caprine species. However, the present phylogenetic tree demonstrated the phylogenetic positions and the close interactions between the observed diplotypes and with ovine and caprine sequences respectively.
A GHRL gene-based phylogenetic tree of the observed 262-bp SSCP banding patterns of Awassi and Karadi sheep and Shami and Iraqi Native goats with other livestocks. Both query variants were colored and highlighted in black, while other livestocks were colored appropriately. All the mentioned numbers referred to GenBank accession number of each reference species. The number “2.0” at the bottom of the tree referred to the degree of scale range amongst the tree-categorized species
Discussion
Recently, genetic polymorphisms at candidate genes affecting economic traits have stimulated research interest because they are considered as an inevitable guide to genetic selection and to mark evolutionary relationships in different livestock breeds [19]. The use of DNA markers to account for genetic variation in metabolism, growth, and appetite controlling genes may provide a tool to assist geneticists in determining their exact role in the inter-species and intra-species relationship though focusing on their variations. The GHRL gene may represent an ideal candidate for this purpose.
Though we have examined two exonic regions spanning over exon 2 and exon 5 through 113 bp and 262 bp respectively, no missense mutations were observed and the structure of the protein does not vary between species. Meanwhile, the only high polymorphic region was found to be restricted on the fourth intron. In consonance with our findings, it was shown that all the observed GHRL SNPs were located in intronic or in the non-coding regions as in the water buffaloes [3], cattle [20], and goats [21]. However, this study reported the highly intronic variations of the GHRL 262-bp fragment and the lack of any polymorphism in the GHRL 113-bp fragment across the ovine and caprine species. Our GHRL 113-bp results were in agreement with earlier studies that have also reported the monomorphic nature of the same investigated locus in all examined ovine populations [5, 22]. Conversely, the highly polymorphic nature of the GHRL gene was successfully highlighted in the present study by designing the 262-bp fragment. The genotyping of the latter fragment has shown that Awassi breed was the most polymorphic one amongst the others since all four SSCP banding patterns were available in Awassi sheep. This observation could be attributed to the high nucleotide diversity ratios that accumulated in Awassi breed around the world [23]. Karadi breed was less polymorphic as being shown only three SSCP banding patterns. In contrary to ovine species, all studied caprine species were shown only two diplotypes irrespective of their breed. Our finding suggests that this deficiency may be due to sheep–goat intronic differences that have been unmasked by utilizing this 262-bp GHRL fragment. As the reported level of GHRL intron 4 variation is not similar in different species, it is rather acceptable to infer that the intronic GHRL gene expression pattern varies from species to species. Our findings of variable alternative patterns of the intronic RNA motifs and splicing of GHRL 262-bp locus may also suggest variable roles of these alternations in the transcriptional modifications [18]. It was shown that the variations of the GHRL gene affected gene expression at the transcriptional level [24]. However, we suggest that GHRL RNA motif alterations might contribute to the differential level of several physiological ghrelin functions in the studied ovine–caprine population through inducing alternative transcriptional modifications. The variability in the RNA motifs has been demonstrated to generate a variety of alternatively spliced transcripts [25], or even altered protein structures [26], which have been shown to exhibit different metabolic activities on their corresponding products. Therefore, the currently observed GHRL mutations have potential motivation to induce a remarkable alteration in the resulting ghrelin function through producing unique RNA motifs in each observed diplotype. For instance, as GHRL gene powerfully increases food intake in diverse species [27], a significant role of the observed GHRL alterations may contribute to the food uptake status between the different species and even within the same species. In the latter case, the exclusive appearance of AB diplotype in Awassi sheep may participate in this phenomenon. Unfortunately, no reliable recording data in all Iraqi ovine and caprine breeds are available to verify this suggestion. Nonetheless, the significance of the discriminative power of the 262-bp fragment may occupy a more important role between sheep and goats. This role has a considerable impact since the feeding strategies have much more noticeable differences between ovine and caprine flocks [8]. Therefore, the absence of BB and AB diplotypes in the caprine DNA can potentially be connected to this natural phenomenon that accommodated in each species.
Our GHRL-based phylogenetic tree has highlighted a particular phylogenetic position for the investigated species which can be recognized easily from other related ovine–caprine species. However, this unique positioning could be attributed to the scarcity of deposited genomic sequences of the Awassi sheep, Karadi sheep, Shami goats, and Iraqi Native goats in the NCBI database. The construction of a phylogenetic tree is necessary in the presently employed GHRL amplicons. This may be attributed to the potential versatile role of the GHRL gene in the feeding strategy differences amongst these species to keep their adaptation to an ever-changing environment. In addition to feeding strategy alteration, several other ghrelin-related physiological characteristics can potentially be linked with the observed SSCP banding patterns in the GHRL 262-bp fragment. Therefore, a phenotypic association study is highly recommended to confirm this highly interesting distribution of SSCP banding patterns.
Conclusions
Based on the results of the present experiments, the conclusion arises that one of the most remarkable features of the GHRL gene in this study is its high degree of the ovine intronic polymorphism particularly within Awassi sheep, while its caprine counterpart has exerted less polymorphic status. This locus has a remarkable ability to evolutionary differentiate between sheep and goats and between Awassi and Karadi sheep breeds too. Besides, the pattern of 262-bp fragment polymorphism may provide an indicator to understand the mechanism(s) by which GHRL variations may influence the gene expression pattern in both ovine and caprine species.
Availability of data and materials
Not applicable
Abbreviations
- acc. no.:
-
Accession number
- exp-Het:
-
Expected heterozygosity
- GHRL :
-
Ghrelin
- HW:
-
Hardy–Weinberg equilibrium
- NCBI:
-
National Center of Biotechnology Information
- NCBI-blastn:
-
NCBI-blast nucleotide
- obs-Het:
-
Observed heterozygosity
- PCR-SSCP:
-
Polymerase chain reaction–single-strand conformation polymorphism
References
Kojima M, Kangawa K (2005) Ghrelin: structure and function. Physiol Rev 85:495–522. https://doi.org/10.1152/physrev.00012.2004
Kita K, Harada K, Nagao K, Yokota H (2005) Characteristics of gene structure of bovine ghrelin and influence of aging on plasma ghrelin. Asian-Aust J Anim Sci 18:723–727. https://doi.org/10.5713/ajas
Gil FM, de Camargo GM, Pablos de Souza FR, Cardoso DF, Fonseca PD, Zetouni L, Braz CU, Aspilcueta-Borquis RR, Tonhati H (2013) Polymorphisms in the ghrelin gene and their associations with milk yield and quality in water buffaloes. J Dairy Sci 96:3326–3331. https://doi.org/10.3168/jds.2012-6362
Menzies M, Seim I, Josh P, Nagaraj SH, Lees M, Walpole C, Chopin LK, Colgrave M, Ingham A (2014) Cloning and tissue distribution of novel splice variants of the ovine ghrelin gene. BMC Vet Res 10:211. https://doi.org/10.1186/s12917-014-0211-x
Tahmoorespur M, Taheri A, Valeh MV, Saghi DA, Ansary M (2010) Assessment relationship between leptin and ghrelin genes polymorphisms and estimated breeding values (EBVs) of growth traits in Baluchi sheep. J Anim Vet Adva 9:2460–2465. https://doi.org/10.3923/javaa.2010.2460.2465
Miraglia del Giudice E, Santoro N, Cirillo G, Raimondo P, Grandone A, D’Aniello A, Di Nardo M, Perrone L (2004) Molecular screening of the ghrelin gene in Italian obese children: the Leu72Met variant is associated with an earlier onset of obesity. Int J Obes Relat Metab Disord 28:447–450. https://doi.org/10.1038/sj.ijo.0802572
Ukkola O, Ravussin E, Jacobson P, Pérusse L, Rankinen T, Tschöp M, Heiman ML, Leon AS, Rao DC, Skinner JS, Wilmore JH, Sjöström L, Bouchard C (2002) Role of ghrelin polymorphisms in obesity based on three different studies. Obes Res 10:782–791. https://doi.org/10.1038/oby.2002.106
Blache D, Martin GB (2009) Focus feeding to improve reproductive performance in male and female sheep and goats: how it works and strategies for using it. In : Papachristou TG (ed.), Parissi ZM (ed.), Ben Salem H. (ed.), Morand-Fehr P. (ed.). Nutritional and foraging ecology of sheep and goats. Zaragoza : CIHEAM / FAO / NAGREF. p. 351-364 (Options Méditerranéennes : Série A. Séminaires Méditerranéens; n. 85), http://om.ciheam.org/article.php?IDPDF = 801028
Bhattacharya TK, Sheikh FD, Sukla S, Kumar P, Sharma A (2007) Differences of ovine butyrophilin gene (exon 8) from its bovine and bubaline counter part. Small Ruminant Res 69:198–202. https://doi.org/10.1016/j.smallrumres.2005.12.006
Gasser RB, Hu M, Chilton NB, Campbell BE, Jex AJ, Otranto D, Cafarchia C, Beveridge I, Zhu X (2006) Single-strand conformation polymorphism (SSCP) for the analysis of genetic variation. Nat Protoc 1:3121–3128. https://doi.org/10.1038/nprot.2006.485
Al-Rawi AA, Al-Haboby AH, Al-Salman MH (1996) Small ruminant breeding and reproductive physiology research and technology transfer in Iraq (W. Mourrani and N. Haddad, ed.). ICARDA-West Asia Regional Program, Amman, Jourdan.
Alkass JE, Juma KH (2005) Chapter 3: Small ruminant breeds of Iraq. In: Iniguez L (ed) Characterization of small ruminant breeds in West Asia and North Africa, vol 1. West Asia. International Center for Agricultural Research in Dry Areas (ICARDA), Aleppo, Syria, pp 63–101. https://doi.org/10.1017/S0021859606006393
Al-Shuhaib MBS (2017) A universal, rapid, and inexpensive method for genomic DNA isolation from the whole blood of mammals and birds. J Genet 96:171–176. https://doi.org/10.1007/s12041-017-0750-6
Al-Shuhaib MBS, Al-Kafajy FR, Badi MA, AbdulAzeez S, Marimuthu K, Al-Juhaishi HAI, Borgio JF (2018) Highly deleterious variations in COX1, CYTB, SCG5, FK2, PRL and PGF genes are the potential adaptation of the immigrated African ostrich population. Comput Biol Med 100:17–26. https://doi.org/10.1016/j.compbiomed.2018.06.019
Byun S, Fang Q, Zhou H, Hickford J (2009) An effective method for silver-staining DNA in large numbers of polyacrylamide gels. Anal Biochem 385:174–175. https://doi.org/10.1016/j.ab.2008.10.024
Yeh FC, Yang R, Boyle T (1999) POPGENE: version 1.31. Microsoft Window–based freeware for population genetic analysis, University of Alberta. Edmonton, AB, Canada.
Zhang Z, Schwartz S, Wagner L, Miller W (2000) A greedy algorithm for aligning DNA sequences. J Comput Biol 7(1-2):203–214. https://doi.org/10.1089/10665270050081478
Huang HY, Chien CH, Jen KH, Huang HD (2006) RegRNA: an integrated web server for identifying regulatory RNA motifs and elements. Nucleic Acids Res 34(Web Server):W429–W434. https://doi.org/10.1093/nar/gkl333
Naval-Sanchez M, Nguyen Q, McWilliam S et al (2018) Sheep genome functional annotation reveals proximal regulatory elements contributed to the evolution of modern breeds. Nat Commun 9(1):859. https://doi.org/10.1038/s41467-017-02809-1
Colinet FG, Portetelle D, Renaville R (2009) Molecular characterization of the bovine GHRL gene. Arch Tierzucht 52:70–84
Jin Q, Xingtang F, Zhang CL, Yang L (2010) A novel SNP of the GHRL gene in goat and its association with growth traits. Small Rum Res 90(1):150–152. https://doi.org/10.1016/j.smallrumres.2010.01.001
Bahrami A, Behzadi S, Miraei-Ashtiani SR, Roh SG, Katoh K (2013) Genetic polymorphisms and protein structures in growth hormone, growth hormone receptor, ghrelin, insulin-like growth factor 1 and leptin in Mehraban sheep. Gene 527:397–404. https://doi.org/10.1016/j.gene.2013.05.066
Pedrosa S, Arranz JJ, Brito N, Molina A, San Primitivo F, Bayon Y (2007) Mitochondrial diversity and the origin of Iberian sheep. Sel Evol 39:91–103. https://doi.org/10.1186/1297-9686-39-1-91
Nakaya HI, Amaral PP, Louro R, Lopes A, Fachel AA, Moreira YB, El-Jundi TA, da Silva AM, Reis EM, Verjovski-Almeida S (2007) Genome mapping and expression analyses of human intronic noncoding RNAs reveal tissue-specific patterns and enrichment in genes related to regulation of transcription. Genome Biol 8:R43. https://doi.org/10.1186/gb-2007-8-3-r43
Shepard PJ, Hertel KJ (2008) Conserved RNA secondary structures promote alternative splicing. RNA 14(8):1463–1469. https://doi.org/10.1261/rna.1069408
Qu W, Cingolani P, Zeeberg BR, Ruden DM (2017) A bioinformatics-based alternative mRNA splicing code that may explain some disease mutations is conserved in animals. Front Genet 8:38. https://doi.org/10.3389/fgene.2017.00038
Tschop M, Smiley DL, Heiman ML (2000) Ghrelin induces adiposity in rodents. Nature 407:908–913. https://doi.org/10.1038/35038090
Acknowledgements
Not applicable
Funding
This work did not receive funding from any organization or institution.
Author information
Authors and Affiliations
Contributions
MBS designed the work, analyzed data, and wrote the manuscript. TM conducted the main experiments. IA and TR participated in conducting some experiments. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The Experimental research on sheep comply with international guidelines and have been approved by the international recommendations for the care and use of animals, and both maintenance and feeding were similar for all animals and remained in accordance with proper animal welfare guidelines for the care and use of sheep (Federation of Animal Science Societies, 2010).
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Al-Shuhaib, M.B.S., Al-Thuwaini, T.M., Fadhil, I.A. et al. GHRL gene-based genotyping of ovine and caprine breeds reveals highly polymorphic intronic sequences in Awassi sheep with several RNA motifs. J Genet Eng Biotechnol 17, 3 (2019). https://doi.org/10.1186/s43141-019-0004-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43141-019-0004-5