Abstract
Background
The infamous multidrug-resistant (MDR) bacterium Acinetobacter baumannii is becoming a nightmare in intensive care units across the globe. Since there are now very few effective antimicrobial agents, it is necessary to explore unconventional resources for novel antimicrobials. This study investigated the potential antimicrobial activity of Origanum majorana L. against A. baumannii employing multiple approaches including antimicrobial susceptibility, fractionation, ultra-performance liquid chromatography–high-resolution mass spectrometry (UPLC-HRMS) dereplication, and in silico analysis for target/ligand identification.
Results
On the extremely pathogenic MDR strain A. baumannii AB5075, O. majorana L. has shown a significant growth inhibitory effect (MIC = 0.675 mg/mL). The polar 50% methanol fraction was the most active (MIC = 0.5 mg/mL). The UPLC-HRMS dereplication of the bioactive fraction detected 29 metabolites belonging to different chemical classes. Justicidin B, one of the identified metabolites, was projected by preliminary in silico analysis to be the most highly scoring metabolite for binding with molecular targets in A. baumannii with a Fit score = 8.56 for enoyl-ACP reductase (FabI) (PDB ID: 6AHE), suggesting it to be its potential target. Additionally, docking, molecular dynamics simulation, and bioinformatics analysis suggested that this interaction is similar to a well-known FabI inhibitor. The amino acids involved in the interaction are conserved among different MDR A. baumannii strains and the effectiveness could extend to Gram-negative pathogens within the ESKAPE group.
Conclusions
Origanum majorana L. extract exhibits antimicrobial activity against A. baumannii using one or more metabolites in its 50% methanol fraction. The characterized active metabolite is hypothesized to be justicidin B which inhibits the growth of A. baumannii AB5075 via targeting its fatty acid synthesis.
Similar content being viewed by others
Background
Healthcare practitioners everywhere give careful consideration to Acinetobacter baumannii as it is a major cause of nosocomial infections. A. baumannii is frequently responsible for outbreaks in intensive care units that show up as urinary tract infections, surgical site infections, pneumonia linked to ventilator use, and sepsis [1]. Based on pooled data from various regions globally, A. baumannii was found to produce approximately 25 hospital-acquired infection cases per 1000 patients; in intensive care units, this ratio rose to 54 cases per 1000 patients [2]. Furthermore, the problem has gotten worse due to its resistance to several antimicrobial agents that are currently available, as well as, its acquired resistance to their alternatives.
Multidrug-resistant (MDR) A. baumannii are increasingly reported worldwide reaching 45% of A. baumannii infections, exceeding the MDR rates reported for all other common nosocomial pathogens [3]. This has placed A. baumannii among the ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, A. baumannii, Pseudomonas aeruginosa, and Enterobacter spp.) [4,5,6]. For multidrug-resistant Gram-negative bacteria, including A. baumannii, colistin and carbapenems like doripenem, imipenem, and meropenem, are typically regarded as the last resort treatments [7,8,9]. Nevertheless, the administration of colistin has been restricted due to its toxicity, including nephrotoxicity and neurotoxicity [10, 11]. The frequency of isolation of carbapenem-resistant A. baumannii (CRAB) is increasing with currently few alternatives for treatment. The Centres for Disease Control and Prevention (CDC) has listed CRAB as an “urgent threat” to human health [12, 13]. In a developing country like Egypt, the rate of isolating CRAB from patients reached 80% and higher [14]. After the COVID-19 pandemic, the reported infections by CRAB increased by 78%, which is highly alarming [13].
The development of novel antibiotics is essential due to the high mortality rates associated with CRAB infections [15, 16]. This has warranted WHO to include A. baumannii, along with other carbapenem-resistant pathogens, P. aeruginosa and Enterobacteriaceae, to be critically prioritized in the research and development of new antibiotics [17].
Plant-produced secondary metabolites, sometimes referred to as phytochemicals, may offer chemical resistance against infectious agents. The abundance of these metabolites, some with efficacies comparable to that of synthetic antibiotics, provides a repertoire of chemicals that could be used to successfully create and deploy new antimicrobial agents against MDR-ESKAPE pathogens [18,19,20].
Origanum majorana L. commonly referred to as Sahtar, Zaatar or sweet marjoram is a photoautotrophic medicinal perennial herb of the Origanum genus, which belongs to the Lamiaceae family [21]. This plant is found broadly all around the Mediterranean region, but especially in Morocco, Algeria, Egypt, Spain, and Portugal [22]. Traditional medicine makes extensive use of it, as a possible treatment for a wide range of illnesses including allergies, respiratory infections, hypertension, diabetes, and stomach pain, as well as, an intestinal antispasmodic, [23]. Pharmacological research revealed that this plant’s essential oils and extracts possessed a variety of biological qualities, including hepatoprotective, antimutagenic, anticancer, antiparasitic, antibacterial, antifungal, antidiabetic, antioxidant, anti-inflammatory, and analgesic effects [24]. The plant extracts’ safety and therapeutic advantages have been verified by a toxicological evaluation [25]. In rats, O. majorana L. extracts did not result in any fatalities, which further confirms their safety [26, 27]. O. majorana L. has previously been found to contain a wide range of bioactive compounds from various chemical classes, including hydroquinone, sterols, terpenoids, tannins, phenolic acids (arbutin and methyl arbutin), fatty alcohols, and flavonoids (diosmetin, orientin, luteolin, apigenin, vitexin, and thymonin) [28]. Thymol, carvacrol, cis-sabinene hydrate, limonene, terpinene, and camphene are among the plant’s abundant volatile oil constituents [28]. It is important to bear in mind that different plant organs, growth stages, and even harvesting techniques can have different effects on the amount and composition of the aforementioned metabolites and volatile oil components within the same species [29]. Antibacterial activity has been demonstrated by the plant against a variety of pathogenic bacteria, including E. faecalis, Bacillus subtilis, Escherichia coli, K. pneumoniae, Serratia sp. and Salmonella choleraesuis [30,31,32].
In search of prospective treatments for MDR A. baumannii, this study aimed to investigate the antibacterial activity of O. majorana L. extract and its fractions. UPLC-HRMS was used to analyse the chemical profile of the active 50% methanol fraction (50% MF) in order to establish a relationship between the antibacterial activity and the bioactive secondary plant metabolites. Justicidin B, a secondary metabolite identified in O. majorana L. bioactive fraction, has been identified as a potential novel antimicrobial agent against A. baumannii and other ESKAPE pathogens, which targets the essential FAS-II fatty acid biosynthesis pathway component FabI.
Methods
Bacterial strains and culture conditions
The multidrug-resistant highly virulent A. baumannii strain AB5075 [33] was obtained from Prof. Dr. Lory, Harvard University, USA as a gift, and was used as the test microorganism in all the experiments. The bacterial strain was typically grown in tryptic soy broth (TSB) with shaking at 180 rpm at 37 °C or plated on tryptic soy agar (TSA) and incubated at 37 °C.
Preparation, extraction and fractionation of O. majorana L. aerial parts
Origanum majorana L. aerial parts were collected from the Experimental Station of the Faculty of Pharmacy, Cairo University, Giza, Egypt. Mrs Therese Labib, Consultant at Orman Botanic Garden, Dokki, Giza, Egypt, kindly authenticated the plant material. Air-dried powder of O. majorana L. aerial parts (0.5 kg) was exhaustively extracted with methanol (5 × 7 L) and evaporated on a rotary evaporator to yield the total methanolic extract (ME, 75 g). Part of the residue (45 g) was suspended in 350 mL of distilled water and then fractionated using dichloromethane (3 × 750 mL), which was then evaporated to get DCM-F (25 g). The mother liquor (ML) was applied on the Diaion-HP20 column and eluted firstly with distilled water (1L, discarded), followed by 50% methanol (1 L), and then 100% methanol (1 L). The fractions were evaporated to yield 50% MF (4 g) and 100% MF (6 g), respectively.
Preliminary screening of O. majorana L. extract for antimicrobial activity
An overnight culture of A. baumannii AB5075 was grown in TSB at 37 °C and 180 rpm. The culture was diluted in TSB to OD600 = 0.1 arbitrary units, then diluted 1:1000 in TSA which was prewarmed to 50 °C. The inoculated TSA (50 mL) was distributed in a 15-cm-wide petri dish. After agar solidification, 10 µL of the extract, or the fraction, were spotted on the agar surface, the spots were dried in a laminar flow cabinet, and the plates were incubated at 37 °C. After 24 h, the plates were inspected visually, and the diameters of the growth inhibition zones were measured using a ruler.
Determination of the minimum inhibitory concentration (MIC)
Following the criteria of the Clinical and Laboratory Standards Institute (CLSI), the MIC was ascertained using the broth microdilution method [34]. The dried extract or fraction was dissolved in DMSO to a final concentration of 100 mg/mL. Then, it was diluted in Muëller-Hinton broth to yield a series of twofold dilutions ranging from 0.001 to 2 mg/mL. One hundred µL aliquots from each concentration were distributed in a 96-well plate. Each well was inoculated with 10 µL (~ 105 CFU as determined spectrophotometrically by measuring absorbance at 600 nm and verified by viable count) of a freshly prepared bacterial suspension. The plates were incubated for 24 h at 37 °C. The MIC was the minimum concentration of the tested crude extract or its fractions that inhibited visible bacterial growth. Muëller-Hinton broth containing the equivalent of DMSO served as a negative control and the experiment was performed in triplicate.
UPLC-HRMS analysis of the active fraction
The O. majorana L. active subfraction (50% MF) was subjected to UPLC-HRMS analysis followed by dereplication (identification of the secondary metabolites present in the extract) using the Dictionary of Natural Products and Reaxys online databases. The dereplication depended on the HRMS isotope profiles, MS/MS fragmentation, and the databases were filtered to include only the plant genus or family.
A Bruker MAXIS II Q-ToF mass spectrometer connected to an Agilent 1290 UHPLC system was used to analyse the samples. The column used was a Phenomenex Kinetex XB-C18 (2.6 mM, 100 × 2.1 mm). The following LC gradient profile was used to achieve separation: 5% MeCN + 0.1% formic acid to 100% MeCN + 0.1% formic acid in 15 min at a flow rate of 0.1 mL/min. MS parameters were: mass range m/z 100–2000, capillary voltage 4.5 kV, nebulizer gas 4.0 bar, dry gas 9.0 L/min, and dry temperature of 250 °C. MS/MS experiments with a step collision energy of 80–200% were carried out using the Auto MS/MS scan mode.
Virtual target identification
PharmMapper was used to characterize potential target(s) of justicidin B [35]. This platform uses an extracted pharmacophore model and stores it as a library ligand dataset in mol2 format. Then, it rates each molecule in the PDB according to how well it fits the model. This approach yields a pure Fit score that is far more reliable and significant than chance pharmacophore matching. The query structure was submitted to the platform in the pdb format, and the results were sorted based on the Fit scores.
Docking study
Using an AutoDock Vina docking machine, the crystal structures of FabI (PDB ID: 6AHE) were utilized for the docking study [36]. The co-crystallized ligand AFN-1252 was used to determine the binding site and the docking grid-box. The coordinates of the grid-box were set to be x = −36.404, y = −8.512, z = −3.293.
A criterion of 2.0 Å was established for the ligand to binding site shape matching root-mean-square deviation (RMSD). The Charmm force field (v.1.02) with a distance-dependent dielectric and a non-bonded cut-off distance of 10.0 was used to obtain the interaction energies. Then, an energy grid was set at 5.0 Å from the binding site [36]. Inside the chosen binding pocket, the studied compound, justicidin B, was energy-minimized. Pymol software was used to edit and visualize the generated binding poses [37].
Molecular dynamics simulation (MDS)
NAMD 3.0.0. software was used to perform MDS [38, 39]. Protein systems were constructed with the VMD software’s QwikMD toolkit [39, 40], and the protonation states of the amino acid residues were set (pH 7.4). The protein structure was examined for any missing hydrogens, and the co-crystalized water molecules were then removed. Following that, the entire structure was immersed in an orthorhombic box of TIP3P water together with 0.15 M Na+ and Cl− ions in a 20 Å solvent buffer. The systems were then energy-minimized and allowed to equilibrate for five nanoseconds. With the help of the VMD plugin Force Field Toolkit (ffTK), the ligand topologies and properties were determined. Afterwards, the produced parameters and topology files were loaded into VMD so that the simulation steps could be carried out and the protein–ligand complexes could be interpreted without errors.
Binding free energy calculations
The molecular mechanics Poisson–Boltzmann surface area (MM-PBSA) method from the AMBER18’s MMPBSA.py module was used to calculate the docked complex’s binding free energy by applying the following equation complex ΔGBinding = ΔGComplex – ΔGReceptor – ΔGInhibitor [41].
Conservation of the FabI sequences
The amino acid sequence of the FabI (PDB ID: 6AHE) enzyme, which showed the best-Fit score with justicidin B, was retrieved from the NCBI database. To confirm the conservation of FabI in different strains of A. baumannii and different strains of ESKAPE pathogens, the protein sequence of FabI was analysed by the BlastP tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins) [42], using a nonredundant protein sequences database against each organism (A. baumannii, E. faecium, S. aureus, K. pneumoniae, P. aeruginosa and Enterobacter spp.) with an expect threshold of 0.001. Proteins were considered conserved if they had a max alignment score of more than 200.
The conservation of amino acids in the predicted binding site for justicidin B was further confirmed by the alignment of the sequences of representative proteins from MDR A. baumannii strains (including those from A. baumannii ATCC 19606 (PDB ID:6AHE) and A. baumannii AB5075) and from representative strains of different ESKAPE pathogens. The sequences of the target proteins were retrieved from the NCBI database. The protein sequences were aligned using the multiple sequence alignment Clustal Omega tool with default settings (https://www.ebi.ac.uk/Tools/msa/clustalo/) [43].
Results
O. majorana L. total methanolic extract and its fractions have potent growth inhibitory effects against A. baumannii
The O. majorana L. total methanolic extract was spotted on the trypticase soy (TS) agar seed inoculated with A. baumannii AB5075. It showed a 14-mm zone of growth inhibition after 24-h incubation (Fig. 1A). Using the broth microdilution method, the minimum inhibitory concentration (MIC) of the total methanolic extract of O. majorana L. was determined at 0.675 mg/mL. Chromatographic techniques were used to prepare subfractions by partitioning with solvents of different polarities and applying them on a Diaion column. The 50% methanol fraction showed considerable antimicrobial activity (Fig. 1B). Upon measuring the MIC for this fraction, it showed a value of 0.5 mg/mL. On the other hand, the non-polar dichloromethane fraction had a higher MIC value of 0.875 mg/mL.
Growth-inhibitory effects of O. majorana L and its fractions against A. baumannii. Ten µl of the total methanolic (TM) extract (A) of O. majorana L or the 50% methanol fraction from Diaion-HP20 (50% MF) (B) were spotted on TSA plates inoculated with A. baumannii AB5075 and the plates were incubated overnight at 37 °C. In both cases, equivalent amounts of the solvent (DMSO) were spotted on the same plates. The experiment was repeated three times, and representative images were presented
UPLC-HRMS analysis of O. majorana L. active fraction reveals multiple metabolites.
To unlock the chemical diversity of the active fraction of the O. majorana L., UPLC-HRMS profiling revealed the presence of 29 metabolites belonging to different structural classes, comprising two terpenes, two alkaloids, one lignan, nine flavonoid aglycones, four flavonoid glycosides, seven phenolic acid derivatives, and two alicyclic derivatives. Moreover, three hits were not identified, indicating new compounds or compounds that were not reported before in the family Lamiaceae (Table 1).
Virtual screening-based target characterization
PharmMapper was employed to suggest a suitable protein target for the identified metabolites. The retrieved results were arranged according to their Fit scores. Only bacterial targets related to the Acinetobacter genus were considered. A threshold Fit score of 7 was set to select the best-scoring hits. As a result, A. baumannii-derived enoyl-ACP reductase (FabI) (PDB ID: 6AHE) was found to be among the top-scoring hits for justicidin B (Fit score = 8.56), where all the remaining bacterial hits were below the cut-off score of 7. Hence, this target (i.e. FabI) was proposed as its potential target, and justicidin B was the only compound among the 29 LC-HRMS-dereplicated metabolites that scored above the threshold Fit score.
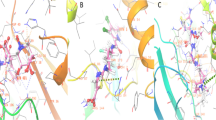
Re-docking justicidin B modelled structure into the FabI active site achieved binding mode and docking score comparable to those of the co-crystalized inhibitor AFN-1252 (docking scores = −10.67 and −11.75 kcal/mol, respectively). Justicidin B was able to establish two H-bonds with TYR-149 and TYR-159. In addition, it was involved in multiple hydrophobic interactions with PHE-96, LEU-102, TYR-149, and ILE-203. Similarly, the co-crystallized inhibitor was involved in the same hydrophobic interactions and formed H-bonds with both TYR-159, as seen in justicidin B, and with ALA-97 (Fig. 2A, B).
Binding modes of justicidin B (brick red-coloured structure) (A) and the co-crystalized inhibitor AFN-1252 (Cyan-coloured structure) (B) inside the binding site of FabI (PDB ID: 6AHE). C RMSDs of both structures (i.e. Justicidin B and AFN-1252) inside the binding site of FabI throughout 100 ns-long MD simulation. The yellow-coloured structure is the co-factor Nicotinamide Adenine Dinucleotide (NAD)
According to subsequent molecular dynamic simulation (MSD) experiments (100 ns-long), the justicidin B structure was able to achieve stable binding mode throughout the simulation, with an average Root Mean Square Deviation (RMSD) of 1.96 Å, which was similar to that of the co-crystallized inhibitor AFN-1252 (average RMSD = 1.65 Å; Fig. 2C). Such stable bindings of both justicidin B and AFN-1252 were translated into stable and significant interaction energies (electrostatic and van der Waals) inside the enzyme’s binding site, where both compounds showed total interaction energies averaged around − 26.16 and − 44.27 kcal/mol, respectively (Fig. 3A, B). Hence, their calculated absolute binding free energies were comparable as well (ΔGBinding = -8.11 and -9.52 kcal/mol, respectively).
According to the previous modelling and MD simulation findings, it could be hypothesized that O. majorana L. extract was able to inhibit the growth of A. baumannii via targeting its fatty acid synthesis by one of its metabolites (i.e. justicidin B).
The conservation of the predicted interaction sites of justicidin B and FabI in A. baumannii and among members of the ESKAPE group
To check if the predicted interaction of justicidin B and FabI could be extended to other strains of A. baumannii, we performed a blast analysis for the enoyl-ACP reductase (FabI) amino acid sequence against A. baumannii strains in the National Centre for Biotechnology Information (NCBI) database. The alignment score was > 200, indicating a high degree of conservation of the overall amino acid sequence of this protein. Upon performing a multiple sequence alignment of the amino acid sequences of the FabI protein in fifteen MDR and CRAB strains, all five amino acids involved in the interaction were conserved (TYR-149, TYR-159, PHE-96, LEU-102, and ILE-203) (Fig. 4A). Moreover, to determine if this interaction could be extended to other members of the ESKAPE pathogens, the FabI amino acid sequence of representative MDR strains from the members of this group was retrieved when another multiple sequence alignment was performed (Fig. 4B). The results of this alignment indicated that all the five amino acids involved in the interaction between justicidin B and FabI are conserved (TYR-149, TYR-159, PHE-96, LEU-102, and ILE-203) in the Gram-negative members: K. pneumoniae, A. baumannii, P. aeruginosa and Enterobacter spp. On the other hand, for the Gram-positive members S. aureus and E. faecium, all the interaction sites were conserved except the non-polar amino acid ILE-203 which was replaced with another non-polar amino acid, valine, and TYR replaced PHE-96 in E. faecium. This change in one amino acid in these two strains could have a minimal impact on justicidin B binding.
Alignment of the protein sequences of the enoyl-ACP reductase (FabI) in (A) MDR and CRAB A. baumannii strains and (B) among ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter spp.) pathogens. The amino acid sequence of each protein was retrieved from the NCBI database and aligned using Clustal Omega software. The alignment visualization was done using Jalview 2.11.3.1. The amino acids involved in justicidin B binding are highlighted
Discussion
The rise of antibiotic-resistant pathogens and the difficulties in developing antimicrobials have aroused concerns among medical professionals for the well-being of humanity. Plants, a rich source of undiscovered metabolites, offer a solution as a natural source of antimicrobial compounds because they are attainable and generally safe to use. O. majorana L. or sweet marjoram is an aromatic, herbaceous, perennial plant in the family Lamiaceae. The plant has been used as a flavouring and herbal spice and is widely reported for its antibacterial activity against multiple pathogens, including E. coli, B. subtilis, B. megaterium, S. aureus, P. aeruginosa, and Proteus vulgaris [44, 45].
In the current study, the methanolic extract of O. majorana L. demonstrated remarkable antimicrobial activity against A. baumannii with a MIC of 0.675 mg/mL. The antimicrobial activity of the methanolic and ethanolic extracts and the decoction of O. majorana L. was reported previously against many Gram-positive and Gram-negative pathogens and Candida spp. [46, 47]. Only one study evaluated and proved its antimicrobial activity against A. baumannii, using the disc diffusion method [48]. The MIC of the methanolic extract was reported previously to be > 100 µg/mL against S. aureus and E. coli [49]. A higher MIC (1.56 mg/mL) was determined for the decoction of O. majorana L. against S. aureus and K. pneumoniae [47]. The ethanolic fraction has MIC between 40 and 80 µg/mL, against E. coli and K. pneumoniae [46]. Additionally, the essential oil fraction of the plant was found to be highly active against different bacteria, fungi and protozoa [31, 49, 50]. However, it was not quantitatively tested against A. baumannii.
To pinpoint the active fraction responsible for the antimicrobial activity against A. baumannii in this study, the plant was fractionated, where the polar 50% methanol fraction was found to be the most active subfraction (MIC = 0.5 mg/mL). Chemical profiling of the bioactive fraction by UPLC-HRMS analysis revealed 29 metabolites belonging to diverse chemical classes. The dominant metabolites were phenolic acids, flavonoids, and lignans [51]. Phenolic acids such as rosmarinic acid, caffeic acid, and chlorogenic acid were previously reported in O. majorana L. [29]. They are beneficial against several chronic diseases such as degenerative diseases, cancer, diabetes, and ageing [52]. However, to the best of our knowledge, no reports are available about the antimicrobial activity of these metabolites from O. majorana L. against A. baumannii. The antimicrobial activity of many of these metabolites against Gram-positive and Gram-negative pathogens, such as naringenin [53], quercetin [54], and caffeic acid [55], was previously reported.
Two alicyclic derivatives, 12-hydroxy jasmonic acid and its 12-O-glucoside, were previously isolated for the first time from Origanum species, i.e. polar extracts of O. vulgare L [56]. Later, they were isolated from O. dictamnus L. Only 12-hydroxy jasmonic acid and not its glucoside exhibited a strong antibacterial effect against A. hemoliticus [57]. When screened for its phytotoxic potential against different phytopathogens, bioguided fractionation of the most active O. majorana L. fraction led to the isolation of different molecules, including a new compound named majoradiol, a carvacrol dimer, which was characterized but was not tested for its bioactivity due to its scarcity [58]. Aristolochic acid I and aristolochic acid II were isolated from O. vulgare and showed potent activity against leukaemia. Additionally, this is the first report to find that aristolochic acid I and aristolochic acid II demonstrated potent antithrombin activity [59]. Justicidin B, an arylnaphthalene lactone, is a plant-derived subclass of lignans which is distributed in many plants and different families. It is the main active component of Phyllanthus piscatorum, exhibiting strong antifungal, antiprotozoal, and anti-proliferative properties [60].
Caffeic acid was isolated from Origanum dictamnus L. and demonstrated a good antibacterial effect against A. hemoliticus and P. aeruginosa [57]. Chlorogenic acid, rosmarinic acid, and rosmarinic acid methyl ester were isolated from the methanol extract of O. dictamnus L. and exhibited weak antibacterial activity against S. aureus [61]. Amburoside A was previously isolated from O. micranthum and exhibited a weak carbonic anhydrase inhibitory effect [62]. Oreganol A and B were isolated from the extract of dried leaves O. vulgare and exhibited strong 1,1-dipehnyl-2-picrylhydrazyl (DPPH) radical scavenging activity [63].
Naringenin was previously reported in O. dictamnus L. and showed a strong antibacterial effect against Acinetobacter hemoliticus [57]. Despite the structural similarity with naringenin, apigenin was also isolated from the same species in the same study by Chatzopoulou and co-workers but did not demonstrate any antibacterial activity [57]. Genkwanin, salvigenin, and 5,6,4'-trihydroxy-7,3'-dimethoxyflavone were isolated from the chloroform extract of glandular hairs of O. x intercedens [64]. Genkwanin is known for its potent antiviral activity against African swine fever viral infection in Vero cells in a dose-dependent manner [65]. Gardenin B, quercitrin, 5-hydroxy-4',6,7-trimethoxyflavone, and aromadendrin were isolated from O. majorana L. with other flavonoid aglycones [58]. Aromadendrin demonstrated moderate anti-inflammatory activity by the inhibition of COX-1 [66]. Apigenin-7-O-glucuronide, luteolin-7-O-glucuronide, acacetin-7-O-rutinoside, and vicenin-2 were isolated from the methanol extract of O. dictamnus. None of them demonstrated antibacterial effects against the Gram-positive or Gram-negative strains tested [61].
It can be challenging to characterize a given molecule’s biological target. The success rate of identifying accurate molecular targets has greatly increased due to the ongoing development of in silico methods like virtual screening and molecular modelling. Nowadays, a plethora of ligand-based or structural search methods are employed in various online target identification platforms. PharmMapper is a dependable platform that makes use of its pharmacophore model to screen and propose the most probable protein targets for a query chemical [35]. Pharmacophore-based screening works on the basic premise that pharmacophore maps, which delineate the spatial arrangement of structural features, are the primary determinants of the binding of certain compounds to their protein targets. Therefore, molecules whose shapes match these pharmacophore maps have the best chance of binding to the same protein target.
PharmMapper software was employed to screen the 29 metabolites detected in the 50% methanol fraction for binding with putative protein targets in A. baumannii. The metabolite justicidin B recorded the highest Fit score (8.56) with the A. baumannii-derived enoyl-ACP reductase (FabI). Justicidin B is an aryl naphthalene lignan previously identified in the Lamiaceae family [67], but it is identified here for the first time in O. majorana L. extract. The antimicrobial activity of justicidin B was previously reported against S. aureus, E. coli, and P. aeruginosa. In addition, justicidin B has been proven to have antiviral, antifungal, and antiparasitic properties. It also exhibits anti-inflammatory, antioxidant and anticancer properties [68]. It was originally isolated through the bioactivity-guided fractionation of the dichloromethane extract of P. piscatorum and demonstrated non-specific cytotoxicity, in addition to potent in vitro fungicidal and antiprotozoal effects [60]. It recently demonstrated strong antiviral potential against Zika virus [69], an effective inhibitor of the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) [70], and strong cytotoxic activity against HeLa cervical cancer cells through targeting key proteins involved in apoptosis regulation [71]. It was even isolated from a marine-derived actinomycete, Nocardia sp. ALAA 2000 and exhibited a broad-spectrum antimicrobial effect against a panel of Gram-positive and Gram-negative strains as well as different fungi [72].
FabI was predicted as a possible binding target for justicidin B. FabI is a bacterial catalytic enzyme that reduces a carbon–carbon double bond in an enoyl moiety covalently linked to an acyl carrier protein. This is the rate-determining step in the elongation cycle of fatty acids used in lipid metabolism and biotin biosynthesis in bacteria [73, 74]. Being a unique bacterial enzyme, FabI has been considered a potential target for developing new antibacterial therapeutics [75, 76]. It has been discovered that FabI is necessary for bacterial viability. Recently, a wide range of compounds has been recognized as FabI inhibitors, such as triclosan, imidazoles, indole naphthyridinones, thiopyridine, and 4-pyridone [77].
Lately, there has been a growing recognition of promising antimicrobial targets involved in fatty acids synthesis pathways [78]. Novel FabI inhibitors are considered promising antimicrobial agents against MDR bacteria which could be identified by using molecular docking and virtual screening with the FabI X-ray crystal structures that are currently accessible [79]. Fabimycin is a previously discovered FabI inhibitor and an antibiotic candidate with in vivo activity against Gram-negative pathogens [80]. Our docking studies and MDS suggested justicidin B as a novel inhibitor against A. baumannii FabI; it binds the enzyme in a pattern similar to its co-crystalized ligand.
The sequence of FabI enzyme that bound to justicidin B was found to be conserved in A. baumannii and ESKAPE pathogens strains available in the NCBI database with alignment scores > 200. The conservation of FabI enzyme among different bacterial species was confirmed previously [81]. The conservation of the amino acids involved in justicidin B binding was confirmed. Only in S. aureus and E. faecium, the amino acid ILE-203 was replaced with valine, and PHE-96 by tyrosine, respectively, which should have minimal impact on justicidin B binding. This predicts justicidin B as a promising antimicrobial agent through FabI inhibition with potential broad-spectrum activity. However, the impact of changes in the amino acid sequence in these two Gram-positive pathogens is yet to be determined.
Finally, the predicted activity of justicidin B (an aryl naphthalene lignan) as a broad-spectrum antimicrobial against A. baumannii and other ESKAPE pathogens through FabI inhibition still needs to be confirmed by different phenotypic and genotypic studies in a step towards the introduction of new antimicrobial compounds. By tracing the aryl naphthalene lignans containing metabolites, we could take a substantial step towards discovering plant-based chemicals as prospective inhibitors of conserved protein targets in MDR-ESKAPE pathogens.
Conclusions
O. majorana L. extract offers a promising source of natural antimicrobial extracts that could be successfully used against the troublesome A. baumannii infections. With further experimental confirmations, justicidin B can be employed as a lead molecule for the development of novel inhibitors against MDR A. baumannii and other ESKAPE pathogens through interaction with its potential target, the FabI, and the subsequent inhibition of lipid metabolism and biotin biosynthesis in bacteria.
Availability of data and materials
All data generated or analysed during this study are included in this published article.
Abbreviations
- DPPH:
-
1,1-Dipehnyl-2-picrylhydrazyl
- 100% MF:
-
100% Methanol fraction
- 50% MF:
-
50% Methanol fraction
- ACP:
-
Acyl carrier protein
- CRAB:
-
Carbapenem-resistant A. baumannii
- FabI:
-
Enoyl-ACP reductase
- ESKAPE pathogens:
-
Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, A. baumannii, Pseudomonas aeruginosa, and Enterobacter spp.
- ffTK:
-
Force field tool kit
- MF:
-
Methanol fraction
- MIC:
-
Minimum inhibitory concentration
- MMPBSA:
-
Molecular mechanics Poisson–Boltzmann surface area
- ML:
-
Mother liquor
- MDR:
-
Multidrug-resistant
- NCBI:
-
National centre for biotechnology information
- NAD:
-
Nicotinamide adenine dinucleotide
- RMSD:
-
Root-mean-square deviation
- SARS-CoV-2:
-
Severe acute respiratory syndrome coronavirus 2
- MSD:
-
Subsequent molecular dynamic simulation
- CDC:
-
The centres for disease control and prevention
- CLSI:
-
The Clinical and Laboratory Standards Institute
- DCM-F:
-
Dichloromethane fraction
- ME:
-
Total methanolic extract
- TSA:
-
Tryptic soy agar
- TSB:
-
Tryptic soy broth
- UPLC-HRMS:
-
Ultra-performance liquid chromatography–high-resolution mass spectrometry
References
Perovic O, Duse A, Chibabhai V, Black M, Said M, Prentice E, Wadula J, Mahabeer Y, Han KSS, Mogokotleng R et al (2022) Acinetobacter baumannii complex, national laboratory-based surveillance in South Africa, 2017 to 2019. PLoS ONE 17:e0271355. https://doi.org/10.1371/journal.pone.0271355
Ayobami O, Willrich N, Harder T, Okeke IN, Eckmanns T, Markwart R (2019) The incidence and prevalence of hospital-acquired (carbapenem-resistant) Acinetobacter baumannii in Europe, Eastern Mediterranean and Africa: a systematic review and meta-analysis. Emerg Microbes Infect 8:1747–1759. https://doi.org/10.1080/22221751.2019.1698273
Giammanco A, Calà C, Fasciana T, Dowzicky MJ (2017) Global assessment of the activity of tigecycline against multidrug-resistant Gram-negative pathogens between 2004 and 2014 as part of the tigecycline evaluation and surveillance trial. Msphere 2:e00310-00316
Sounouvou HT, Toukourou H, Catteau L, Toukourou F, Evrard B, Van Bambeke F, Gbaguidi F, Quetin-Leclercq J (2021) Antimicrobial potentials of essential oils extracted from West African aromatic plants on common skin infections. Sci Afr 11:e00706. https://doi.org/10.1016/j.sciaf.2021.e00706
Nabi M, Tabassum N, Ganai BA (2022) Phytochemical screening and antibacterial activity of Skimmia anquetilia N.P. Taylor and Airy Shaw: A first study from Kashmir Himalaya Front. Plant Sci. 13:937946. https://doi.org/10.3389/fpls.2022.937946
Chipenzi T, Baloyi G, Mudondo T, Sithole S, Fru Chi G, Mukanganyama S (2020) An evaluation of the antibacterial properties of tormentic acid congener and extracts from callistemon viminalis on selected ESKAPE pathogens and effects on biofilm formation. Adv Pharmacol Pharm Sci 2020:8848606. https://doi.org/10.1155/2020/8848606
Li J, Nation RL, Turnidge JD, Milne RW, Coulthard K, Rayner CR, Paterson DL (2006) Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect Dis 6:589–601. https://doi.org/10.1016/s1473-3099(06)70580-1
Palombo M, Bovo F, Amadesi S, Gaibani P (2023) Synergistic Activity of Cefiderocol in Combination with Piperacillin-Tazobactam, Fosfomycin, Ampicillin-Sulbactam, Imipenem-Relebactam and Ceftazidime-Avibactam against Carbapenem-Resistant Gram-Negative Bacteria. Antibiotics (Basel) 12:858. https://doi.org/10.3390/antibiotics12050858
Du X, Xu X, Yao J, Deng K, Chen S, Shen Z, Yang L, Feng G (2019) Predictors of mortality in patients infected with carbapenem-resistant Acinetobacter baumannii: a systematic review and meta-analysis. Am J Infect Control 47:1140–1145
Hartzell JD, Neff R, Ake J, Howard R, Olson S, Paolino K, Vishnepolsky M, Weintrob A, Wortmann G (2009) Nephrotoxicity associated with intravenous colistin (colistimethate sodium) treatment at a tertiary care medical center. Clin Infect Dis 48:1724–1728. https://doi.org/10.1086/599225
Cheng CY, Sheng WH, Wang JT, Chen YC, Chang SC (2010) Safety and efficacy of intravenous colistin (colistin methanesulphonate) for severe multidrug-resistant Gram-negative bacterial infections. Int J Antimicrob Agents 35:297–300. https://doi.org/10.1016/j.ijantimicag.2009.11.016
(CDC), U.C.f.D.C.a.P. Antibiotic Resistance Threats in the United States 2019 report. CDC.: Atlanta, GA: U.S. Department of Health and Human Services, 2019.
(CDC), U.C.f.D.C.a.P. Covid-19 impact on antimicrobial resistance. CDC.: Atlanta, GA: U.S. Department of Health and Human Services, 2022.
Elwakil WH, Rizk SS, El-Halawany AM, Rateb ME, Attia AS (2023) Multidrug-resistant Acinetobacter baumannii Infections in the United Kingdom versus Egypt: trends and potential natural products solutions. Antibiotics 12:77. https://doi.org/10.3390/antibiotics12010077
Min EK, Yim SH, Choi MC, Lee JG, Joo DJ, Kim MS, Kim DG (2023) Incidence, mortality, and risk factors associated with carbapenem-resistant Acinetobacter baumannii bacteremia within 30 days after liver transplantation. Clin Transplant 37:e14956. https://doi.org/10.1111/ctr.14956
Kim T, Park KH, Yu SN, Park SY, Park SY, Lee YM, Jeon MH, Choo EJ, Kim TH, Lee MS et al (2019) Early intravenous colistin therapy as a favorable prognostic factor for 28-day mortality in patients with CRAB bacteremia: a multicenter propensity score-matching analysis. J Korean Med Sci 34:e256. https://doi.org/10.3346/jkms.2019.34.e256
(WHO), W.H.O. WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. Availabe online: https://www.who.int/news-room/detail/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 02–12–2022).
Suroowan S, Jugreet BS, Mahomoodally MF (2019) Endemic and indigenous plants from Mauritius as sources of novel antimicrobials. S Afr J Bot 126:282–308. https://doi.org/10.1016/j.sajb.2019.07.017
Sieberi BM, Omwenga GI, Wambua RK, Samoei JC, Ngugi MP (2020) Screening of the dichloromethane: methanolic extract of Centella asiatica for Antibacterial Activities against Salmonella typhi, Escherichia coli, Shigella sonnei, Bacillus subtilis, and Staphylococcus aureus. Sci World J 2020:6378712. https://doi.org/10.1155/2020/6378712
Idris FN, Nadzir MM (2023) Multi-drug resistant ESKAPE pathogens and the uses of plants as their antimicrobial agents. Arch Microbiol 205:115. https://doi.org/10.1007/s00203-023-03455-6
Vasudeva N (2015) Origanum majorana L.–Phyto-pharmacological review. Indian J Nat Prod Resour 6:261–267
Ennaji H, Chahid D, Aitssi S, Badou A, Khlil N, Ibenmoussa S (2020) Phytochemicals screening, cytotoxicity and antioxidant activity of the Origanum majorana growing in Casablanca. Morocco Open Biol Sci J 5:053–059
Bouyahya A, Chamkhi I, Benali T, Guaouguaou F-E, Balahbib A, El Omari N, Taha D, Belmehdi O, Ghokhan Z, El Menyiy N (2021) Traditional use, phytochemistry, toxicology, and pharmacology of Origanum majorana L. J Ethnopharmacol 265:113318
Morshedloo MR, Ahmadi H, Yazdani D (2018) An over review to Origanum vulgare L. and its pharmacological properties. J. Med. Plants 17:15–31
Seoudi D, Medhat A, Hewedi I, Osman S, Mohamed MK, Arbid M (2009) Evaluation of the anti-inflammatory analgesic and anti-pyretic effects of Origanum majorana ethanolic extract in experimental animals. J. Radiat. Res. Appl. Sci 2:513–534
Elsherbini DMA, Almohaimeed HM, El-Sherbiny M, Mohammedsaleh ZM, Elsherbiny NM, Gabr SA, Ebrahim HA (2022) Origanum majorana L. extract attenuated benign prostatic hyperplasia in rat model: effect on oxidative stress, apoptosis, and proliferation. Antioxidants 11:1149
Selim SA, Aziz MHA, Mashait MS, Warrad MF (2013) Antibacterial activities, chemical constitutes and acute toxicity of Egyptian Origanum majorana L., Peganum harmala L. and Salvia officinalis L. essential oils. Afr. J. Pharm. Pharmacol 7:725–735
Muqaddas RAK, Nadeem F, Jilani MI (2016) Essential chemical constituents and medicinal uses of Marjoram (Origanum majorana L.)–A comprehensive review. Int J Chem Biochem Sci 9(56):62
Sellami IH, Maamouri E, Chahed T, Wannes WA, Kchouk ME, Marzouk B (2009) Effect of growth stage on the content and composition of the essential oil and phenolic fraction of sweet marjoram (Origanum majorana L.). Ind Crops Prod 30:395–402
Busatta C, Vidal R, Popiolski A, Mossi A, Dariva C, Rodrigues M, Corazza F, Corazza M, Oliveira JV, Cansian R (2008) Application of Origanum majorana L. essential oil as an antimicrobial agent in sausage. Food Microbiol 25:207–211
Hajlaoui H, Mighri H, Aouni M, Gharsallah N, Kadri A (2016) Chemical composition and in vitro evaluation of antioxidant, antimicrobial, cytotoxicity and anti-acetylcholinesterase properties of Tunisian Origanum majorana L. essential oil. Microb Pathog. 95(86):94
Amor G, Caputo L, La Storia A, De Feo V, Mauriello G, Fechtali T (2019) Chemical composition and antimicrobial activity of Artemisia herba-alba and Origanum majorana essential oils from Morocco. Molecules 24:4021
Jacobs AC, Thompson MG, Black CC, Kessler JL, Clark LP, McQueary CN, Gancz HY, Corey BW, Moon JK, Si Y et al (2014) AB5075, a highly virulent isolate of Acinetobacter baumannii, as a model strain for the evaluation of pathogenesis and antimicrobial treatments. MBio 5:e01076-01014. https://doi.org/10.1128/mBio.01076-14
CLSI (2018) Reference Method for Broth Microdilution Antibacterial Susceptibility Testing; Approved Standard-11th edition. In CLSI document M07-A11, Clinical and Laboratory Standards Institute: Wayne, PA
Wang X, Shen Y, Wang S, Li S, Zhang W, Liu X, Lai L, Pei J, Li H (2017) PharmMapper 2017 update: a web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res 45:W356–W360
Huey R, Morris GM, Forli S (2012) Using AutoDock 4 and AutoDock vina with AutoDockTools: a tutorial. Scr Res Inst Mol Gr Lab 10550:1000
Yuan S, Chan HS, Hu Z (2017) Using PyMOL as a platform for computational drug design. Wiley Interdiscip. Rev. Comput. Mol. Sci. 7:e1298
Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K (2005) Scalable molecular dynamics with NAMD. J Comput Chem 26:1781–1802
Ribeiro JV, Bernardi RC, Rudack T, Schulten K, Tajkhorshid E (2018) QwikMD-gateway for easy simulation with VMD and NAMD. Biophys J 114:673a–674a
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph Model 14:33–38
Miller BR III, McGee TD Jr, Swails JM, Homeyer N, Gohlke H, Roitberg AE (2012) MMPBSA. py: an efficient program for end-state free energy calculations. J Chem Theory Comput 8:3314–3321
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Madeira F, Pearce M, Tivey AR, Basutkar P, Lee J, Edbali O, Madhusoodanan N, Kolesnikov A, Lopez R (2022) Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res 50:W276–W279. https://doi.org/10.1093/nar/gkac240
Charai M, Mosaddak M, Faid M (1996) Chemical composition and antimicrobial activities of two aromatic plants: Origanum majorana L. and O. compactum Benth. J. Essent. Oil Res. 8:657–664
Leeja L, Thoppil J (2007) Antimicrobial activity of methanol extract of Origanum majorana L. (Sweet marjoram). J. Environ. Biol. 28:145
Abdel-Massih R, Abdou E, Baydoun E, Daoud Z (2010) Antibacterial activity of the extracts obtained from Rosmarinus officinalis, Origanum majorana, and Trigonella foenum-graecum on highly drug-resistant Gram-negative bacilli. J Bot 2010:464087. https://doi.org/10.1155/2010/464087
Gomes F, Dias MI, Lima Â, Barros L, Rodrigues ME, Ferreira IC, Henriques M (2020) Satureja montana L. and Origanum majorana L. decoctions: antimicrobial activity, mode of action and phenolic characterization. Antibiotics 9:294. https://doi.org/10.3390/antibiotics9060294
Kozlowska M, Laudy AE, Starosciak BJ, Napiórkowski A, Chomicz L, Kazimierczuk Z (2010) Antimicrobial and antiprotozoal effect of sweet marjoram (Origanum majorana L.). Acta Sci. Pol. Hortorum Cultus 9:133–141
Ghazal TSA, Schelz Z, Vidács L, Szemerédi N, Veres K, Spengler G, Hohmann J (2022) Antimicrobial, multidrug resistance reversal and biofilm formation inhibitory effect of Origanum majorana extracts, essential oil and monoterpenes. Plants 11:1432
Vági E, Simándi B, Suhajda A, Hethelyi E (2005) Essential oil composition and antimicrobial activity of Origanum majorana L. extracts obtained with ethyl alcohol and supercritical carbon dioxide. Food Res Int 38:51–57
Wink M (2003) Evolution of secondary metabolites from an ecological and molecular phylogenetic perspective. Phytochemistry 64:3–19
Dröge W (2002) Free radicals in the physiological control of cell function. Physiol Rev 82:47–95. https://doi.org/10.1152/physrev.00018.2001
Duda-Madej A, Stecko J, Sobieraj J, Szymańska N, Kozłowska J (2022) Naringenin and its derivatives—Health-promoting phytobiotic against resistant bacteria and fungi in humans. Antibiotics 11:1628
Nguyen TLA, Bhattacharya D (2022) Antimicrobial activity of quercetin: an approach to its mechanistic principle. Molecules 27:2494
Panda L, Duarte-Sierra A (2022) Recent advancements in enhancing antimicrobial activity of plant-derived polyphenols by biochemical means. Horticulturae 8:401
Koukoulitsa C, Karioti A, Bergonzi MC, Pescitelli G, Di Bari L, Skaltsa H (2006) Polar constituents from the aerial parts of Origanum vulgare L. ssp. hirtum growing wild in Greece. J Agric Food Chem 54:5388–5392. https://doi.org/10.1021/jf061477i
Chatzopoulou A, Karioti A, Gousiadou C, Lax Vivancos V, Kyriazopoulos P, Golegou S, Skaltsa H (2010) Depsides and other polar constituents from Origanum dictamnus L. and their in vitro antimicrobial activity in clinical strains. J Agric Food Chem 58:6064–6068. https://doi.org/10.1021/jf904596m
Cala A, Salcedo JR, Torres A, Varela RM, Molinillo JM, Macías FA (2021) A study on the phytotoxic potential of the seasoning herb marjoram (Origanum majorana L.) leaves. Molecules 26:3356
Goun E, Cunningham G, Solodnikov S, Krasnykch O, Miles H (2002) Antithrombin activity of some constituents from Origanum vulgare. Fitoterapia 73:692–694. https://doi.org/10.1016/S0367-326X(02)00245-9
Gertsch J, Tobler RT, Brun R, Sticher O, Heilmann J (2003) Antifungal, antiprotozoal, cytotoxic and piscicidal properties of Justicidin B and a new arylnaphthalide lignan from Phyllanthus piscatorum. Planta Med 69:420–424. https://doi.org/10.1055/s-2003-39706
Paloukopoulou C, Tsadila C, Govari S, Soulioti A, Mossialos D, Karioti A (2023) Extensive analysis of the cultivated medicinal herbal drug Origanum dictamnus L. and antimicrobial activity of its constituents. Phytochemistry 208:113591. https://doi.org/10.1016/j.phytochem.2023.113591
Rauf A, Raza M, Saleem M, Ozgen U, Karaoglan ES, Renda G, Palaska E, Orhan IE (2017) Carbonic anhydrase and urease inhibitory potential of various plant phenolics using in vitro and in silico methods. Chem Biodivers 14:e1700024. https://doi.org/10.1002/cbdv.201700024
Matsuura H, Chiji H, Asakawa C, Amano M, Yoshihara T, Mizutani J (2003) DPPH radical scavengers from dried leaves of oregano (Origanum vulgare). Biosci Biotechnol Biochem 67:2311–2316. https://doi.org/10.1271/bbb.67.2311
Bosabalidis A, Gabrieli C, Niopas I (1998) Flavone aglycones in glandular hairs of Origanum x intercedens. Phytochemistry 49:1549–1553. https://doi.org/10.1016/S0031-9422(98)00186-1
Hakobyan A, Arabyan E, Kotsinyan A, Karalyan Z, Sahakyan H, Arakelov V, Nazaryan K, Ferreira F, Zakaryan H (2019) Inhibition of African swine fever virus infection by genkwanin. Antiviral Res 167:78–82. https://doi.org/10.1016/j.antiviral.2019.04.008
Zhang X, Hung TM, Phuong PT, Ngoc TM, Min B-S, Song K-S, Seong YH, Bae K (2006) Anti-inflammatory activity of flavonoids from Populus davidiana. Arch Pharmacal Res 29:1102–1108. https://doi.org/10.1007/BF02969299
Sreeja, P.S.; Arunachalam, K.; de Oliveira Martins, D.T.; da Silva Lima, J.C.; Balogun, S.O.; Pavan, E.; Saikumar, S.; Dhivya, S.; Kasipandi, M.; Parimelazhagan, T. Sphenodesme involucrata var. paniculata (CB Clarke) Munir.: chemical characterization, anti-nociceptive and anti-inflammatory activities of methanol extract of leaves. J. Ethnopharmacol. 2018, 225, 71–80.
Hemmati S, Seradj H (2016) Justicidin B: a promising bioactive lignan. Molecules 21:820. https://doi.org/10.3390/molecules21070820
Ferraz AC, Gomes PWP, da Silva Menegatto MB, Lima RLS, Guimarães PH, Reis JDE, Carvalho ARV, Pamplona SDGSR, Muribeca ADJB, de Magalhães JC (2024) Exploring the antiviral potential of justicidin B and four glycosylated lignans from Phyllanthus brasiliensis against Zika virus: a promising pharmacological approach. Phytomedicine 123:155197. https://doi.org/10.1016/j.phymed.2023.155197
Toth G, Horvati K, Kraszni M, Ausbuttel T, Pályi B, Kis Z, Mucsi ZN, Kovacs GM, Bosze S, Boldizsár I (2023) Arylnaphthalene Lignans with Anti-SARS-CoV-2 and Antiproliferative Activities from the Underground Organs of Linum austriacum and Linum perenne. J Nat Prod 86(672):682. https://doi.org/10.1021/acs.jnatprod.2c00580
Tajuddeen N, Muyisa S, Maneenet J, Nguyen HH, Naidoo-Maharaj D, Maharaj V, Awale S, Bringmann G (2024) Justicidin B and related lignans from two South African Monsonia species with potent activity against HeLa cervical cancer cells. Phytochem Lett 60:234–238. https://doi.org/10.1016/j.phytol.2023.09.007
El-Gendy M, Hawas UW, Jaspars M (2008) Novel bioactive metabolites from a marine derived bacterium Nocardia sp. ALAA 2000. J. Antibiot. 61(379):386
Parker EN, Cain BN, Hajian B, Ulrich RJ, Geddes EJ, Barkho S, Lee HY, Williams JD, Raynor M, Caridha D et al (2022) An iterative approach guides discovery of the FabI Inhibitor Fabimycin, a late-stage antibiotic candidate with in vivo efficacy against drug-resistant gram-negative infections. ACS Cent Sci 8:1145–1158. https://doi.org/10.1021/acscentsci.2c00598
Yao J, Rock CO (2016) Resistance mechanisms and the future of bacterial enoyl-acyl carrier protein reductase (FabI) antibiotics. Cold Spring Harb Perspect Med 6:a027045
Rao NK, Nataraj V, Ravi M, Panchariya L, Palai K, Talapati SR, Lakshminarasimhan A, Ramachandra M, Antony T (2020) Ternary complex formation of AFN-1252 with Acinetobacter baumannii FabI and NADH: crystallographic and biochemical studies. Chem Biol Drug Des 96:704–713
Narasimha Rao K, Lakshminarasimhan A, Joseph S, Lekshmi SU, Lau MS, Takhi M, Sreenivas K, Nathan S, Yusof R, Abd Rahman N (2015) AFN-1252 is a potent inhibitor of enoyl-ACP reductase from Burkholderia pseudomallei—crystal structure, mode of action, and biological activity. Prot Sci 24(832):840
Lu X, Lv M, Huang K, Ding K, You Q (2012) Pharmacophore and molecular docking guided 3D-QSAR study of bacterial enoyl-ACP reductase (FabI) inhibitors. Int J Mol Sci 13:6620–6638
Yang X, Lu J, Ying M, Mu J, Li P, Liu Y (2017) Docking and molecular dynamics studies on triclosan derivatives binding to FabI. J Mol Model 23:1–13
Seefeld MA, Miller WH, Newlander KA, Burgess WJ, DeWolf WE, Elkins PA, Head MS, Jakas DR, Janson CA, Keller PM (2003) Indole naphthyridinones as inhibitors of bacterial enoyl-ACP reductases FabI and FabK. J Med Chem 46:1627–1635
Abraham A, Philip S, Jacob MK, Narayanan SP, Jacob CK, Kochupurackal J (2015) Phenazine-1-carboxylic acid mediated anti-oomycete activity of the endophytic Alcaligenes sp. EIL-2 against Phytophthora meadii. Microbiol Res 170:229–234. https://doi.org/10.1016/j.micres.2014.06.002
Rana P, Ghouse SM, Akunuri R, Madhavi Y, Chopra S, Nanduri S (2020) FabI (enoyl acyl carrier protein reductase)-A potential broad-spectrum therapeutic target and its inhibitors. Eur J Med Chem 208:112757
Acknowledgements
Not applicable.
Statement regarding plants
Origanum majorana L. aerial parts were collected from the Experimental Station of the Faculty of Pharmacy, Cairo University, Giza, Egypt. Mrs Therese Labib, Consultant at Orman Botanic Garden, Dokki, Giza, Egypt, kindly authenticated the plant material. Voucher specimen No. (PHARM-16.12.2020) was kept at the Herbarium of the Department of Pharmacognosy, Faculty of Pharmacy, Cairo University.
Funding
This work was supported by the Institutional Links Grants / 2017 IL6 July Newton-Mosharafa Institutional Links (Grant ID: 351952191 by the British Council UK and Grant ID: 30863 by The Science, Technology, and Innovation Funding Authority (STIFA), Egypt awarded to Mostafa E. Rateb and Ahmed S. Attia, respectively.
Author information
Authors and Affiliations
Contributions
Conceptualization, A.S.A., A.M.E. and M.E.R.; methodology, N.H.M., A.E.M.S., A.M.S., R.A.E., N.M.E., M.Y, W.M., M.T.K.; software, A.E.M.S., A.M.S., R.A.E., M.E.R.; validation, A.S.A., A.M.E., A.S.A. and M.E.R.; formal analysis, N.H.M., A.E.M.S., R.A.E., M.Y., W.M., N.M.E., A.M.S.; investigation, N.H.M., A.E.M.S., R.A.E.; resources, A.S.A. and M.E.R.; data curation, A.S.A., A.M.E. and M.E.R.; writing—original draft preparation, N.H.M., A.E.M.S., R.A.E., A.M.S., N.M.E., M.T.K., W.M., M.Y.; writing—review and editing, A.S.A., A.M.E. and M.E.R.; supervision, A.S.A., A.M.E. and M.E.R.; project administration, A.S.A., A.M.E. and M.E.R.; funding acquisition, A.S.A. and M.E.R.. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mahdally, N.H., Salih, A.E.M., El-Shiekh, R.A. et al. Exploring the antimicrobial activity of Origanum majorana L. against the highly virulent multidrug-resistant Acinetobacter baumannii AB5075: UPLC-HRMS profiling with in vitro and in silico studies. Futur J Pharm Sci 10, 71 (2024). https://doi.org/10.1186/s43094-024-00641-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43094-024-00641-1