Abstract
Background
Inflammatory diseases are the vast array of disorders caused by inflammation. During most inflammatory events, many cytokines expressions were modulated, and one such cytokine is tumor necrosis factor-alpha (TNF-α). TNF-α is mainly secreted by monocytes and macrophages. Notably, it has been proposed as a therapeutic target for several diseases. The anti-TNF biology approach is mainly based on monoclonal antibodies. The fusion protein and biosimilars are prevalent in treating inflammation for decades. Only a few small molecule inhibitors are available to inhibit the expression of TNF-α, and one such promising drug was thalidomide. Therefore, the study was carried out to design thalidomide-based small molecule inhibitors for TNF-α. The main objective of our study is to design thalidomide analogs to inhibit TNF-α using the insilico approach.
Results
Several thalidomide analogs were designed using chemsketch. After filtration of compounds through ‘Lipinski rule of 5’ by Molinspiration tool, as a result, five compounds were selected. All these compounds were subjected to molecular docking, and the study showed that all five compounds had good binding energy. However, based on ADMET predictions, two compounds (S3 and S5) were eliminated.
Conclusions
Our preliminary results suggest that S1, S2, S4 compounds showed potential ligand binding capacity with TNF-α and, interestingly, with limited or no toxicity. Our preliminary investigation and obtained results have fashioned more interest for further in vitro studies.
Similar content being viewed by others
Background
Inflammation is a response to tissue injury or infection. It is characterized in its acute phase by increasing vascular permeability by plasma extravasation, resulting in an accumulation of fluid, leucocytes, and mediators to the inflammatory site [1, 2]. Inflammatory diseases are the vast array of disorders and conditions caused by inflammation, including allergy, asthma, autoimmune diseases, coeliac disease, glomerulonephritis, hepatitis, inflammatory bowel disease, reperfusion injury, and transplant rejection, etc. [3]. During most of these events, many cytokines expressions were modulated, and one such cytokine is tumor necrosis factor alpha (TNF-α). TNF-α is among one of the pleiotropic cytokines, mainly secreted by monocytes and macrophages, TNF-α has been shown to play a pivotal role in orchestrating the cytokine cascade in many inflammatory diseases, and it is known to be “master-regulator” of inflammatory cytokine production. Notably, it has been proposed as a therapeutic target for several diseases [4]. TNF-α is a multidimensional cytokine protein that modulates various cellular functions, including survival, proliferation, and cell death [5]. The anti-TNF biology approach is mainly based on monoclonal antibodies (mAbs); the fusion protein and biosimilars are prevalent in treating inflammation for a decade. Small molecule inhibitors are likely to be cheaper to produce than mAbs or Ig-Fc-based fusion proteins [6]. Currently, few small molecule inhibitors have come to light. However, not even a single small molecule antagonist for TNF-α is released into the market. Therefore, the main objective of the present study is to design small molecules to inhibit TNF-α using the insilico method and evaluation of drug likeness.
Before designing a new molecule, thoroughly reviewed and studied all the existing small molecule drugs, inhibiting TNF-alpha directly and indirectly. After a thorough review, thalidomide was selected. Thalidomide derivative treats multiple diseases like scleroderma, Crohn's disease, and refractory multiple myeloma cutaneous lupus [7, 8]. Thalidomide (2-2,6-dioxo-3-piperidyl)isoindoline-1,3-dione) was a synthetic glutamic acid derivative and first synthesized by “Chemie Grunenthal” a German pharmaceutical company, in 1954 to treat morning sickness in pregnant women [9]. But due to its teratogenic effects, thalidomide was withdrawn from the world market [10]. Thalidomide was reintroduced to the market in 1998 and started clinical research to understand anti-inflammatory and immunomodulatory roles [11, 12]. Thalidomide was a well-recognized inhibitor of TNF-α expression [13, 14]. The overexpression of TNF-α leads to change in physiological conditions in various inflammatory diseases. Thalidomide blocks the production of TNF-α in many disease conditions without inducing immune suppression compared with glucocorticoid and cyclosporine [15]. The long-term objective of our study is to address the exiting demerits associated with thalidomide molecules for being used as drug molecule in the management of diseases. In the present study, novel thalidomide analogs were designed having better binding activities, improved drug potency and conversely reducing the side effects. The study also reports designed small molecule inhibitors have reduced teratogenic effect and more drug effectiveness with greater specificity. Further, our studies are under progress for In vitro validation.
Methods
Structure preparation of TNF-α
The crystal structure of TNF-α (PDB ID: 2az5) was downloaded from Protein Data Bank (PDB) [16]. Further, characterization and validation, stereochemistry, and structural geometry were accomplished using structure analysis and verification server (SAVES) (https://servicesn.mbi.ucla.edu/SAVES/).
Active site prediction of TNF-α
Active sites of targeted protein TNF-α were analyzed using PDBsum of RCSB server (www.ebi.ac.uk/pdbsum/) [17]. In this approach, the 2az5 protein structure was studied to identify amino acid residues, which were interacting with the inhibitory compound SPD-304 as revealed by PDBsum from the protein–ligand interaction map.
Referral drug selection
Referral drugs were selected through extensive literature survey and analyzed through TTD (Therapeutic Target Database), Uniprot, and drug bank databases. Only those drugs that are capable of inhibiting TNF-α associated with inflammatory diseases are selected for the study. In this approach, Thalidomide and SPD-304 were selected because they can inhibit the expression of TNF-α [18, 19].
Ligand preparation
Several analogs were designed manually based on the scaffold structure of Thalidomide using ACD (Advanced Chemistry Development)/ChemSketch [20] software and saved in mol format. The pharmacophore and pharmacokinetics properties of ligand molecules were studied using Hyperchem Professional 7.0 [21] and molinspiration (https://www.molinspiration.com/cgi-bin/properties). Further, the resultant ligand molecules were used for molecular docking studies.
Molecular docking
AutoDock Tool 1.5.6 program was used to perform molecular docking studies [22, 23]. A grid box was prepared to cover all the active sites/ligand binding sites of the protein. For running docking and molecular simulation, the Lamarckian genetic algorithm is used. Each simulation is carried out ten times, which ultimately yielded ten docking conformations. Further, during simulation, the best binding conformations are selected based on the least energy conformations.
ADMET analysis
ADMET (Absorption, Distribution, Metabolism, Excretion, and Toxicity) are calculated by using admetSAR (http://lmmd.ecust.edu.cn/admetsar/) web server [24]. It provides various parameters such as blood–brain barrier (BBB +), human intestinal absorption (HIA+), Caco-2 permeability (Caco2-), P-glycoprotein Substrate (logpGI), P-glycoprotein inhibitor (logGI), aqueous solubility (logs), and Probability of Caco-2 permeability (log Papp, cm/s) as well as toxicity also calculated as AMES toxicity, carcinogenicity and lethal dose 50 (LD50) in the rat.
Results
Structure preparation of TNF-α
The 3D structure of TNF-α with PDB ID: 2az5 was fetched from Protein Data Bank with a resolution of 3.0 Å and is ligand binded PDB structure. This was selected after inspecting all available TNF-α PDB structures. A Co-crystalized TNF-α trimer with a ligand binded structure was not found and also having mutations in structures. The TNF-α dimer structure was found binded with ligand SPD-304. This structure provides active sites for insilico work. Using the discovery studio tool, the protein was prepared for molecular docking study by removing ligand and water molecules. SAVES online server predicts protein structure quality; in the Ramachandran plot, 89.6% of residues are in the favored region.
Active site prediction of TNF-α
PDBsum of RCSB server identified the active pockets of TNF-α molecule. The interacting amino acid residues with the ligand are selected as active sites by the ligplot shown in Fig. 1. The blue-colored chemical structure is a ligand molecule (SPD-304). The eyelids-like structures are active site amino acids.
Referral drug selection
Through literature survey thalidomide and SPD-304 potential inhibitors were selected for the study. Thalidomide indirectly inhibits the expression of TNF-α, and SPD-304 directly binds to TNF-α trimer and disassociating its subunits into dimer and monomer [16]. Those binding residues considered as active sites to build the grid box to study docking. The Referral drugs structures are shown in Fig. 2.
Ligand preparation
Several new molecules were designed based on the referral drug (Thalidomide), manually changing the functional groups, adding and deleting chemical bonds, benzene rings, etc., through ACD/ChemSketch. Canonical smiles and molecular formulas are also generated. These canonical smiles are used to predict the ‘Lipinski rule of 5’ [25] from molinspiration. Based on these rules, the designed compounds are filtered. As a result, a total of five compounds were selected, which are shown in Table 1.
Molinspiration online tool was used to calculate ‘Lipinski rule of 5’ of ligand molecules, and it creates Pharmacophore properties of logarithm of octanol–water partitioning coefficient (LogP) − 5 to 5, hydrogen bond Donor (HBD) ≤ 5, hydrogen bond acceptor (HBA) ≤ 10, number of rotatable bonds (NROTB) ≤ 12, molecular weight (MW) ≤ 500 Dalton, topological polar surface area (TPSA) ≤ 1000, and other properties like number of atoms (nAtoms), molecular volume (MV), number of violations of Lipinski’s rule of five (nviolations). These values help to understand molecular descriptors of drug-likeness shown in Table 2. The table illustrates that all five compounds obey the properties of the Lipinski rule of 5 with 0 violations. Both referral drugs had also undergone this step thalidomide shows 0 violations, but SPD-304 shows 2 violations. Further, new compounds are proceeding for the energy minimization step.
The energy minimizations of compounds are lead to optimize ligand molecules by hyperchem professional 7.0. [21] It was minimizing the energy of compounds by using AMBER (Assisted Model Building with Energy Refinement) force field. The energy minimization will change the molecular geometry to lower the compound's energy and yield a much more stable conformation. In minimization progression, it will search for a molecular structure in that energy will not modify with infinitesimal changes in geometry. This means that the derivative of energy regarding all Cartesian coordinates, called the gradient, is near zero. This is called a stationary point of potential energy surface [26]. Table 3 illustrated the values of before and after energy minimization. We can predict the stable structure conformation energy of all the five compounds from those values and the gradient values near zero. After energy minimization, these compounds are subjected to docking studies.
Docking result
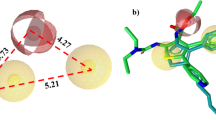
The five thalidomide analogs and referral drug (SPD-304) were docked against the TNF-α cytokine to check the binding energy between them using Autodock 1.5.6. SPD-304 docking results demonstrated in Table 4 and Fig. 3. They show its binding mode with one hydrogen bond between Gly121 and binding energy − 5.37 kcal/mol. The five compounds are showing good binding energy. However, S2, S3, S4, and S5 are showing hydrogen bond interactions. The S2 shows four hydrogen bond interactions between leu120, Gly121, Tyr151, and Ser60 with binding energy − 6.24 kcal/mol, S3 shows two hydrogen bond interactions between Ser60 and Leu120 with binding energy − 6.79 kcal/mol, S4 shows two hydrogen bond between Tyr119 and Lys98 with binding energy − 7.53 kcal/mol, and S5 shows three hydrogen bond interaction with Ser60, Leu120, Tyr151 with binding energy − 7.13 kcal/mol. Comparatively with referral drug, all five compounds that show less binding energy with more hydrogen bonds were detected. Docking results are illustrated in Table 4 and Fig. 4. Both three-dimensional (3D) and two-dimensional (2D) docked images illustrate the binding pose of the compounds in which the green dotted lines denotes hydrogen bond interactions between ligand and receptor.
The figure shown in three-dimensional interaction represented in stick format, visualized using discovery studio showing the interaction residues labeled in grey color and the two-dimensional interaction of ligand SPD-304 with TNF-α shows conventional hydrogen bond with GLY121 shown in green dotted line
The 2D structure shows conventional hydrogen bond interaction in green dotted lines. The 3D images shows stick structure, green color represents Ligands and grey color residues are targeted protein. H-bond plot was also shown, pink color indicates hydrogen bond donor, and green color indicates hydrogen bond acceptors
ADMET analysis
Insilico ADMET prediction is a very important step in drug discovery, whereas the earlier detection of poor pharmacokinetics property and toxicity issues is very helpful for further, and it reduces the time of drug development process. The pharmacokinetic properties of the five compounds are shown in Table 5. The central nervous system (CNS) was concluded by the effectiveness of the blood/brain partition co-efficient. This concluded activity of CNS was determined from the − 3 (inactive) to + 1 (active) scale; all compounds are within the acceptable range. The blood–brain barrier plays an important role in managing homeostasis of the CNS, splitting circulating blood from the brain [27]. Cell permeability Caco-2 range is from − 1 (poor) to + 1 (good), and all five compounds are within the range. The human intestinal absorption (HIA) of drug permeability and drug metabolism with intestinal membrane was calculated from 0 (poor) to 1 (great). All five compound are within the acceptable range of HIA. P-glycoprotein (logPGI) substrate and non-inhibitor study are depending on drug–drug interaction within different tissues. This alters xenobiotics of effective reduction in drug absorption and intensifies the removal of drugs and moves into the liver and kidney [28]. The P-gp substrate range was − 5 (poor) to + 1 (good), and the P-gp non-inhibitor range was 0 to 1. The aqueous solubility (PlogS) range was − 6.5 to − 0.5. P-gp of substrate, non-inhibitors, and PlogS are within range. The range of (logpapp) probability of Caco-2 cell permeability was − 1 to 1. Except S3, other componds are within the acceptable range. AMES toxicity test was to identify the compound is mutagenic or not. In this, S3 and S5 compounds are showing AMES toxic. All five compounds are noncarcinogenic and predicted lethal dose 50 (LD 50) in rats [29].
Discussion
Thalidomide was introduced as an inhibitor of TNF-α synthesis in 1991 [19], and it is used to treat numerous inflammatory diseases such as ulcerative colitis, ENL (Erythema Nodosum Leprosum), arthritis, Crohn's disease, multiple myeloma [30, 31]. Thalidomide can inhibit TNF-α through increasing mRNA degradation. Consequently, the TNF-α expression is reducing [18]. As a result, new analogs were synthesized and evaluated, aiming to enhance drug potency and reduce side effects, often referred to as IMiDs (Immunomodulatory imide drugs). Thalidomide’s first analogs were produced in 1996 [32]. Thalidomide and its analogs have anti-proliferation, anti-angiogenic and anti-inflammatory activities [33]. Thalidomide analogs are 3,6’- dithiothalidomide [34], lenalidomide [35, 36], pomalidomide [35, 37], apremilast [38], adamantly phthalimidine and noradamantyl-type phthalimidine [39, 40], etc. These analogs indirectly inhibit the TNF-α as they will reduce TNF-α protein synthesis, and this interaction between analogs and TNF-α was studied through in vitro and in vivo methods. Thalidomide and its analogs are having teratogenic and neurotoxic effects [40, 41]. Because of the reported limitation of Thalidomide and its analogs, it is necessary to build new analogs of Thalidomide having less or no toxicity. In our study, we prepared different new Thalidomide analogs to behave like an antagonist for TNF-α. The direct inhibition can be accessed through the insilico method. Most of the existing Thalidomide and its analogs inhibit TNF-α production via binding to the IKK (IKB kinase) complex and several other proteins. In contrast, our designed Thalidomide analogs directly bind to TNF-α and bring the inhibition. Since the binding of Thalidomide to the IKK complex and other proteins may indirectly inhibit several other molecular pathways and may result in unknown effects. Newly designed analogs have specificity for binding and inhibiting TNF-α. Most of these analyses were conducted using CADD (Computer-Aided Drug Design).
CADD is one of the important insilico methods for drug designing. It demonstrated that the drug designing tool is more efficient, saving time and money for the drug development process. The novelty of the present work is direct inhibition of TNF-α by insilico approach, and such inhibition is not being reported yet. In the present work, the insilico approach was adopted to build analogs of Thalidomide. TNF-α protein dimer structure (2AZ5) was retrieved from PDB, which binded to ligand SPD-304. These ligand binded residues are considered as active site amino acids. SPD-304 analogs were not generated because present study is to design analogs with main moiety, but SPD-304 contains toxic moiety of 3-alkylindole which is metabolized by cytochrome P450 enzymes via a dehydrogenation pathway same as potent pneumotoxin 3-methylindole, generating reactive electrophilic iminium material that could react with protein and DNA targets. Due to this reason, thalidomide was selected as referral drug for new compounds and thalidomide analogs were generated and filtered based on drug-likeness properties. Protein and ligands were proceeding to molecular docking. Many parameters were generated due to primarily binding energy, which shows affinity and strength of interaction between receptor and ligand. Lower binding energy will have stronger interaction. According to our molecular docking analysis, all the screened compounds show good binding energy, but S3 and S5 show AMES toxicity. The pharmacokinetic properties of ligands were analyzed earlier to reduce the further timeline burden to drug development. The finalized molecules S1, S2, and S4 show less binding energy, and S2, S4 are showing good binding energy with hydrogen bond interaction compared with SPD-304. These ligands are showing potent inhibitory activity against TNF-α. Further, in the future, these molecules will be chemically synthesized and subjected to in vitro validation studies on a cell-based model system.
Conclusions
To prevent inflammatory disease, the available TNF-α inhibitors are antibodies and fusion proteins used for decades. Still, small-molecule inhibitors are cheaper to produce as compared with antibodies. In the present work, we designed some new analogs of Thalidomide to inhibit TNF-α. The study shows that five compounds are showing good binding energy compared with SPD-304. Among the five, four compounds (S2, S3, S4, and S5) have hydrogen bond interactions 4, 2, 2, and 3, respectively; compared to referral drugs, these compounds have more hydrogen bond acceptors and hydrogen bond donor atoms in their structure due to this feature the compounds get more hydrogen bonds. Based on the ADMET prediction, the S1, S2, and S4 molecules have values within the acceptable range for all the parameters. This study suggests that S1, S2, and S4 compounds show highest potential with respect to referral drug SPD-304. S1, S2, and S4 show Binding energy − 6.33, − 6.24, and − 7.53 kcal/mol, respectively (SPD-304: − 5.37 kcal/mol). Hence, these molecules are carried forward for further in vitro validation studies.
Availability of data and materials
Data and material are available upon request.
Abbreviations
- TNF-α:
-
Tumor necrosis factor-alpha
- ADMET:
-
Absorption, distribution, metabolism, excretion, toxicity
- admetSAR:
-
ADMET structure activity relationship
- mAbs:
-
Monoclonal antibodies
- Ig-Fc:
-
Immunoglobulin-fragment crystallizable
- PDB:
-
Protein data bank
- SAVES:
-
Structure analysis and verification server
- PDBsum:
-
Pictorial database of 3D structures in the Protein Data Bank
- RCSB:
-
Research collaboratory for structural bioinformatics
- TTD:
-
Therapeutic target database
- ACD:
-
Advanced chemistry development
- BBB:
-
Blood–brain barrier
- HIA:
-
Human intestinal absorption
- LD50:
-
Lethal dose50
- HBD:
-
Hydrogen bond Donor
- HBA:
-
Hydrogen bond acceptor
- MW:
-
Molecular weight
- NROTB:
-
Number of rotatable bonds
- TPSA:
-
Topological polar surface area
- nAtoms:
-
Number of atoms
- MV:
-
Molecular volume
- Nviolations:
-
Number of violations
- AMBER:
-
Assisted model building with energy refinement
- RMSD:
-
Root mean square deviation
- 3D:
-
Three dimensional
- 2D:
-
Two dimensional
- CNS:
-
Central nervous system
- ENL:
-
Erythema nodosum leprosum
- IMiDs:
-
Immunomodulatory imide drugs
- IKK:
-
IKB kinase
- CADD:
-
Computer-aided drug design
References
Guo L, Ghassemian M, Komives EA, Russell P (2012) Cadmium-induced proteome remodeling regulated by Spc1/Sty1 and Zip1 in fission yeast. Toxicol Sci Off J Soc Toxicol 129(1):200–212. https://doi.org/10.1093/toxsci/kfs179
Patil RH, Naveen Kumar M, Kiran Kumar KM, Nagesh R, Kavya K, Babu RL, Ramesh GT, Chidananda Sharma S (2018) Dexamethasone inhibits inflammatory response via down regulation of AP-1 transcription factor in human lung epithelial cells. Gene 645:85–94. https://doi.org/10.1016/j.gene.2017.12.024
Acton QA (2013) Autoimmune diseases: new insights for the healthcare professional, 2013th edn. ScholarlyEditions, Atlanta
Parameswaran S, Balasubramanian S, Babai N, Qiu F, Eudy JD, Thoreson WB, Ahmad I (2010) Induced pluripotent stem cells generate both retinal ganglion cells and photoreceptors: therapeutic implications in degenerative changes in glaucoma and age-related macular degeneration. Stem cells (Dayton, Ohio) 28(4):695–703. https://doi.org/10.1002/stem.320
Wang Q, Wang L (2008) New methods enabling efficient incorporation of unnatural amino acids in yeast. J Am Chem Soc 130(19):6066–6067. https://doi.org/10.1021/ja800894n
Sedger LM, McDermott MF (2014) TNF and TNF-receptors: from mediators of cell death and inflammation to therapeutic giants—past, present and future. Cytokine Growth Factor Rev 25(4):453–472. https://doi.org/10.1016/j.cytogfr.2014.07.016
Pearson JM, Vedagiri M (1969) Treatment of moderately severe erythema nodosum leprosum with thalidomide: a double-blind controlled trial. Lepr Rev 40(2):111–116. https://doi.org/10.5935/0305-7518.19690022
Rutgeerts P, Van Assche G, Vermeire S (2004) Optimizing anti-TNF treatment in inflammatory bowel disease. Gastroenterology 126(6):1593–1610. https://doi.org/10.1053/j.gastro.2004.02.070
Randall T (1990) Thalidomide has 37-year history. JAMA 263(11):1474. https://doi.org/10.1001/jama.1990.03440110028006
Ito T, Ando H, Handa H (2011) Teratogenic effects of thalidomide: molecular mechanisms. Cell Mol Life Sci 68(9):1569–1579. https://doi.org/10.1007/s00018-010-0619-9
Spilker B, Fitzsimmons S, Horan MJDDR (1999) US drug and biologic approvals in 1998. 48
Tseng S, Pak G, Washenik K, Pomeranz MK, Shupack JL (1996) Rediscovering thalidomide: a review of its mechanism of action, side effects, and potential uses. J Am Acad Dermatol 35(6):969–979. https://doi.org/10.1016/s0190-9622(96)90122-x
Noman AS, Koide N, Hassan F, IE-Khuda I, Dagvadorj J, Tumurkhuu G, Islam S, Naiki Y, Yoshida T, Yokochi T (2009) Thalidomide inhibits lipopolysaccharide-induced tumor necrosis factor-alpha production via down-regulation of MyD88 expression. Innate Immun 15(1):33–41. https://doi.org/10.1177/1753425908099317
Yagyu T, Kobayashi H, Matsuzaki H, Wakahara K, Kondo T, Kurita N, Sekino H, Inagaki K, Suzuki M, Kanayama N, Terao T (2005) Thalidomide inhibits tumor necrosis factor-α-induced interleukin-8 expression in endometriotic stromal cells, possibly through suppression of nuclear factor-κB activation. J Clin Endocrinol Metab 90(5):3017–3021. https://doi.org/10.1210/jc.2004-1946%JTheJournalofClinicalEndocrinology&Metabolism
Majumder S, Sreedhara SR, Banerjee S, Chatterjee S (2012) TNF α signaling beholds thalidomide saga: a review of mechanistic role of TNF-α signaling under thalidomide. Curr Top Med Chem 12(13):1456–1467. https://doi.org/10.2174/156802612801784443
He MM, Smith AS, Oslob JD, Flanagan WM, Braisted AC, Whitty A, Cancilla MT, Wang J, Lugovskoy AA, Yoburn JC, Fung AD, Farrington G, Eldredge JK, Day ES, Cruz LA, Cachero TG, Miller SK, Friedman JE, Choong IC, Cunningham BC (2005) Small-molecule inhibition of TNF-alpha. Science (New York, NY) 310(5750):1022–1025. https://doi.org/10.1126/science.1116304
Laskowski RA, Jabłońska J, Pravda L, Vařeková RS, Thornton JM (2018) PDBsum: structural summaries of PDB entries. Protein Sci Publ Protein Soc 27(1):129–134. https://doi.org/10.1002/pro.3289
Moreira AL, Sampaio EP, Zmuidzinas A, Frindt P, Smith KA, Kaplan G (1993) Thalidomide exerts its inhibitory action on tumor necrosis factor alpha by enhancing mRNA degradation. J Exp Med 177(6):1675–1680. https://doi.org/10.1084/jem.177.6.1675
Sampaio EP, Sarno EN, Galilly R, Cohn ZA, Kaplan G (1991) Thalidomide selectively inhibits tumor necrosis factor alpha production by stimulated human monocytes. J Exp Med 173(3):699–703. https://doi.org/10.1084/jem.173.3.699
Spessard GO (1998) ACD Labs/LogP dB 3.5 and ChemSketch 3.5. J Chem Inf Comput Sci 38:1250–1253
Froimowitz M (1993) HyperChem: a software package for computational chemistry and molecular modeling. Biotechniques 14(6):1010–1013
Goodsell DS, Olson AJ (1990) Automated docking of substrates to proteins by simulated annealing. Proteins 8(3):195–202. https://doi.org/10.1002/prot.340080302
Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem 19(14):1639–1662
Cheng F, Li W, Zhou Y, Shen J, Wu Z, Liu G, Lee PW, Tang Y (2012) admetSAR: a comprehensive source and free tool for assessment of chemical ADMET properties. J Chem Inf Model 52(11):3099–3105. https://doi.org/10.1021/ci300367a
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46(1–3):3–26. https://doi.org/10.1016/s0169-409x(00)00129-0
Iman M, Davood A (2014) Homology modeling of lanosterol 14α-demethylase of Candida albicans and insights into azole binding. Med Chem Res 23(6):2890–2899. https://doi.org/10.1007/s00044-013-0769-z
Ntie-Kang F, Lifongo LL, Mbah JA, Owono LC, Megnassan E, Mbaze LMA, Judson PN, Sippl W, Efange SMN (2013) In silico drug metabolism and pharmacokinetic profiles of natural products from medicinal plants in the Congo basin. In Silico Pharmacol 1:12–12. https://doi.org/10.1186/2193-9616-1-12
Lin JH, Yamazaki M (2003) Role of P-glycoprotein in pharmacokinetics: clinical implications. Clin Pharmacokinet 42(1):59–98. https://doi.org/10.2165/00003088-200342010-00003
Ganeshan M, Polachi N, Nagaraja P, Subramaniyan BJ (2015) Antiproliferative activity of N-butanol floral extract from Butea monosperma against hct 116 colon cancer cells; drug likeness properties and in silico evaluation of their active compounds toward glycogen synthase kinase-3β/axin and β-catenin/T-cell factor-4 protein complex. Asian J Pharm Clin Res. 8:134–141
Haslett PA, Roche P, Butlin CR, Macdonald M, Shrestha N, Manandhar R, Lemaster J, Hawksworth R, Shah M, Lubinsky AS, Albert M, Worley J, Kaplan G (2005) Effective treatment of erythema nodosum leprosum with thalidomide is associated with immune stimulation. J Infect Dis 192(12):2045–2053. https://doi.org/10.1086/498216
Sheskin J (1965) Thalidomide in the treatment of lepra reactions. Clin Pharmacol Ther 6:303–306. https://doi.org/10.1002/cpt196563303
Corral LG, Muller GW, Moreira AL, Chen Y, Wu M, Stirling D, Kaplan G (1996) Selection of novel analogs of thalidomide with enhanced tumor necrosis factor alpha inhibitory activity. Mol Med (Cambridge, Mass) 2(4):506–515
Zeldis JB, Knight R, Hussein M, Chopra R, Muller G (2011) A review of the history, properties, and use of the immunomodulatory compound lenalidomide. Ann N Y Acad Sci 1222:76–82. https://doi.org/10.1111/j.1749-6632.2011.05974.x
Belarbi K, Jopson T, Tweedie D, Arellano C, Luo W, Greig NH, Rosi S (2012) TNF-α protein synthesis inhibitor restores neuronal function and reverses cognitive deficits induced by chronic neuroinflammation. J Neuroinflammation 9(1):23. https://doi.org/10.1186/1742-2094-9-23
Fujimoto H, Noguchi T, Kobayashi H, Miyachi H, Hashimoto Y (2006) Effects of immunomodulatory derivatives of thalidomide (IMiDs) and their analogs on cell-differentiation, cyclooxygenase activity and angiogenesis. Chem Pharm Bull 54(6):855–860. https://doi.org/10.1248/cpb.54.855
Aragon-Ching JB, Li H, Gardner ER, Figg WD (2007) Thalidomide analogues as anticancer drugs. Recent Pat Anticancer Drug Discov 2(2):167–174. https://doi.org/10.2174/157489207780832478
Schey SA, Fields P, Bartlett JB, Clarke IA, Ashan G, Knight RD, Streetly M, Dalgleish AG (2004) Phase I study of an immunomodulatory thalidomide analog, CC-4047, in relapsed or refractory multiple myeloma. J Clin Oncol Off J Am Soc Clin Oncol 22(16):3269–3276. https://doi.org/10.1200/jco.2004.10.052
Fala L (2015) Otezla (apremilast), an oral PDE-4 inhibitor, receives FDA approval for the treatment of patients with active psoriatic arthritis and plaque psoriasis. Am Health Drug Benefits 8(Spec Feature):105–110
Luo W, Tweedie D, Beedie SL, Vargesson N, Figg WD, Greig NH, Scerba MT (2018) Design, synthesis and biological assessment of N-adamantyl, substituted adamantyl and noradamantyl phthalimidines for nitrite, TNF-α and angiogenesis inhibitory activities. Bioorg Med Chem 26(8):1547–1559. https://doi.org/10.1016/j.bmc.2018.01.032
Jung YJ, Tweedie D, Scerba MT, Greig NH (2019) Neuroinflammation as a factor of neurodegenerative disease: thalidomide analogs as treatments. Front Cell Dev Biol 7:313. https://doi.org/10.3389/fcell.2019.00313
Benjamin E, Hijji YM (2017) A novel green synthesis of thalidomide and analogs. J Chem 2017:6436185. https://doi.org/10.1155/2017/6436185
Acknowledgements
Authors wish to acknowledge DBT-BIF, Govt. of India and K-FIST L1, VGST, Govt. of Karnataka for providing facility for molecular Docking and also wish to acknowledge Karnataka State Akkamahadevi Women’s University for providing infrastructure facility.
Funding
The authors of this research did not receive any funding concerning this research.
Author information
Authors and Affiliations
Contributions
BS contributed to preparation of ligands and target and molecular docking analysis; SSC contributed to prediction of ADMET property of all the designed small molecule inhibitors; SSJ contributed to preparation of manuscript and proofreading work; BRL contributed to research supervisor, hypothesis design, guidance for the entire project and manuscript preparation. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The author does not have any competing interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shivaleela, B., Srushti, S.C., Shreedevi, S.J. et al. Thalidomide-based inhibitor for TNF-α: designing and Insilico evaluation. Futur J Pharm Sci 8, 5 (2022). https://doi.org/10.1186/s43094-021-00393-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43094-021-00393-2