Abstract
Background
Barely Hordeum vulgare L. is considered one of the most important cereal crops with economic and industrial importance in the world, but its productivity is affected by climate change and abiotic stresses. One of the most recent and important microbiological promising aspects is the use of associated microorganisms, especially the endophytic bacteria producers for non-ribosomal peptides which play an important role in promoting plant growth, productivity, and tolerance to biotic and abiotic stresses. This work aims to identify vertically transferred or inherited endophytic bacterial communities in barely seeds, detect the presence of non-ribosomal peptides from these isolated endophytic strains and study their effect on protein patterns as a response to salinity stress.
Results
From two different tolerant (Giza 126) and sensitive (Giza 123) barely seeds cultivars, six different endophytic bacterial strains were isolated and identified using 16S rRNA. Bacterial strains belonged to Bacillus, Paenibacillus, Staphylococcus and Acinetobacter genera. Three of them have been isolated from both sensitive and tolerant barely cultivar (Uncultured Staphylococcus, Acinetobacter and Priestia endophytica or Bacillus endophyticus), while the other three endophytes have been isolated uniquely from the tolerant barely cultivar (Paenibacillus glucanolyticus, Bacillus cereus and Bacillus sp.). Non-ribosomal peptide synthetases genes NRPs of two lipopeptide families; surfactins and kurstakins have been detected using both bioinformatic analysis and degenerate primers. On the other hand, fragments similar to NRPs genes might be considered new NRPS molecules in Paenibacillus glucanolyticus, Acinetobacter and Priestia endophytica which have been detected using degenerate primers and required whole genome sequencing. The effect of soaking barely seeds exposed to 2.5% NaCl using SDS-PAGE electrophoresis revealed the presence of 24 bands, 10 of them were monomorphic with 41.5%, and 14 were polymorphic with 58.5% polymorphism.
Conclusion
The overnight soaking and co-cultivation of isolated endophytic strains with barely seeds before planting proved their capability in conferring salt stress tolerance to barely seedlings which appeared in protein patterns. We could consider these barely seeds endophytic among the PGPR strains promising to improve plant growth during abiotic stresses.
Similar content being viewed by others
1 Background
Barely Hordeum vulgare L. is considered one of the most important cereal crops in the world, used in bread and beverage industries and provides human health with nutrients that enhance heart health and help prevent cancer [1]. One of the most important factors of climate change is salinity which has negative effects on plant growth and crop productivity. Around the world, more than 800 million hectares are affected by salinity [2]. Plant-associated bacteria or endophytic bacteria can live inside plant tissues without harming and have an important role in promoting plant growth and tolerance to abiotic and biotic stress [3]. One of the most microbiological promising roles of endophytic bacteria is to interact with plants by settling in ecosystem restoration processes. One or more endophytic bacterial species might be harbored by each. The presence and the density of endophytic bacterial species depend on the variables of the bacterial species, host genotypes and environmental conditions [4]. Endophytic bacteria can excrete natural products benefits to plants in reducing diseases severity, promoting growth, and inducing plant defense mechanisms [5, 6]. Among these natural products, lipopeptides are produced by the non-ribosomal peptide synthesis mechanism using a multi-enzymes function synthetases system [7]. Surfactins, iturins and fengycins are three different families of non-ribosomal lipopeptides which were discovered between 1949 and 1986 from Bacillus spp. [8]. Seed-associated endophytic bacteria role is not well understood yet, but many studies have reported that diverse plant seed microorganisms are critical to seed and plant health [9, 10]. Recently, the role of seed endophytic community has been studied using multi-omics techniques which improved and significantly increased their understanding role. Seed endophytic isolates have been reported mostly as Proteobacteria, especially γ-Proteobacteria, followed by the Actinobacteria, Firmicutes and Bacteroidetes phyla [11]. The seed endophytes genera that are often detected and passed on to the next generation are Bacillus, Pseudomonas, Paenibacillus, Micrococcus, Staphylococcus, Pantoea and Acinetobacter. The presence of these endophytes genera has been reported in rice seeds [12], and Crotalaria pumila seeds during three consecutive years, respectively [13]. Plant growth-promoting (PGP) properties have been detected for barely seed endophytes and other plants in vitro and in vivo [12,13,15]. To our knowledge, there are not enough studies on barely seed endophytes till now.
The interactions between genotypes genes and the environment affected protein and plant response to stress [16, 17] which could be understanding using the SDS-PAGE technique tool to explain the genetic diversity and relationships among different crop species/varieties such as wheat and barley genotypes [18, 19] and may provide information for the use of genotypes in different breeding experiments used as a rapid, easy, inexpensive evaluation of genetic diversity, because of that it became accepted valuable tool [20, 21].
One of the most important techniques employed in biological analysis is the SDS-PAGE technique which can determine shifts in protein bands that could be proteins or enzymes produced due to bio-stress [21,22,23,24,26]. Electrophoretic pattern changes have been detected in the soluble proteins of different cultivars grown in different environments. Plants, when exposed to certain types of environmental stress conditions, can help organisms to tolerate such stresses by activation of stress genes to produce stress proteins.
This work aims to identify the vertically transferred or inherited endophytic bacterial communities in barely seeds, detect the presence of non-ribosomal peptides from these isolated endophytic strains and study their effect on protein patterns as a response to salt stress.
2 Methods
2.1 Plant samples
Sensitive (Giza 123) and tolerant (Giza 126) barely seeds were obtained from Field Crops Research Institute, Agricultural Research Center (ARC), Giza, Egypt.
2.2 Bacterial strains isolation and medium
About 20 seeds of each cultivar sensitive (Giza 123) and tolerant (Giza 126) were surface sterilized according to [27]. The attached dust was removed by washing with tap water, then with ethanol 70% for 1 min and 5 min with 5% sodium hypochlorite. After that, the seeds were rinsed with sterilized distilled water three times, and then, seeds were sterilized one more time with ethanol 70% for 10 s and rinsed with sterilized distilled water three times. After that, soaking seeds in 25 ml sterilized distilled water overnight was recommended to become soft and easy to mash. This mixture was incubated on two different cultures media YPDA (20 g peptone, 10 g yeast extract, 20 g dextrose and 15 g agar in 1litertotal volume, pH 6.5) and 1/10 869 [0.1 g glucose, 0.5 g Nacl, 1 g tryptone, 0.5 g yeast extract and 15 g agar in 1-L total volume, pH 6.5 [28]. Three replicates for each cultivar were prepared in addition to one negative control (without surface sterilization) and positive control (100 µl of the last rinsed water) at 28 C until the appearance of colonies. Each resulting colony was cultured on LB solid plate separately at 30 C overnight till growth. Physiological identification for strains followed by DNA extraction and 16srRNA isolation has been performed.
2.3 Identification of barely seeds fungi
A sterilized needle was used to obtain fungal tissues from a petri dish. Fungal tissues were placed on a clean microscopic slide. Fungal spores were photographed with a fixed camera attached to a compound microscope at 200 × magnification (Olympus microscope camera, Japan). Fungal key identification texts such as [29] were consulted for aiding in accurate identification.
2.4 Identification of endophytic bacteria isolated from barely
The grown colonies were total genomic DNA extracted by a DNA extraction kit (Qiagen, USA). Bacterial 16srDNA universal primers were used as described by using primers 27F (AGAGTTTGATCMTGGCTCAG) and 1525R (AAGGAGGTGWTC-CARCC) [30] in a Biometra thermocycler. The PCR amplification was programmed for 30 cycles as follows: an initial denaturation for 5 min at 95◦C, a denaturation step for 1 min at 95◦C, an elongation step for 1 min at 55◦C and finally, an extension step for 1.30 min at 72◦C. The final extension was at 72◦C for 10 min. The PCR product was purified by a PCR purification kit (Qiagen, USA) and sent to Macrogen Inc. (Seoul, South Korea) for sequencing. The closest match sequences were identified using BLAST search of the NCBI GenBank website.
2.5 Detection of NRPS encoding genes for identified endophytic strains by bioinformatics tools
Sequenced genomes of related identified endophytic strains were obtained from the National Center for Biotechnology Information (USA) NCBI and analyzed with the Anti-Smash website to detect the secondary products and the gene clusters. PKS-NRPS website (http://nrps.igs.umaryland.edu/nrps) was used to identify the NRPS genes, annotation, description of modules and domains, specificity prediction of adenylation (A) domains, potential primary structure of the peptide. The last step was to compare the predicted peptides structures obtained by PKS/NRPS website with the Norine database (http://bioinfo.lifl.fr/norine) which includes a database of 1185 non-ribosomal peptides to know whether the predicted peptide is an existing molecule or a new one.
2.6 NRPS synthetases genes detection by degenerate primers
NRPS synthetases genes were detected using five sets of primers (Table 1) which were designed by (31; 32) from the conserved sequences of adenylation and thiolation domain of some NRPS synthetases genes of plipastatin (Ap1-F/ Tp1-R), fengycin (Af2-F/ Tf1-R), mycosubtilin (Am1-F/ Tm1-R), surfactins (As1-F/ Ts2-R) and kurstakin (Aks-F/Tks-R). The PCR conditions were performed at 94 °C for 2 min as initial denaturation step, followed by 35 cycles of three steps; denaturation for 30 s at 94 °C; annealing step for 30 s at 43 °C with (As1-F/Ts2-R), at 58 °C with (Ap1-F/Tp1-R) and at 44.4 °C with (Aks-F/Tks-R); and an extension step of 45 s at 72 °C except with Ap1-F/Tp1-R primers (75 s at 72 °C) and (Aks-F/Tks-R). A final extension step was lanced at 72 °C for 5 min.
The amplified fragments were analyzed by agarose gel electrophoresis (1.2%), and then gels were visualized by the Gel Doc system (Bio-Rad). Fragments were purified from gels by a PCR purification kit (Qiagen, USA) and sent to Macrogen Inc.(Seoul, South Korea) for sequencing.
2.7 Phylogenetic tree
The sequences were aligned to the 16S rDNA database on GenBank using BlastN, and the closest sequences were used to construct the phylogenetic tree which was performed by Mega (Version 6.0) with a neighbor-joining method [33].
2.8 Bacteria preparation for salinity experiment
The isolated endophytic bacteria strains from barely seeds (B1, B2, B3, B4, B5 and B6) as shown in Table 3 were grown in LB liquid media overnight culture at 30 °C. Barley (Hordeum Vulgare) verity Giza 138 salinity sensitive was used to select the best bacteria-treated seeds under salinity stress. The barely seeds were soaked in overnight culture bacteria, 10 seeds with three replicates for each treatment B1, B2, B3, B4, B5, B6, B1.3, B 2.4.5.6, B1.2.3.6 and B1.2.3.4.5.6 for 24 h. The barley-treated grains with bacteria were germinated in pots. The growing seedling plants were treated with 2.5% NaCl after two weeks of germination for 48 h the control and treatments were collected for protein extraction.
2.9 SDS-PAGE protein banding patterns
Samples preparations and extraction of total proteins were performed according to [34] and analyzed according to the method of [35]. Separating gel of 15% were prepared, whereas protein fractionations were performed exclusively on vertical slab gel (19.8 cm × 26.8 cm × 0.2 cm) using the vertical electrophoresis apparatus manufactured by Cleaver, UK. The gel was photographed and scanned by Gel Doe Bio-Rad System (Gel Pro analyzer V.3) followed by protein bands analysis, by the Total Lab program.
3 Results
3.1 Isolation and identification of endophytes from sensitive and tolerant barely seeds cultivars
Fifty colonies were grown on both YPDA and 1/10 diluted 869 media which were screened physiologically under microscope after staining with gram. After the screening, six different bacterial endophytes were selected for identification, three of them were isolated from both sensitive (Giza 123) and tolerant barely cultivar (Giza 126) (Uncultured Staphylococcus, Acinetobacter and Priestia endophytica or Bacillus endophyticus), while the other three endophytes have been isolated uniquely from the tolerant barely cultivar (Paenibacillus glucanolyticus, Bacillus cereus and Bacillus sp.). The identification using 16 s rRNA revealed six different strains Acinetobacter johnsonii, Bacillus cereus, Bacillus sp., Paenibacillus glucanolyticus, priestia endophytica and Staphylococcus pasteuri.
Two experiments 1 (without plant extract) and 2 (with plant extract) using YPDA and 1/10 diluted 869 media have proved its significance; using 1/10 diluted 869 (rich) media have delivered the highest diversity of endophytes, as well as the highest numbers of cultivable endophytes, while the addition of plant extract to the medium composition significantly increased the numbers of isolated bacteria but had small effect on the cultivable endophytes diversity (Table 2).
3.2 Phylogenetic analysis
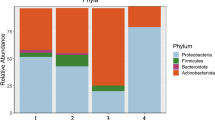
The phylogenetic tree revealed six different strains belonging to two phyla Firmicutes (Staphylococcus pasteuri, Paenibacillus glucanolyticus, Bacillus cereus, Bacillus sp. and priestia endophytica) and Proteobacteria (Acinetobacter johnsonii). Among the Firmicutes, 3 families were found including: Bacillaceae (3 strains), Paenibacillaceae (2 strains) and Staphylococcaceae (one strain). Five strains showed high similarity from 98 to 96% except strain no. 6 which revealed 93% with Staphylococcaceae spp. (Fig. 1). The isolated barely endophytic strains have been submitted to GenBank, and accession numbers have been obtained (Table 3).
3.3 Identification of barely seeds fungi
The identification of barely cultivated fungi based on colony color and conidial shape revealed the presence of three different fungal species: Alternaria alternate, Aspergillus niger and Rhizopus stolonifera.
3.4 Detection of secondary metabolite genes involved in some isolated endophytic strains from barely seeds by Anti-smash
The identified barely endophytic bacterial strains-related genomes were analyzed by Anti-smash which allows the rapid analysis of secondary metabolite biosynthesis gene clusters of bacterial and fungal genomes by genome-wide identification and annotation. Between the six identified endophytic isolates, three of them (two Paenibacillus glucanolyticus and Bacillus cereus) were analyzed by Anti-smash due to the availability of their complete genome sequences on GenBank (NCBI), while the other three isolates were analyzed depending on their encoded protein data on GenBank followed by annotation and NRPS synthetases domains and modules detection using PKS-NRPS website. Two endophytic strains: Staphylococcus and Acinetobacter, have shown the absence of NRPS genes, while two strains: Bacillus sp. (In bacteria) and Priestia endophytica, have shown the presence of NRPS genes. Bacillus sp. (In bacteria) have shown the presence of bacillibactin synthetase gene, and two partial genes involved in surfactin biosynthesis, while Priestia endophytica encoded protein analysis revealed the presence of bacillibactin synthetase gene, six genes cluster of unknown NRPS product and three genes cluster responsible for the biosynthesis of kurstakin lipopeptide (Fig. 2 and Table 4). Anti-smash analysis of Paenibacillus glucanolyticus genome sequence (accession no: CP028366.1) revealed 14 regions of identified secondary metabolites, four of which are NRPS and PKS products. Bacillibactin, PKS-NRPS hybrid unknown product, unknown PKS product and one synthetase gene encoded for tryptophan (Fig. 3 and Table 4). Bacillus cereus complete genome sequence analysis by Anti-smash revealed the presence of 9 regions of identified secondary metabolites, three of which are NRPS products. In bacillibactin, two regions were identified as fengycin with a similarity of 40%, which was well analyzed by PKS-NRPS and identified as kurstakin (Fig. 4 and Table 4).
Bacillus cereus and Priestia endophytica are found to harbor kurstakin synthetases genes encoded for kurstakin lipopeptide which consists of three gene cluster heptapeptide as shown in Fig. 4.
Three gene clusters were identified from Priestia endophytica. One NRP (Fig. 2A) consists of one gene encoded for bacillibactin. The gene cluster revealed in Fig. 2B consists of five genes and is a Type I PKs-NRPs hybrid BGC (unknown NRPs) with a size of approximately 30 kb. The PKs module consists of a ketosynthase (KS) domain, an acyltransferase (AT) domain, an acyl carrier protein (ACP) domain and a thioesterase (TE) domain which incorporates the polyketide moiety of malonyl-CoA, while the NRPs modules incorporate six amino acid residues. Based on anti-SMASH analysis, this unknown molecule shows a low similarity of 28% to the paenilamicin BGC, synthesized by pam BGC from Paenibacillus larvae DSM25430 which shows antibacterial and antifungal activity. This pam cluster has a size of ∼60 kb and consists of five NRPs genes, two Type I PKs genes and two Type I PKs-NRPs hybrid genes. Additionally, B. endophyticus FH5 has been found to harbor Type I PKs-NRPs hybrid BGC but consists of only three NRPS genes and one Type I PKS gene and differs from the pam cluster of Paenibacillus larvae DSM25430. Other NRPs of 25 kb total size (Fig. 2C) consist of three genes encoding for 24 domains: seven condensation (C) domains, seven adenylation (A) domains, seven thiolation (T) domains, two epimerization (E) domains and one thioesterase (TE) domain. This BGC catalyzes the primary formation of a lipopeptide product highly similar to kurstakin.
3.5 NRPs synthetase genes detection in endophytic isolates strains by degenerate primers
Kurstakin degenerate primers amplified fragments of 1162 bp with uncultured Staphylococcus, Paenibacillus glucanolyticus and Priestia endophytica (Fig. 5). These results agree with bioinformatic detection of kurstakin synthetase genes in Paenibacillus glucanolyticus and Priestia endophytica, while it seems different with bioinformatic detection of kurstakin synthetase genes in Bacillus cereus which may refer to that this strain belongs to another Bacillus species or by the absence of kurstakin synthetase genes.
Plipastatin degenerate primers amplified no specific fragments for plipastatin synthetase genes which indicates the absence of this lipopeptide in all isolated strains from barely (Fig. 5).
Surfactin degenerate primers led to the amplification of surfactin synthetase partial gene fragment with the expected size of 431 bp in both strains Bacillus cereus and Bacillus sp., while it led to the detection of fragments similar to a NRPs gene in Paenibacillus glucanolyticus, Acinetobacter and Priestia endophytica (Fig. 6). These strains are highly recommended for whole genome sequencing because they may harbor new NRPS molecules.
Bioinformatic analysis results of related complete genomes of endophytic isolates from barely seeds match with degenerate primers results in all isolated strains except in Bacillus cereus and Uncultured Staphylococcus which appear to need more analysis by sequencing their complete genomes to verify their NRPs presence. Uncultured Staphylococcus strain has no related complete genome sequence on NCBI to detect the NRPs synthetases genes by bioinformatics, while the use of degenerate primers led to the detection of kurstakin synthetases genes in the strain. Bioinformatic analysis of Bacillus cereus-related complete genome sequence on NCBI revealed the presence of bacillibactin synthetase gene and kurstakin synthetase genes cluster, while the use of degenerate primers has proven the absence of kurstakin and the presence of surfactin synthetase genes cluster (Table 5).
3.6 Soluble Proteins electrophoresis
Protein profiling assessment by SDS-PAGE for barley samples of different treatments showed different banding patterns. Many alterations in protein patterns have been observed based on the relative mobility of gel proteins in barley leaves treated with different endophytic bacteria isolated from barley seeds B1, B2, B3, B4, B5, B6, B1.3, B2.4.5.6, B1.2.3.6 and B1.2.3.4.5.6 under control and 2.5% NaCl, whereas after 48 h of treatment 24 total bands appeared; ten of them are monomorphic bands no 4, 11, 12, 15, 17, 18, 19, 20, 21 and 24 with molecular weights MW; 22, 80, 52, 50, 42, 35, 30, 25, 20, 19 and 11 KDa, respectively, and 41.5% percentage, while fourteen polymorphic bands were revealed with 58.5% percentage no. 1, 3, 5, 6, 7, 8, 9, 10, 13,16, 22 and 23 with MW 245, 100, 76, 72, 69, 63, 60, 56, 48, 38, 18 and 17 KDa. There were positive and negative unique bands. Two unique bands at MW 180 and 45 KDa appeared in samples no. 2 and 17 (2.5% NaCl and B2.4.5.6), respectively. The highest number of total and polymorphic with unique bands were revealed in sample 22 (B1.2.3.4.5.6 + 2.5% NaCl) with 21 and 11 bands, respectively. On the other hand, samples no. 2 and 8 (2.5% NaCl and B3 + 2.5% NaCl) were the lowest total and polymorphic with unique bands 12 and 2 bands, respectively, as shown in Tables 6, 7 and Figs. 7 and 8.
4 Discussion
In this work, we study the role of barely seeds endophytic bacteria vertically transferred or inherited in tolerance to salinity stress and focus on the role of a group of natural products secreted by these bacteria called non-ribosomal peptides NRPs which consider alternatives to traditional chemical substances. The use of 1/10 diluted 869 (rich) media has delivered the highest diversity of endophytes, as well as the highest numbers of cultivable endophytes, while the addition of plant extract to the medium composition significantly increased the numbers of isolated bacteria but had a small effect on the cultivable endophytes diversity [36]. These endophytes have been found to belong to three main families: Bacillaceae, Paenibacillaceae and Staphylococcaceae. These results agreed with [36] who reported that highland barley seeds harbored the genus Bacillus. The use of Anti-Smash in detecting secondary metabolites from bacterial complete genomes facilitates and economizes the time of researchers [37]. Here, we analyzed three complete genome sequences of Paenibacillus glucanolyticus, Bacillus cereus and Priestia endophytica endophytes using Anti-Smash which revealed the presence of gene clusters of bacillibactin and kurstakin in addition to the use of PKS-NRPS website which detected the presence of gene clusters of bacillibactin and surfactin in Bacillus spp. endophytic strain. These results are agreed with [36, 38] who identified and characterized Bacillus endophytes species which are widely used as producers of growth-promoting substances, bioactive compounds and metabolites with antimicrobial effects.
To our knowledge, endophytes in barely seeds and consequently the detection of NRPs genes have not been studied to date, but kurstakin synthesis has been reported in several studies from Bacillus thuringiensis and Bacillus cereus group. Also, Paenibacillus spp. included in the group of plant growth-promoting rhizobacteria and biofertilizers [8] which produced a wide spectrum of NRPs as: polymyxins, fusaricidins, tridecaptins and other secondary metabolites weapons against Gram-positive, Gram-negative and fungal competitors [39, 40]. Detecting NRPS genes by using degenerate primers was reported before by [32, 33] who confirmed the detection of kurstakin synthetase genes in Bacillus thuringiensis using kurstakin AKs-F/TKs-R primers pair and detection of surfactin synthetase genes in Bacillus subtilis using surfactin degenerate As1-F/Ts2-R primers pair, respectively. We here detected the presence of fragments similar to an NRPs gene in Paenibacillus glucanolyticus, Acinetobacter and Priestia endophytica using surfactin degenerate As1-F/Ts2-R primers pair, which considered in agreement with [33] who detected fragments similar to an NRPs gene in B. thuringiensis berliner 1915 using the same primers.
Salinity treatment of barely seeds previously soaked in different endophytic strain suspensions has revealed many alterations in protein profile using SDS-PAGE. The results were in agreement with those of [21] who used SDS-PAGE to study the genetic diversity between wheat genotypes and scored 31 total bands among the genotypes and maximum polymorphism of 95.8%. Also, SDS-PAGE was used to identify twelve durum wheat Genotypes and detected a total number of 22 bands, 18 of them were polymorphic bands with 81.8% polymorphism, including two unique bands [41]. A total number of 36 bands have been detected by [42] arranged between 124 to 12 kDa, 8 bands of which were polymorphic under salt stress in barley. In addition, upon transition from control to stress environment, [43] detected that band numbers increased in both cultivars and compared to the susceptible cultivar, resistant cultivar showed more number of bands. The use of all the six bacterial strains might play an important role in plant growth promotion by plant pathogens growth inhibition, and/or by producing several secondary metabolites that can help in improving crop yield. These results agreed with those of [44] who used it as an eco-friendly alternative tool in the agricultural sector for the detection of plant defense to assist the host in sustaining different abiotic and biotic stresses.
We observed that the best bacterial treatment under salinity stress 2.5% NaCl was the co-cultivation between all barely isolated endophytic strains B 1.2.3.4.5.6 which revealed a synergistic effect by the interaction between the endophytic bacteria. It is known that the plant-associated beneficial rhizosphere and/or endosphere bacteria are considered biocontrol agents that protect their host plants against pathogens directly or by promoting the plant’s induced systemic resistance (ISR) [3, 45] which depend on increasing the chemical or physical barrier of the host plant. Indeed, several elicitors involved in ISR have been identified in Bacillus subtilis GB03 [18, 29] including antibiotics [8], organic volatile compounds and biosurfactants [lipopeptides families such as; surfactin] in Paenibacillus polymyxa C13 [46]. Our results revealed a significant effect of the use of Bacillus genus members (strains B2, B4, B5 and B6) in addition to Staphylococcus (B1 strain) and Acinetobacter (B3 strain) members in conferring salt stress tolerance to barely seedlings. These results agree with [47] which reported that Bacillus amyloliquefaciens NBRI-SN13 (SN13) plant growth-promoting rhizobacteria (PGPR), confer salt stress tolerance to rice seedlings and are known to modulate endogenous level of phytohormones in plants as a member of Bacillus genus [47, 48] and a Bacillus producing lipopeptide families (surfactin, fengycin or plipastatin and iturin). According to these results, we could consider barely seeds endophytic strains among the PGPR which are promising to improve plant growth during abiotic stress.
5 Conclusion
The use of endophytic bacteria is considered an alternative method to improve plant stress tolerance. According to this work, the overnight soaking and co-cultivation of these endophytic strains harbor NRPs synthetase clusters of two different lipopeptide families, surfactin and kurstakin with barely seeds before planting proved their capability in conferring salt tolerance to barely seedlings which appeared in protein patterns. We could consider these barely seeds endophytic among the PGPR strains which are promising in improving plant growth during abiotic stresses.
Availability of data and materials
All the data we generated or analyzed in this study are included in this published article.
Abbreviations
- ACP:
-
Acyl carrier protein domain
- AT:
-
Acyltransferase domain
- A:
-
Adenylation domain
- BLAST:
-
Basic Local Alignment Search Tool
- BGC:
-
Bacterial Gene Cluster
- C:
-
Condensation domain
- E:
-
Epimerization domain
- ISR:
-
Induced Systemic Resistance
- KS:
-
Ketosynthase domain
- NCBI:
-
National Center for Biotechnology Information
- NRPs:
-
Non-ribosomal peptides
- PGPR:
-
Plant growth-promoting rhizobacteria
- PKS:
-
Polyketide Synthase
- SDS-PAGE:
-
SDS polyacrylamide gel electrophoresis
- TE:
-
Thioesterase domain
References
Meng G, Rasmussen SK, Christensen CSL, Fan W, Torp AM (2023) Molecular breeding of barley for quality traits and resilience to climate change. Front Genet 13:1–8. https://doi.org/10.3389/fgene.2022.1039996
Pour-Aboughadareh A, Mehrvar MR, Sanjani S, Amini A, Nikkhah-Chamanabad H, Asadi A (2021) Effects of salinity stress on seedling biomass, physiochemical properties, and grain yield in different breeding wheat genotypes. Acta Physiol Plant 43:1–14
Vandana UK, Rajkumari J, Singha LP, Satish L, Alavilli H, Sudheer PD, Pandey P (2021) The endophytic microbiome as a hotspot of synergistic interactions with prospects of plant growth promotion. Biol 10(2):101
Chebotar VK, Malfanova NV, Shcherbakov AV, Ahtemova GA, Borisov AY, Lugtenberg B, Tikhonovich IA (2015) Endophytic bacteria in microbial preparations that improve plant development (review). Appl Biochem Microbiol 51:271–277
Ongena M, Jourdan E, Adam A, Paquot M, Brans A, Joris B, Arpigny JL, Thonart P (2007) Surfactin and fengycin lipopeptides of Bacillus subtilis as elicitors of induced systemic resistance in plants. Environ Microbiol 9:1084–1090
Hussein W, Awad H, Fahim S (2016) Systemic resistance induction of tomato plants against ToMV virus by surfactin produced from Bacillus subtilis BMG02. Americ J Microbiol Res 4(5):153–158
Marahiel MA (1997) Protein templates for the biosynthesis of peptide antibiotics. Chem Biol 4:561–567
Lopes R, Tsui S, Gonçalves PJRO, de Queiroz MV (2018) A look into a multifunctional toolbox: endophytic Bacillus species provide broad and underexploited benefits for plants. World J Microbiol Biotechnol 34:94. https://doi.org/10.1007/s11274-018-2479-7
Jacques P (2011) Surfactin and other lipopeptides from Bacillus spp. In: Soberon-Chavez G (ed) Biosurfactants Microbiology Monographs. Springer, Cham, pp 57–91
Grumet R, Gifford F (1998) Plant biotechnology in the United States: issues and challenges in route to commercial production. HortScience 33:187–192. https://doi.org/10.1023/A:1008693614060
Gitaitis R, Walcott R (2007) The epidemiology and management of seedborne bacterial diseases. Annu Rev Phytopathol 45:371–397. https://doi.org/10.1146/annurev.phyto.45.062806.094321
Truyens W, Cuypers V (2015) Bacterial seed endophytes: genera, vertical transmission and interaction with plants. Environ Microbiol Rep 7:40–50. https://doi.org/10.1111/1758-2229.12181
Kaga H, Mano H, Tanaka F, Watanabe A, Kaneko S, Morisaki H (2009) Rice seeds as sources of endophytic bacteria. Microbes Environ 24:154–162. https://doi.org/10.1264/jsme2.ME09113
Sánchez-López AS, Pintelon I, Steven V, Imperato V, Timmermans JP, González-Chávez C et al (2018) Seed endophyte microbiome of Crotalaria pumila unpeeled: identification of plant-beneficial methylobacteria. Int Mol Sci 19:291. https://doi.org/10.3390/ijms19010291
Rahman MM, Flory E, Koyro HW, Abideen Z, Schikora A, Suarez C et al (2018) Consistent associations with beneficial bacteria in the seed endosphere of barley (Hordeum vulgare L.). Syst Appl Microbiol 41:386–398. https://doi.org/10.1016/j.syapm.2018.02.003
Rios-Galicia B, Villagómez-Garfias C, De la Vega-Camarillo E, Guerra-Camacho JE, Medina-Jaritz N, Arteaga-Garibay RI et al (2021) The Mexican giant maize of Jala landrace harbour plant-growth-promoting rhizospheric and endophytic bacteria. Biotech 3(11):447. https://doi.org/10.1007/S13205-021-02983-6
Johansson E, Malik AH, Hussain A, Rasheed F, Newson WR, Plivelic T, Hedenqvist M, Gällstedt M, Kuktaite R (2013) Gluten protein structures: Variation in wheat grain and for various applications. In: He, Z., Wang, D. eds., Proceeding: 11th International Gluten Workshop. Beijing, China, August 12–15, 2012, Mexico : International Maize and Wheat Improvement Center.
Ling HQ, Zhang A, Wang D, Liu D, Wang JY, Sun H, Fan HJ, Li ZS, Zhao Y, Wang DW, Zhang KP, Yang YS, Wang JJ, Dong L. Wheat A genome sequencing and its application for quality modification (2012) In: He, Z., Wang, D. eds., Proceeding: 11th International Gluten Workshop. Beijing, China, August 12–15, 2012, Mexico : International Maize and Wheat Improvement Center.
Miháliková D, Galova Z, Petrovičová L, Chňapek M (2016) Polymorphism of proteins in selected slovak winter wheat genotypes using SDS-PAGE. J Central Eur Agric 17(4):970–985
Banta N, Singh R, Singh N (2021) Comparative protein profile analysis by SDS-PAGE of different grain cereals. Pharma Innov J 10(9):104–108
Kakaei M, Kahrizi D (2011) Evaluation of seed storage protein patterns of ten wheat varieties using SDS-PAGE. Biharean Biologist 5(2):116–118
Hlozakova TK, Gregova E, Vivodik M, Gálová Z (2016) Genetic diversity of European cultivars of common wheat (Triticum aestivum L.) based on RAPD and protein markers. J Central Europ Agricu 17(4):957–969. https://doi.org/10.5513/JCEA01/17.4.1798
Ghasempour HR, Anderson EM, Gianello RD, Gaff DF (1998) Growth inhibitor effects on protoplasmic drought tolerance and protein synthesis in leaf cells of resurrection grass. S stapfianus J Plant Growth Regul 24:179–183
Gianello RD, Kuang J, Gaff DF, Ghasempour HR, Blomstedt CK, Hamill JD (2000) Proteins correlated with desiccation tolerance in a resurrection grass S stapfianus. Scientific Publishers, Oxford, pp 161–166
Ghasempour HR, Anderson EM, Gaff Donald F (2001) Effects of growth substances on the protoplasmic drought tolerance of leaf cells of the resurrection grass, Sporobolus stapfianus. Aust J Plant Physiol 28:1115–1120
Ghasempour HR, Kianian J (2002) Drought stress induction of free proline, total proteins, soluble sugars and its protein profile in drought tolerant grass Sporobolus elongates. J Sci Teacher Training Univ 1:111–118
Ghasempour HR, Maleki M (2003) A survey comparing desiccation tolerance in resurrection plant Nothelaena vellea and studying its protein profile during drought stress against a non-resurrection plant Nephrolepsis sp. Iranian J Biol 15:43–48
Wen CY, Yen ZG, Wang KX, Chen JG, Shen SS (2011) Structural analysis of surfactin produced by endophytic strain Bacillus subtilis EBS05 and its antagonistic activity against Rhizoctonia cerealis. Plant Pathol 27(4):342–348
Mergeay M, Nies D, Schlegel HG, Gerits J, Van CP, Gijsegem F (1985) Alcaligenes entrophus CH34 is a facultative chemolitotroph with plasmid-bound resistance to heavy metals. J Bacteriol 162:328–334
Robinson M (2011) Pictorial atlas of soil and seed fungi: morphologies of cultured fungi and key to species. Ref Rev 25(4):43–44. https://doi.org/10.1108/09504121111134098
DeLong EF (1992) Archaea in coastal marine environments. PNAS 89:5685–5689. https://doi.org/10.1073/pnas.89.12.5685
Tapi A, Chollet-Imbert M, Scherens B, Jacques P (2010) New approach for the detection of non-ribosomal peptide synthetase genes in Bacillus strains by polymerase chain reaction. Appl Microbiol Biotechnol 85:1521–1531
Abderrahmani A, Tapi A, Nateche F, Chollet M, Leclère V, Wathelet B, Hacene H, Jacques P (2011) Bioinformatics and molecular approaches to detect NRPS genes involved in the biosynthesis of kurstakin from Bacillus thuringiensis. Appl Microbiol Biotechnol 92:571–581
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:6800685
Stegmann H (1979) Electrophoresis and focusing in slabs using the Pantaphor apparatus for analytical and preparative separations in gel (Polyacrylamide, Agarose, Starch, Sephadex).Messeweg 11, D03300, Braunschweig Institute of Biochemistry, West Germany pp: 1029
Chen Y, Liang J, Zia A, Gao X, Wang Y, Zhang L, Xiang Q, Zhao K, Yu X, Chen Q, Penttinen P, Nyima T, Gu Y (2022) Culture dependent and independent characterization of endophytic bacteria in the seeds of highland barley. Front Microbiol 13:981158. https://doi.org/10.3389/fmicb.2022.981158
Blin K, Shaw S, Kautsar SA, Medema MH, Weber T (2021) The antiSMASH database version 3: increased taxonomic coverage and new query features for modular enzymes. Nucleic Acids Res 49(D1):D639–D643. https://doi.org/10.1093/nar/gkaa978.PMID:33152079;PMCID:PMC7779067
Timmusk S, Wagner EG (1999) The plant-growth-promoting rhizobacterium Paenibacillus polymyxa induces changes in Arabidopsis thaliana gene expression: a possible connection between biotic and abiotic stress responses. Mol Plant-Microbe Interact MPMI 12:951–959
Cochrane SA, Findlay B, Bakhtiary A, Acedo JZ, Rodriguez-Lopez EM, Mercier P, Vederas JC (2016) Antimicrobial lipopeptide tridecaptin A 1 selectively binds to Gram-negative lipid II. Proc Natl Acad Sci 113:11561–11566
Kajimura Y, Kaneda M (1996) Fusaricidin A, a New Depsipeptide Antibiotic Produced by Bacillus polymyxa KT-8 Taxonomy, Fermentation, Isolation, Structure Elucidation and Biological Activity. J Antibiot (Tokyo) 49:129–135
Osman SA, Ramadan WA (2020) Characterization of Egyptian durum Wheat Genotypes using biochemical and molecular Markers. Jordan J Biolog Sci 13(4):419–429
Metwali EMR, Fuller MP, Gowaye SMH, Almaghrabi OA, Mosleh YY (2013) Evaluation and selection of barley genotypes under optimum salt stress condition using tissue culture techniques and SDS-PAGE gel electrophoresis. J Food Agric Environ 11:1386–1394
Vyomesh SP, Pitambara Shukla YM (2018) Proteomics study during root knot nematode (Meloidogyne incognita) infection in tomato (Solanum lycopersicum L.). J Pharmacognosy Phytochem 7(3):1740–1747
Kumar A, Kumar R, Kumari M, Goldar S (2020) Enhancement of plant growth by using PGPR for a sustainable agriculture: a review. Int J Curr Microbiol Appl Sci 9:152–165
Berg G, Smalla K (2009) Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. F EMS microbiol ecol 68(1):1–13
Lee B, Farag MA, Park HB, Kloepper JW, Lee SH, Ryu CM (2012) Induced resistance by a longchain bacterial volatile: elicitation of plant systemic defence by a C13 volatile produced by Paenibacillus polymyxa. PLoS ONE 7:e48744
Nautiyal CS, Srivastava S, Chauhan PS, Seem K, Mishra A, Sopory SK (2013) Plant growth-promoting bacteria Bacillus amyloliquefaciens NBRISN13 modulates gene expression profile of leaf and rhizosphere community in rice during salt stress. Plant Physiol Biochem 66:1–9
Srivastava AK, Kumar S, Kaushik R, Singh P (2014) Diversity analysis of Bacillus and other predominant genera in extreme environments and its utilization in Agriculture. Technical report 414402/C30026.
Acknowledgements
Not applicable
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
WH performed all isolation, identification of endophytic communities, bioinformatic analysis and NRPs detection by PCR, and WAR and HFI performed all salinity stress and protein pattern experiments and data analysis. WH, WAR and HFI wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors report there are no competing interests to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hussein, W., Ramadan, W.A. & Ibrahim, H.F. Isolation and identification of associated endophytic bacteria from barely seeds harbour non-ribosomal peptides and enhance tolerance to salinity stress. Beni-Suef Univ J Basic Appl Sci 13, 27 (2024). https://doi.org/10.1186/s43088-024-00483-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43088-024-00483-z