Abstract
Background
In some β-thalassemia intermedia patients, hydroxyurea (HU) increases hemoglobin and HbF levels. However, HUs’ effects molecular mechanism is still unclear.
Methods
In this study, a weighted gene co-expression network analysis was conducted on the GSE109186 dataset. The genes LIN28A, COL4A5, SP1, BCL2, and IGF2BP3 were identified as hub genes involved in the γ-gene switching process. The effect of HU treatment at doses of 50, 100, and 150 μM for 12, 24, and 48 h on the K562 cell line was examined by using qRT-PCR to measure the expression levels of these hub genes.
Results
SP1 gene expression decreased after treatment with 50, 100, and 150 μM HU for 12, 24, and 48 h. The expression of the LIN28A gene was tripled at three concentrations of 50, 100, and 150 μM for 12 h. The IGF2BP3 gene expression was doubled after 24 and 48 h at a concentration of 150 μM HU. Regarding COL4A5 gene expression, except at 12 h after treatment at a concentration of 50 μM, a significant increase was observed in other concentrations and times. The BCL2 gene expression pattern at all concentrations decreased significantly after 12 h. The γ gene showed a significant increase compared to the control group after 24 and 48 h at the different concentrations.
Conclusion
The results showed that in HU-treated cells, changes in the expression of LIN28A, COL4A5, SP1, and IGF2BP3 genes were accompanied by an increase in γ-gene expression. By elucidating precisely the mechanism of γ-to-β gene switching, we can hope for less complicated drugs.
Similar content being viewed by others
Introduction
Hemoglobinopathy is a group of disorders related to the abnormal structure or production of the hemoglobin molecule. More than 330,000 babies are born with these disorders each year in developing countries [1]. These disorders affect various organs and are associated with high morbidity and mortality, making them a severe public health problem. Regular blood transfusions with iron chelation and bone marrow transplantation are practical therapeutic approaches in beta-thalassemia (β-thal) [2, 3].
Today, pharmacologically induced human γ-globin gene expression in the intermediate β-thal treatment is a new therapeutic strategy [3, 4]. Drugs that reactivate the γ-globin gene can reduce the complications of ineffective erythropoiesis by increasing γ-globin and fetal hemoglobin (HbF) synthesis [1, 4, 5]. To date, drugs such as HU, demethylating agents such as 5-azacitidine, and short-chain fatty acids have been studied to reactivate fetal hemoglobin [3, 5]. In clinical practice, HU promotes HbF production in intermedia β-thal. In addition, previous research has shown that the HU's beneficial effects on HbF production diminish over time due to its possible side effects [6]. Modulation of soluble guanylate cyclase, cyclic adenosine monophosphate (cAMP), and cyclic guanosine monophosphate (cGMP)-dependent protein kinase (PKG), among other signaling pathways, are associated with HU-regulated γ-globin production [7,8,9].
cGMP suppresses the activity of cAMP-specific phosphodiesterase 3 (PDE3), which occurs when there is an increase in cAMP levels inside cells. This leads to the activation of PKA, which acts as a negative control for the expression of the γ-globin gene [7]. In K562 cells, the p38 mitogen-activated protein kinase (MAPK) pathway and ERK/MAPK pathway function as activating and inhibitory mechanisms, respectively, for inducing γ-globin gene expression through histone deacetylase inhibitors like butyrate [7, 8]. Hyroxyurea (HU) inhibits erythroid growth by inducing c-Jun expression in transcriptional and post-transcriptional stages [8]. HU has cytotoxic effects on the bone marrow and reduces the absolute number of neutrophils, reticulocytes, and platelets [5]. Success in treating beta-thalassemia with HU has different consequences in different populations. Still, it is usually associated with reducing the need for blood transfusions and preventing side effects such as viral infections and iron deposition in tissues [6, 10]. However, the specific process by which HU triggers an increase in HbF levels is still unclear.
Systems biology has added a fresh perspective to pathophysiology by examining the levels of gene expression related to particular diseases [11]. Genes with different expressions (DEG) and their expression levels and regulatory mechanisms can be identified through transcriptome analysis [12]. This research utilized the weighted gene co-expression network analysis (WGCNA) algorithm to identify and assess the levels of gene expression in cells treated with HU. The likely path of HbF expression change was explored using bioinformatics studies and online databases, and essential genes involved in this process were found. Finally, changes in the target genes expression have been studied to investigate the action of HU on erythroid cells.
Materials and methods
Transcriptome analysis of β-thal RNA-seq dataset
This study was approved by the ethics review board of Birjand University of Medical Sciences and also followed the ethical guidelines of the Declaration of Helsinki (IR.BUMS.REC.1400.022). For the in silico study, we used the GSE90878 RNA-seq gene expression dataset from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE90878). This dataset contains raw data from 12 fetal liver erythroblast and 12 bone marrow erythroblast. First, differential gene expression (DEG) analysis was conducted and are considered as DEGs; they have the following criteria: FDR < 0.001 and |log2FC|≥ 2.89. Next, the gene co-expression network of fetal liver erythroblast and adult bone marrow erythroblast (control group) was reconstructed using the WGCNA algorithm. The expression profiles for each module were condensed into eigenvectors that were then compared to the first principal component of the overall expression matrix. This was done using the module eigengene (ME) technique, which helped identify modules that displayed strong correlations with certain clinical features. Individual genes' associations with the β-thal were calculated using gene significance (GS) values. In this study, the membership of each module was determined using both the ME correlation profile and gene expression. The genes that showed higher levels of GS and MM compared to the control sample were considered hub genes.
Cell culture and viability determination
The K562 cell line, a type of CML (chronic myelogenous leukemia) human cells, was grown in RPMI-1640 medium (Gibco-Invitrogen, USA), containing 1% penicillin/streptomycin and 10% heat-inactivated fetal calf serum (provided by Gibco-Invitrogen, USA), at a temperature of 37°C with 5% CO2. The cells were passaged every three days. Cell viability and growth kinetics were evaluated using a hemocytometer and a trypan blue dye exclusion method. An equivalent volume of cell suspension with 4% trypan blue solution were mixed, and the cells were counted using a hemacytometer under a light microscope. The cells were passaged twice a week and then were divided into control (untreated) and HU-treated groups. The cells were cultured at 50, 100, and 150 μM HU concentrations for 12, 24, and 48 h at 37 °C with 5% CO2 in three replications.
MTT assay
MTT assay was performed to evaluate the cell viability after treating HU at different concentrations. K562 cells were seeded in a 96-well plate at a density of 10 × 103 cells/well in 50, 100, and 150 μM HU for 12, 24, and 48 h in five replications and incubated at 37°C with 95 percent humidity and 5% CO2. Next, 20 μl of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) solution (5 mg/ml) was introduced into each well and left to incubate at 37°C for four h. After removing the supernatant, 100 μl of DMSO was added to dissolve the formazan crystals. Using a microplate reader, the optical density at wavelengths of 570–630 nm was measured for each well (Bio-Rad Laboratories, Inc.).
RNA extraction and cDNA synthesis
After cell harvesting, total genomic RNA was extracted using the Gene All kit (Hybrid R-South Korea) according to the manufacturer's procedure. The isolated RNA's optical density (OD 260/280) was measured using spectrophotometry to determine its quantity (BioTek, Epoch-USA). The extracted RNA was stored in a – 80 °C freezer. Complementary DNA (cDNA) was synthesized according to a commercial kit (Pars Tous manufacturer, Iran). The thermal cycle conditions (Eppendorf Germany) were as follows: 10 min at 25 °C, 60 min at 47 °C, 5 min at 85 °C, and holding at 4 °C. To perform qPCR, the generated cDNA was stored at 20 °C.
Quantitative RT-PCR (Q-RT-PCR)
Table 1 displays the primer sequences utilized for measuring the expression of γ-globin, BCL2, SP1, IGF2BP3, LIN28A, and COL4A5 genes. The qRT-PCR technique was used to evaluate the levels of gene expression. All samples were performed in triplicate. qRT-PCR was carried out in a reaction volume of 20 μl containing: 10 μl of Master Mix SYBR Green (Amplicon A325402 2X qRT-PCR Master Mix Green high Rox), 7μl DEPC, one μl (10 pm concentration) each of forward and reverse gene-specific primers (Table 1), and one μL of cDNA sample. Thermal cycling conditions for 40 cycles are presented in Table 2. The ABI StepOne software was used to analyze the cycle threshold (Ct), amplification plot, and melting curve of each product. The effectiveness of each gene primer was assessed by utilizing the standard curve from a series of diluted cDNAs that were synthesized. We used the GAPDH primers for normalizing the data in a real-time instrument (Applied Biosystem, StepOne, USA). The 2−ΔΔCt technique was used to calculate the relative fold change in gene expression in the treatment groups compared with the untreated control group.
Statistical analysis
Statistical analysis was conducted in GraphPad Prism 8.4.3.686 to examine alterations in gene expression, using a one-way ANOVA test and Tukey’s test for the mean ± standard deviation data (p < 0.05).
Results
Transcriptome analysis using WGCNA
In 93 published studies, there was an evaluation of DEG differences between fetal liver and bone marrow erythroblasts, with 65 genes showing increased activity and 28 genes showing decreased activity. These DEGs were subsequently used to identify hub genes. According to the range of expression values, WGCNA included a total of 4000 genes. The findings from WGCNA showed that the blue module (r = 0.99, p = 1.00 e-200) had the strongest positive correlation with erythroblast evaluation. Genes that had the highest MM and GS scores in the blue module were subsequently compared to the DEG list. Any genes that displayed similarities were identified as ultimate hub genes. Finally, the LIN28A, COL4A5, SP1, BCL2, and IGF2BP3 genes were selected as the hub genes in the γ-globin gene switching process.
Experimental validation of Silico analysis
MTT assay was conducted in order to determine the survivability of cells treated with HU at 50, 100, and 150 μM. After 12 h, there was no notable difference in cell viability between the control group and those treated with 50 or 100 μM of HU (p > 0.05). However, there was a significant decrease in viability at the concentration of 150 μM (p < 0.05). Evaluation of the viability of K562 cells in the different concentrations of HU after 24 and 48 h showed a significant change compared to the control group (p < 0.05) (Fig. 1).
Genes expression levels
To assess the impact of HU on the reactivation of fetal hemoglobin, we examined the levels of SP1, LIN28A, IGF2BP3, COL4A5, BCL2, and γ-globin genes in K562 cells treated with varying concentrations of HU. The expression of these genes was measured at three concentrations of 50, 100, and 150 μM HU at three time points of 12, 24, and 48 h. SP1 gene expression levels at 50, 100, and 150 μM HU after 12, 24, and 48 h were significantly decreased compared to the control group (p < 0.05) (Fig. 2) (reduce by half).
LIN28A gene expression pattern at 50, 100, and 150 μM HU after 12 h revealed a threefold increase compared to the control group (p < 0.05) (Fig. 3). There were no significant differences at 50 and 100 μM HU concentrations after 24 and 48 h compared to the control group (p > 0.05). But at 150 μM, it increased significantly compared to the control group (Fig. 3).
The results did not show a significant difference in the expression pattern of the IGF2BP3 gene at 50, 100, and 150 μM HU concentrations after 12 h, also at 50 and 100 μM HU after 24 and 48 h compared to the control group. In contrast, the expression level of the IGF2BP3 significantly increased at 150 μM HU concentration after 24 and 48 h (p < 0.05) (Fig. 4).
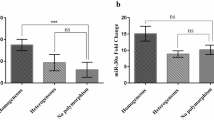
There was no significant difference in COL4A5 gene expression pattern at 50 μM HU concentration after 12 h compared to the control group (p > 0.05), while at 100 and 150 μM HU increased significantly (p < 0.05). In the expression pattern at 50, 100, and 150 μM HU concentrations after 24 and 48 h, a significant increase was observed compared to the control group (p < 0.05) (Fig. 5).
Evaluation of COL4A5 gene expression in K562 cells treated with concentrations of 50, 100, and 150 μM HU at three times of 12–24-48 h. The data are presented as a relative expression with the internal control gene GAPDH and repeated three times (ns = non-significant, p = 0.004**, p = 0.0002***, p < 0.0001****)
BCL2 gene expression pattern at all concentrations decreased significantly after 12 h (p < 0.05). There was no significant expression at 50 μM HU and significantly increased expression at concentrations of 100 and 150 μM after 24 h. It showed a decrease at 50 and 100 μM HU after 48 h (p < 0.05) (Fig. 6).
Evaluation of BCL2 gene expression in K562 cells treated with concentrations of 50, 100, and 150 μM HU at three times of 12, 24, and 48 h. The data are presented as a relative expression with the internal control gene GAPDH and repeated three times (ns = non-significant, p < 0.0001****, p = 0.0004***, p = 0.02*, p = 0.004**)
Evaluation of γ-globin gene expression showed no significant difference at 50, 100, and 150 μM HU after 12 h compared to the control group (p > 0.05). But, the expression pattern at 50, 100, and 150 μM HU concentrations showed a 15-fold increase after 24 and 48 h compared to the control group (p < 0.05) (Fig. 7).
Evaluation of γ-globin gene expression in K562 cells treated with concentrations of 50, 100, and 150 μM HU at three times of 12, 24, and 48 h. The data are presented as a relative expression with the internal control gene GAPDH and repeated three times (ns = non-significant, p < 0.0001****, p = 0.0009***, p = 0.02*, p = 0.008**)
Discussion
Thalassemia is a prevalent genetic disorder worldwide that results from abnormal production of α/β globin due to mutations during mRNA translation [1]. The unpaired globin is seen in erythrocytes as cytoplasmic inclusion bodies and causes the shortening the erythrocytes' lifespan and occurrence of chronic hemolytic anemia. In intermediate β-thal and some hemoglobinopathies, elevation of HbF levels in adult erythroblasts is an important therapeutic target [1, 13, 14]. The mechanism by which HU generates her HbF is unknown. The discovery of associations between genes affected by HU and those involved in HbF switching may help explain some of these mechanisms.
In this study, using transcriptome analysis by WGCNA method, six genes were selected and the expression of these genes (IGF2Bp3, LIN28A, COL4A5, BCL2, and SP1) under HU treatment and their relationship with γ-globin switching were evaluated.
According to the findings, the level of expression for the LIN28A gene was elevated following HU treatment. The discovery of this gene initially occurred in the Caenorhabditis elegans nematode, and it is responsible for regulating the timing and progression of developmental processes [15]. In humans, two similar versions of the LIN28 gene, known as LIN28A and LIN28B, have been identified. These genes are involved in promoting pluripotency in stem cells and the differentiation of skeletal muscle cells. They also contribute to various physical developments such as changes in height, puberty timing, and age at menopause [15]. The LIN28 gene alters the let-7 family of miRNAs, and its expression plays a role in blocking let-7 [15]. In mature red blood cells, LIN28 protein has been found to reduce let-7 miRNA levels and potentially increase HbF expression [16]. Lee et al. confirmed that a higher level of LIN28B expression in mature erythroid cells can lead to down-regulation of let-7 expression and a significant increase in HbF expression level. Furthermore, they found that increased expression of the HbF in mature CD34 + cells coincided with a decrease of mature globin expression [17]. These findings align with Lee et al.’s results that LIN28A, homologous to LIN28B, also increases its expression under the influence of HU.
The results showed that SP1 gene expression decreased with the time series and dose in HU-treated cells. In other studies, they have been shown that the effect of some microRNAs, such as miR-23a and miR-27a, on SP1 mRNA, inhibits γ-to-β switching [18, 19]. Moreover, HU inhibits the ERK pathway by activating the MAPK pathway, inhibiting SP1 and fetal hemoglobin increases [20]. The ERK pathways are highly regulated cascades responsible for fundamental cellular processes including cell proliferation and differentiation [21]. The normal maturation of erythrocytes is linked to heightened levels of transcriptional activators GATA-1 and SP1 [22]. GATA-1 plays a significant role in promoting the development of red blood cells by regulating various genes specific to this process, such as globin. Additionally, research has demonstrated that GATA-1 and SP1 collaborate at the γ-globin promoter [22]. Mnika et al. showed that SP1 is a main target of miR-23a, which inhibits switching γ-globin to β-globin. KLF3, a negative regulator of erythrocytes, is also inhibited explicitly by miR-27a [23]. The results of this study are in line with the study of Mnika et al. which showed that HU prevents gamma-to-beta switching by inhibiting SP1.
The results demonstrated that IGF2BP3 gene expression increased after HU treatment. The IGF2BP3 gene is highly expressed in the early stages of development of most tissues and declines after birth. This protein induces erythroblast differentiation and maturation. Also, IGF2BP3 plays an important role in the differentiation process at the early stages of embryogenesis, by binding to three IGF2 leader mRNA and blocking its expression [15]. Jaira et al. showed that induction of IGF2BP1 expression increased γ-globin gene. IGF2BP3 is evolutionarily regulated in human erythroid tissues by being silenced during fetal–adult transition. However, the effect of IGF2BP3 on HbF levels was lower than that of IGF2BP1 in cultured adult erythroblasts [24]. The results of this study are consistent with Jaira et al.’s study that the expression of IGF2BP3, like IGF2BP1, increases γ-globin gene expression [24].
The results demonstrated that COL4A5 gene expression was increased in K562 cells exposed to HU. Hydroxyurea is known to induce the expression of γ-globin by miRNA-mediated silencing of BCL11A, a gene involved in switching from γ to β globin gene expression during erythropoiesis in adulthood. The gene expression modulating effects of HU can be mediated through three main mechanistic actions, including epigenetic modifications, post-transcriptional effects, and signaling mediators, including MAPK, PI3K/Akt, and MYB [25]. In this study, we demonstrated that HU treatment can be associated with the up-regulation of COL4A5 along with the induction of γ-globin expression. It is currently unknown whether a similar HU-mediated regulatory pathway controls the expression of COLA45 and γ-globin, warranting more research in this area. It has been noted that COL4A5 can activate phosphatidylinositol 3-kinase PI3K/protein kinase B (AKT) signaling pathway [26], contributing to the expression of γ-globin [27]. There is a need for further evaluations to divulge other possible roles of COL4A5 in HU-mediated cellular and molecular functions.
The results demonstrate that BCL2 gene expression decreased significantly after 12 h, but increased again at 24 and 48 h. BCL2 proto-oncogene inhibits the apoptosis pathway. HU is a compound that acts on ribonucleoside diphosphate reductase and inhibits DNA synthesis. Caspase-3 activation is an important factor in the activation of the apoptosis cascade. HU increased caspase-3 activity 20 to 30 times higher than the untreated cells. Therefore, there is a correlation with this reagent-induced apoptosis [28]. In a study, it was reported that HU induced apoptotic death of K562 cells through BCL2 protein down-regulation and Bax protein up-regulation and degraded the PARP protein in a p53-independent pathway [29]. The results are consistent with other studies showing that HU induces apoptosis through down-regulation of the anti-apoptotic protein BCL2, inhibiting apoptosis and keeping cells alive.
The findings demonstrated that HU notably increased the expression of the γ-globin gene after 24 and 48 h. Pule et al. found that when HU down-regulated BCL11A, MYB, and KLF-1, it resulted in an increase in γ-globin expression. Their study revealed a connection between the expression patterns of miR-15a, miR16-1, miR-210, miR-29a, miR-96, and the genes MYB, BCL11A, and KLF-1 in response to HU treatment. They proposed a direct interaction between miR-26b and the MYB 3′-UTR as a possible explanation for the up-regulation of γ-globin through the MYB/KLF-1/BCL11A pathway [30]. Consequently, blocking the c-MYB pathway results in an increase of HbF production. According to Barbarani and colleagues, a heightened HbF expression in cultured red blood cells was linked to decreased c-MYB levels [31]. Although these results may help clarify the molecular mechanism of the γ-globin gene switching process, further in vivo and clinical studies are still needed to find alternative therapies for β-thal patients.
Conclusion
The results showed that changes in the expression of LIN28A, COL4A5, SP1, and IGF2BP3 genes were associated with an increase in the γ-gene in HU-treated cells. By elucidating the mechanism of more precise switching from the γ- gene to β-gene, we can hope for new, less complicated treatments for thalassemia patients.
Availability of data and materials
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
References
Modell B, Darlison M (2008) Global epidemiology of haemoglobin disorders and derived service indicators. Bull World Health Organ 86:480–487. https://doi.org/10.2471/blt.06.036673
Aldemir Ö (2018) The genetic aspect of thalassemia: from diagnosis to treatment. In: Thalassemia and other hemolytic anemias, pp 11, BoD–Books on Demand
Fard AD et al (2013) Evaluation of novel fetal hemoglobin inducer drugs in treatment of β-hemoglobinopathy disorders. Int J Hematol Oncol Stem Cell Res 7:47
Cokic VP et al (2003) Hydroxyurea induces fetal hemoglobin by the nitric oxide–dependent activation of soluble guanylyl cyclase. J Clin Investig 111:231–239
Tayebi B et al (2017) Modulation of microRNAs expression in hematopoietic stem cells treated with sodium butyrate in inducing fetal hemoglobin expression. Artif Cells Nanomed Biotechnol 45:146–156
Musiałek MW, Rybaczek D (2021) Hydroxyurea—the good, the bad and the ugly. Genes 12:1096
Inoue A et al (2004) Negative regulation of γ-globin gene expression by cyclic AMP-dependent pathway in erythroid cells. Exp Hematol 32:244–253
Tang DC et al (2005) The hydroxyurea-induced small GTP-binding protein SAR modulates γ-globin gene expression in human erythroid cells. Blood 106:3256–3263
Ikuta T et al (2001) Mechanism for fetal globin gene expression: role of the soluble guanylate cyclase–cGMP-dependent protein kinase pathway. Proc Natl Acad Sci 98:1847–1852
Timson J (1975) Hydroxyurea. Mutation Res Rev Genet Toxicol 32:115–131
Derakhshani A et al (2020) Gene co-expression network analysis for identifying modules and functionally enriched pathways in vitiligo disease: a systems biology study. Iran J Allergy Asthma Immunol 19:517–528
Cui Y, Paules RS (2010) Use of transcriptomics in understanding mechanisms of drug-induced toxicity. Pharmacogenomics 11:573–585. https://doi.org/10.2217/pgs.10.37
Li YY et al (2016) gamma-globin inductive therapy of beta-thalassemia and its relationship with microrna. Zhongguo Shi Yan Xue Ye Xue Za Zhi 24:627–631. https://doi.org/10.7534/j.issn.1009-2137.2016.02.060
Zhu X et al (2017) Hydroxyurea differentially modulates activator and repressors of gamma-globin gene in erythroblasts of responsive and non-responsive patients with sickle cell disease in correlation with Index of Hydroxyurea Responsiveness. Haematologica 102:1995–2004. https://doi.org/10.3324/haematol.2017.175646
de Vasconcellos JF et al (2017) IGF2BP1 overexpression causes fetal-like hemoglobin expression patterns in cultured human adult erythroblasts. Proc Natl Acad Sci U S A 114:E5664-e5672. https://doi.org/10.1073/pnas.1609552114
de Vasconcellos JF et al (2014) LIN28A expression reduces sickling of cultured human erythrocytes. PLoS ONE 9:e106924
Lee YT et al (2013) LIN28B-mediated expression of fetal hemoglobin and production of fetal-like erythrocytes from adult human erythroblasts ex vivo. Blood 122:1034–1041
Ismail NA et al (2019) The relation between serum hepcidin, ferritin, hepcidin: ferritin ratio, hydroxyurea and splenectomy in children with β-thalassemia. Open Access Macedonian J Med Sci 7:2434
Finotti A et al (2015) Recent trends in the gene therapy of β-thalassemia. J Blood Med 6:69
Shah F, Dwivedi M (2020) Pathophysiology and recent therapeutic insights of sickle cell disease. Ann Hematol 99:925–935
Guo YJ et al (2020) ERK/MAPK signalling pathway and tumorigenesis. Exp Ther Med 19:1997–2007
Bacon ER et al (1995) Hemoglobin switching in humans is accompanied by changes in the ratio of the transcription factors, GATA-1 and SP1. Mol Med 1:297–305. https://doi.org/10.1007/BF03401554
Mnika K et al (2019) Hydroxyurea-induced miRNA expression in sickle cell disease patients in Africa. Front Genet 10:509
de Vasconcellos JF et al (2016) IGF2BP3 moderately increases fetal hemoglobin levels in human adult erythroblasts. Blood 128:2464. https://doi.org/10.1182/blood.V128.22.2464.2464
Pule GD et al (2016) Hydroxyurea down-regulates BCL11A, KLF-1 and MYB through miRNA-mediated actions to induce γ-globin expression: implications for new therapeutic approaches of sickle cell disease. Clin Transl Med 5:15. https://doi.org/10.1186/s40169-016-0092-7
Zhao N et al (2022) MicroRNA-221-5p Promotes Ricin Toxin-induced Inflammation via PI3K/Akt signaling pathway by targeting COL4a5. Toxicon 212:11–18. https://doi.org/10.1016/j.toxicon.2022.03.010
Xie Y et al (2019) PI3K/Akt signaling transduction pathway, erythropoiesis and glycolysis in hypoxia (Review). Mol Med Rep 19:783–791. https://doi.org/10.3892/mmr.2018.9713
Olmos G et al (2005) Involvement of Bax, Bcl-2 and caspase 3 in hydroxyurea-or etoposide-induced apoptosis of mouse interleukin-3-dependent lymphoma cells. Anticancer Res 25:999–1007
Lee YJ et al (2001) The expression of hydroxyurea induced apoptosis associated proteins on chronic myelogenous leukemic K562 Cells. Korean J Clin Pathol 21:98–103
Pule GD et al (2016) Hydroxyurea down-regulates BCL11A, KLF-1 and MYB through miRNA-mediated actions to induce γ-globin expression: implications for new therapeutic approaches of sickle cell disease. Clin Transl Med 5:1–15
Barbarani G et al (2021) Physiological and aberrant γ-globin transcription during development. Front Cell Dev Biol 9:563
Acknowledgements
The Research Deputy of Birjand University of Medical Sciences Grant, Birjand, Iran, supported this work. The authors would like to thank Mohammad Eini for his helpful assistance and all the staff of the University Research Laboratory.
Funding
No funding was received for this research.
Author information
Authors and Affiliations
Contributions
E.A., H.S., and E.M.M. designed the experiments. A.A.F and E.A. performed the experiments. A.A.F. and H.S. carried out the data analysis. A.A.F, E.A., and H.S. wrote the manuscript. E.M.M. revised the manuscript. All authors reviewed, considered, and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the ethical review board of Birjand University of Medical Sciences and followed the ethical guidelines of the Declaration of Helsinki (IR.BUMS.REC.1400.022).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Fashami, A.AA., Alemzadeh, E., Safarpour, H. et al. Effect of hydroxyurea on SP1, LIN28B, IGF2BP3, COL4A5, BCL2, gamma globin genes expression: an in vitro study. Egypt J Med Hum Genet 24, 72 (2023). https://doi.org/10.1186/s43042-023-00452-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43042-023-00452-8