Abstract
Background
Flavin monoamine oxidase gene encodes a protein (MAOB) that forms a part of the flavin monoamine oxidase family in the outer membrane of mitochondria. It plays a role in the tissue metabolism of neuroactive and vasoactive amines as well as the oxidative deamination of xenobiotic and biogenic amines. However, overexpression of the receptor reduced apoptosis in cells, resulting in the progress of prostate sarcoma. Therefore, various kinds of MAOB antagonists are often used to fix an apoptosis mechanism that makes it hard to get rid of cancer from live tissues. Moreover, chemical compounds that have been discovered to be MAOB inhibitors to date exhibit side effects that are causing problems in chemotherapy treatment. The study aims to discover new purchasable compound that induces apoptosis by allowing caspases to operate at their maximum efficiency and is low toxic.
Methods
With the assistance of virtual screening, molecular docking, and molecular dynamics simulation (MD), a structure-based pharmacophore model of the protein active site cavity was made. Twenty hits were found, and then a molecular docking strategy was used to choose four molecules to study in more depth. MD simulations were used to check the stability of the four compounds, and they were all shown to be stable when bound to the target protein.
Results
Four newly discovered compounds, included with ZINC ID Such as ZINC12143050, ZINC08301324, ZINC16743012, and ZINC64165826 with binding scores of − 11.7, − 11.4, − 11.2 and − 11.1 kcal/mol, respectively, may serve as lead compounds for the treatment of prostate cancer associated with MAOB; however, further evaluation through wet lab is needed to determine the compounds effectiveness.
Conclusion
A structure-based model was initially developed, followed by molecular docking, ADMET analysis, and MD simulation. The top four natural compounds identified in the A-to-Z virtual screening process could serve as lead molecules in the fight against prostate cancer.
Similar content being viewed by others
Background
Prostate cancer has been common chorionic disease, especially in male in the world especially in the developed countries. After lung cancer, it is the second-greatest cause of cancer mortality in the United States [1]. However, prostate cancer killed approximately one man among the forty-one in the world. The incidence of prostate cancer is rising rapidly, although the mechanism of origination and progress of prostate cancer is still not fully understand [2]. Therefore, the investigation of prostate cancer related gene may be provide better therapeutic solution for the treatment. Some of the things used to stop the growth of cancer cells are curcumin (CUR), ursolic acid (UA), epigallocatechin-3-gallate (EGCG), resveratrol (RES), sulforaphane (6-SHO), and 6-shogaol [3].
The enzyme monoamine oxidase B that is encoded by the MAOB gene in the humans. It belongs to flavin monoamine oxidase family and located in the outer mitochondrial membrane [4]. Monoamine oxidase B has a hydrophobic bipartite elongated cavity that occupies a combined volume approximately to 700 Å3. The first cavity of hMAO has designated the entrance cavity (290 Å3) where the second substrate cavity or active site cavity was (~ 390 Å3) [5]. The inhibitor specificity of hMAO B has shown open or closed form that depending on the substrate or bound inhibitor. The active site has adjusted along with FAD coenzyme for favorable amine binding about the flavin involving two nearly parallel tyrosyl (398 and 435) residues that form of aromatic cage [6]. Monoamine oxidase B sn b pacifically has degraded benzylamine and phenethylamine, whereas monoamine oxidase A (MAOA) degrades dopamine in the favorable environment. However, dopamine and tyramine were the substrates for MAO-B. In the Parkinson’s disease, selegiline (L-deprenyl) was the first compound for treating diseases in the last 40 years [7]. The functional MAO-B polymorphism contains in a single-base variation (A or G) in the intron. Moreover, MAOB enzyme was responsible for accelerating in cancer cell into the body. The upregulation and down regulation of MAOB were found in gliomas and nut-associated cancer. MAO-B is not only participate for storing regulation, and concentrations of biogenic amines in the synaptic cleft but also it generated reactive oxygen species (ROS) from the monoamine substrate oxidation. The toxicity synthesis may occur form the excess amount of oxidation into the body [8]. MAOB expression patterns in three pairs of fibroblasts from the same patient showed that MAOB expression was higher in the stroma of clinical samples with primary PC than in normal prostatic tissues. Higher levels of MAOB in the stroma were linked to higher Gleason scores, resistance to castration, survival, and the expression of CHGA in the surrounding epithelia, which shows metabolic differentiation [9]. Proliferation, migration, and invasion of cancer cells were all accelerated when MAOB-overexpressing PrSC cells were co-cultured with various human PC cell lines. In contrast, these cancer cell phenotypes were inhibited when MAOB-knockdown PrSC cells were present. Additionally, C4-2 tumor xenografts grew much smaller in mice when co-inoculated with MAOB-knockdown PrSC. In comparison with controls, there was a decrease in the Ki-67 index and the expression of the reactive stromal marker SMA in the tumor and stromal compartments, respectively [10]. In mice, the major enzymes in monoamine metabolism, MOAB, have an effect on the prostate and prostate basal progenitor cells. Controlling the function of prostatic progenitor cells and maintaining the prostate gland are two new areas of responsibility for MAOB [11].
To identify biologically active compounds, new hits or leads can be defined using theoretical and computational drug design methods. Computer-aided drug design (CADD) tools, such as pharmacophore modeling, virtual screening, molecular docking, and dynamic simulation, are used a lot in drug discovery, development, and analysis to narrow down the list of possible candidates to the most promising compounds [12]. The structure-based pharmacophore model used to detect similar active molecules against to specific target protein. In-silico molecular docking technique was used to observe the large scale compound for binding affinity. Docking analysis shows the molecular docking-based scoring function and interaction [13]. However, it is easy and time-consuming process and reasonable to predict compound against specific protein. The absorption, distribution, metabolism, and excretion (ADME) was screening to measure the toxicity and efficacy of compounds. The drug's stability against the target protein was verified using molecular dynamic modeling. When it comes to predicting a compound's effectiveness and toxicity, computational approaches have made great strides, with MD simulation confirming the stability of a therapeutic candidate for the targeted protein [14]. However, it has been difficult to address the need for new drugs, so there is a need to progress strong investigation for the invention of bioactive compounds by targeting novel protein classes [15]. To find a natural antagonist against the 1S3B protein for cancer treatment, study has turned to structure-based drug discovery strategies such as pharmacophore modeling, virtual screening, ADMET, molecular docking, and dynamic simulation. The newly discovered compounds may have the ability to inhibit these endogenous inhibitors, allowing caspases to function more effectively. By blocking caspase inhibitors, the compounds may shift the balance in favor of apoptosis induction.
Methods
Structure-based pharmacophore modeling and virtual screening
Structure-based pharmacophore modeling
The methods has been utilized for a developing of pharmacophore based structural features against to the target protein. The possible active sites generation and co-crystallized ligand will be estimated in the process. The active antagonists of Monoamine oxidase B (MAOB) protein were identified from the any target annotations that are present by using ChEMBL database and highly related to broad works [16]. However, twenty active antagonists were identified for generating a structure-based pharmacophore model (Additional file 1: Table S1). The similar compounds were observed from the ChEMBL (https://www.ebi.ac.uk/chembl/) website and literature search. PyRx AutoDock Vina software were utilized for docking and selected compound based on the scoring function [17]. The high-scored compound were selected from the maximum binding affinity (kcal/mol) for generating model. The natural compounds retrieval hits were considered for the observing of interaction between top scoring compounds and MAOB protein [18]. The molecular design software LigandScout 4.4 Essential was used to prepare a structure-based pharmacophore model. LigandScout 4.4 software was used to document the ways in which inhibitors interacted with essential amino acids at protein-active sites. Hydrogen bond donor, charge transfer, hydrophilic and hydrophobic areas, and hydrogen bonds were found in the pharmacophore, distinct from the ligand-receptor complex. On the other hand, sequential algorithms can be used to figure out the number of aromatic rings, the state of hybridization, the binding pattern, and the distance between the receptor molecule and the binding site. Therefore, hydrophilic properties were not only deleted from the protein (MAOB) but also were excluded or included features to the active site through the LigandScout 4.4 essential software [19].
Pharmacophore model validation
Potential features from active and inactive compounds that can be obtained with specific protein–ligand interaction were identified during the pharmacophore validation process. The popular web-based database DUD-E decoys was used for protein–ligand complex validation and thereby it distinguishes active compounds and performance through the screening of a set of 20 known actives and their 7132 correspondent decoy compounds [20]. The twenty antagonists selected by using ChEMBL (https: //www.ebi. ac.uk/chembl/) database that was remarked as active against MAOB protein. Compounds have chosen for further experiment after experimental validation. The ligandScout 4.4 was used for converting DUD-E to.idb format before screening and opened “create screening database” menu and assessed the Güner-Henry (GH) score and early enrichment factors (EF).
Dataset generation for pharmacophore-base screening
Active and new compounds were found by completing a virtual screening that relied heavily on the pharmacophore model. The chemical database ZINC (https://zinc.docking.org/) was used to discover possible lead compounds [21]. The compound was retrieved from the database by entering either its structure, its name, or its chemical ID. The physical and chemical properties were looked at. This included figuring out the 2D and 3D structures, figuring out the melting and boiling points, and writing down this information. The desired compound identified through the securing of molecular weight, crystal structure, and biological information. The priority has given the most similar features compounds which can easily interact with the target protein. The query of the highest matches compounds have selected by the counting of possible hit. Consequently, ZINC natural product library namely ZINCPharmer (http://zincpharmer.csb.pitt.edu/pharmer.html) server has been utilized for screening initially target MAOB.
Pharmacophore‑based virtual screening
The validated structure-based pharmacophore feature used to create database through the ZINCPharmer. However, the 3D model of protein–ligand interaction was documented by Ligand Scout 4.4 Essential and file format change to specific (idb) file. All of compounds have taken from the database for generating pharmacophore feature based on the virtual screening [22]. The selected relative pharmacophore feature used to get few function by omitting some features more than two feature. The pharmacophore feature score utilized to fitted hit compound that was subject to further validation.
Molecular docking based virtual screening
Protein and ligand preparation
In-silico drug design process was used to convert molecular structure for more suitable in the perspective of protein preparation. The protein crystal structure was constructed prior to the docking mechanism with additional and improved hydrogen bonds, the elimination of atomic collisions, and other processes not included in the X-ray crystal structure refining process. The 3D structure of the MOAB protein was determined experimentally and confirmed using the X-ray diffraction technique with a resolution of 1.65, an R-value free score of 0.223, and a standard value of 0.204. It was then taken from the protein data bank (PDB ID: 1S3B) [16]. To construct the desired X-ray crystallography protein, water, metal ions, and cofactors were removed. Polar hydrogen bonds and non-polar hydrogen bonds were then inserted, and the Gasteiger change was computed using AutoDockTools (ADT). The retrieved selected hit compounds from Ligand Scout 4.4 Essential and minimized and optimized energy along with bond angle by selecting default of the Universal Force Field (UFF) for each ligand [23].
Active site identification and grid generation
The identification of active sites from the specific protein is a key strategy to treat a particular disease. On the other hand, improper binding or attachment of the protein–ligand interaction may have the side effect into the body or have the higher possibility of toxicities. The chemical's H bonds, hydrophobic or hydrophilic interactions, ionization, and zinc compound chelation all affect the chemical's binding affinities [24]. The BIOVA Discovery Studio Visualizer Tool 16.1.0 pinpoints the binding sites or active pocket of the target protein. Moreover, the PrankWeb server (https://prankweb.cz/) was used to confirm a putative protein binding location. The server was able to anticipate ligand binding sites from a given protein structure using a machine learning-based approach. When the protein was chosen, a receptor grid was generated using the PyRx program.
Molecular docking
The PyRx virtual screening program was used to perform molecular docking on the compounds that were chosen from the virtual screening. The well-known program PyRx was chosen for the virtual scoring of several medication designs against various ailments. For molecular docking, it included the Lamarckian Genetic Algorithm (LGA) as a scoring function with AutoDock and AutoDock Vina [25]. In this investigation, the interaction between ligands and proteins was facilitated using the PyRx tool AutoDock Vina. Following that, the complicated binding postures were evaluated using the BIOVA Discovery Studio Visualization Tools [26].
ADME analysis
ADME properties is an important criteria to develop any effective and successful drug candidate. In the early stage of drug design process, without analysis of ADME no one can think about the drug design because of the negative effect of compound. It can predict or reduce the failure rate in the clinical trial and save money and time [27]. The drug's elimination in urine and feces has a direct effect on the ADME profile in people [28]. Hydrophobicity, lipophilicity, the GI tract, and the blood–brain barrier were all directly affected by the ejection process. The Swiss-ADME server (http://www.swiss-adme.ch/) was used to test the chosen drugs' ADME parameters, such as their solubility profile, GIT absorption, and bioavailability profile. Moreover, 3D-QSAR (three-dimensional quantitative structure–activity relationship) methods are widely used in drug design to understand the relationship between the three-dimensional structure of a drug molecule and its biological activity. These methods involve computational techniques to model, analyze, and predict the activity of potential drug candidates. CoMFA is used 3D-QSAR technique that involves building a three-dimensional grid around a set of aligned drug molecules. The grid represents steric and electrostatic fields surrounding the molecules. Then, the activity data of the molecules is used to develop a quantitative relationship between the field values and the observed activity. CoMFA can be used to identify important regions in the molecule that contribute to the activity and guide modifications for improving the potency of the drug.
Toxicity test
The toxicity test is performed to determine the safety profile of the intended substances for human consumption. Alternatively, it may be detrimental to the production of small molecules as a therapeutic candidate. In silico toxicity tests give the idea of figuring out the range of mutagenicity, carcinogenicity, LD50 value, and immunotoxicity and analyzing it quantitatively and qualitatively. To test the hazardous impact of the selected chemicals, the free server ProTox-II (http://tox.charite.de/protox II) was utilized [29]. The Toxicity Estimation Software Tool (TEST) was selected to assess the toxicity of the study's substance of interest without the need of any additional software. Quantitative structure–activity relationship (QSAR) approaches have been used to make estimates for the selected compounds. Pathways on this site range from those involved in nuclear receptor signaling to those involved in the body's reaction to stress [30].
MD simulation
To determine the stable interactions of the ligands and binding pockets of the receptors, the Desmond v3.6 program in Schrödinger was used. The stability of selected compounds with target proteins was evaluated using 100-ns MD simulations. The automated simulation used free energy perturbation (FEP) computing to estimate the equation of state (EOS), which was then paired with a variety of temperatures [13]. The system was employing a predetermined TIP3P water model in which an orthorhombic periodic boundary box shape with a value of 10 was used to allocate a particular volume to both sides. This technique electronically neutralized stable ions such as Na + and Cl with a salt concentration of 0.15 M. After generating the solvated system containing the protein in combination with the ligand, the system may be optimized using the OPLS-2005 force field [31]. The molecular dynamic result was verified and assessed with the use of the simulation interaction diagram (SID) from the Schrodinger software. The stability of the complex was calculated using the root-mean-squared deviation (RMSD), root-mean-squared free energy (RMSF), protein–ligand (P-L) interactions, and hydrogen bond interactions, all of which were extracted from the trajectory.
Results
Structure-based pharmacophore modeling and virtual screening
Pharmacophore model generation
A pharmacophore is not only a set of common steric features, but also a set of electronic chemical features that show how a compound works at the active site of a specific biological macromolecule. Pharmacophore characteristics have been shown to be useful in observing ligand–protein interactions and in screening huge chemical databases for novel scaffolds that might serve as lead small molecules. The PDB entry 1s3b in association with chemicals was used to identify the lead drug against prostate cancer, and a pharmacophore model was then constructed. A small-molecule medication candidate based on the previously-identified antagonist need not be chemically synthesized; a compound library can be purchased instead. Twenty chemically synthesized active antagonists of the Monoamine oxidase B (MAOB) protein were identified using the well-known databases ChEMBL and an extensive literature search (Additional file 1: Table S1). PyRx performed molecular docking, and the antagonist CHEMBL3938629 had the highest binding score (PubChem CID: 56,961,657). The most significant binding was − 8.7 kcal/mol (Table 1). The overall schematic diagram for our research work has been provided by the flow chart (Fig. 1).
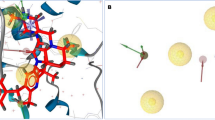
It is an essential to determine 3D structure of protein in the drug design and need to validate protein structure from the various protein data bank. The desire crystal X-ray structure of protein MAOB (PDB: 1S3B) was documented against antagonist and a structure-based pharmacophore model to the enzymatic cavity was generated. The ligands interaction with MAOB protein were determined experimentally and validated through X-ray diffraction method. The active site of MAOB protein have regulated by the binding of the inhibitor from the overall expression. The reliability of protein interaction with inhibitor ensured by the proper binding efficacy. Therefore, the active sites of inhibitor determined or examined to observe the sufficient interaction for getting more biological activity compared to the existing one. The key chemical features generated through the LigandScout 4.4 essential advance molecular design software based on pharmacophore model (Fig. 2).
A The X-ray crystal structure of MAOB protein (PDB ID: 1s3b) was used to generate a 3D structure-based pharmacophore model of MAOB protein in combination with 46,781,908 (CID) ligands. B A number of pharmacophore features were generated following complex interaction; these include four yellow spheres, representing hydrophobic interaction; a shaded pink star shape, representing the positive ignitable with tolerance 2; three red arrows and spherical shapes, representing H bond acceptors with tolerance 1.5; and five hydrogen bond donors, represented by a green spherical or arrow shape
Fourteen distinct chemical characteristics were identified. As an example of a protein–ligand complex interaction, we showed that there were 43 characteristics, of which 33 were hydrophobic, 2 were positively ionizable bonds, 23 were H bond acceptors, 8 were H bond donors, and 42 had an exclusion volume (Fig. 2). Certain aspects have been left out of the pharmacophore model development process to ensure that the pharmacophore's optimal features are preserved.
The pharmacophore characteristics exhibited were formed by the protein–ligand interaction. The hydrophobic interactions generated with the amino acid residues of the chosen protein are prominent. There are several hydrogen bond donor interacted with protein whereas nitrogen atom in the benzene ring interact with HOH622A, HOH68OA, THR426A, HOH609A, SER15A, ARG42A and HOH607A (Fig. 3).
The 2D structure of our chosen MAOB protein showed interactions with amino acid residues and hydrophobic contacts, which were shown in yellow color. The most common aspects of ligand–protein interactions, hydrogen bond donors (HBDs), were illustrated in red color, while HBAs' interactions with the oxygen and nitrogen atoms in the benzene ring and its various side chains were shown in blue color. The form and location of the binding pocket, which is maintained by hydrogen atoms and a constrained region, are not shown in the figure
Pharmacophore model validation
Validation is an important part for the authentication of pharmacophore analysis as well as the quality of molecular model. The structure-based pharmacophore model generated before database screening because of models are able to differences with active compounds from the decoy set. Twenty active known MAOB antagonists with correspondences 600 decoy compounds in supplementary file (Additional file 2: Table S2) validated by enhance Database of Useful Decoys (DUDe). The IC50 values were merged with the decoy compounds and initially compound screened was track to validated model. The AUC value and EF value were estimated through the receiver operating curve (ROC). It was expressed the performance of a classification model that can give an idea about degree of reparability whereas AUC was utilized to describe the summery of the model performance. The higher AUC value proved the better predictability and it ranges between 0 and 1. However, the model shows the 100% correct prediction regarding of AUC value. Moreover, the early enrichment factor (EF1%) was 100 with an excellent AUC that was proved, the model has ability to distinguish true actives from the decoy compounds (Fig. 4).
Dataset generation for pharmacophore-base screening
The identification of best lead molecules was an important part through the dataset generation during screening process. In the study, ZINC dataset utilized for collecting of commercially available chemical compounds. The information supplied included molecular weight, chemical structure, physical and chemical characteristics, and biologically active macromolecules. The ZINC data collection contains over 230 million chemical substances in 3D format that are publicly accessible via the website and ready to dock. Other compound information was also gathered from the ZINC data collection, such as Ambinter, which serves as a natural compound database library. The virtual screening database based on pharmacophores was recorded, and the pharmacophore model developed for each active chemical was uploaded to ZINCPharmer. Initially, the hits were calculated using the ZINC database of "ZINC natural products and ZINC natural derivatives," which contained millions of drug-like small molecules, natural products, and FDA-approved medications. The expected values for the RMSD sphere center fed into ZINCPharmer were 0.5Å, and a total of 11,000 compounds were returned for further screening. Finally, the hit was counted and save as well as downloaded for further screening.
Pharmacophore-based virtual screening
The protein–ligand complex structure has documented through the purchasable compounds library for the generation of pharmacophore feature. The relative pharmacophore has been used for scoring function during the screening process and omitted maximum four features for all query features. It is a quiet hard to query all features during the screening process, for this reasons, few features omitted to enhance the ability of pharmacophore fit score. The higher score can be sowed the better activity against the targeted macromolecules and fitted with the desire environment. As a result, the ROC curve displays the pharmacophore fit value for geometric feature fit to the 3D-structure-based pharmacophore model (Fig. 4). The protein with the highest match score to the verified pharmacophore model demonstrates action against our desired MAOB protein. As a result, the chemical was labeled a cornuted hit, recovered, and stored for future analysis.
Molecular docking based virtual screening
The desired MAOB protein has two ligands attached to it (PDB ID: 1S3B). The protein pockets have several points of attachment and a variety of shapes for ligand binding. From this particular protein, the binding location of the complex structure's active site has been obtained and determined. The active site (AS) of the protein is formed by combining several AA residues in a specific area that has the ability to form a transient connection with the substrate, known as the binding site. The protein's active site serves as the chemical substrate for the reaction that is being catalyzed. The protein's binding site can identify ligands and form a strong binding connection with the protein in order to stabilize intermediate reactions. The CASTpi server was used to identify the MAOB protein active sites, and the combined binding positions of the active sites were then obtained. The protein's estimated active pocket helps to recover the binding site residue. Total twelve active sites identified from the MAOB protein and further confirmed through the PrankWeb (https://prankweb.cz/) server. The active site pocket was analyzed and binding site position at ASN117, ASP114, MET 125, GLU142, TRP 143, CYS156, LEU151, LYS149, SER488, VAL489, PRO490 and ARG494 that have been depict by the different color such as red, deep green, green, yellow, deep pink, pink, orange, deep blue, mild blue, blue, black, and white, (Fig. 5). The server-identified binding sites were used to build a receptor grid during the molecular docking simulation process, with grid box dimensions X = 98.39, Y = 102.46, and Z = 69.21 in angstrom (Å).
Molecular docking
As a vital part of the drug discovery process, molecular docking is being used in this work to evaluate how well the hit compounds attach to the target MAOB protein. Using the assumptions of the pharmacophore model as a guide, the PyRx tools Autodock Vina were used to dock drugs onto MAOB and assess their binding affinity. Four compounds were identified to have higher binding affinities than the MAOB antagonist ZINC ID 8215434 (− 9.2 kcal/mol), which was employed in the primary pharmacophore model creation (ZINC ID 12143050, ZINC ID 08301324, ZINC ID 16743012, and ZINC ID 64165826; Table 2). All of the hit chemicals in Additional file 2: Table S2 have slightly different binding affinities. Interestingly, it was hypothesized that drugs with higher docking scores would bind the target protein more effectively.
Interpretation of protein–ligands interactions
The interpretation of protein–ligands interaction showed in the ZINC ID: 12,143,050 formed three conventional hydrogen bond interactions with TYR435, SER59 and TYR60, seven van der Waals interaction with PHE168, LEU328, LYS296, PHE343, GLN206, ARG42, GLY58, two pi carbon hydrogen bond with GLY434 and ILE199, two pi sigma CYS172 and ILE 198, one pi sulfur bond with CYS 397 and three Pi-alkyl bonds with TYR398, GLY57 and TYR326 with desire MAOB protein (Figs. 5A and 6A). In the case of ZINC08301324 van der walls bonds were predominantly formed with GLY11, GLU34, VAL10, PRO234, ILE272, LYS271, LEU268, ALA263, SER394, GLY13, SER15, THR43, MET436, THR426, TYR435, GLY434, TYR398, and CYS397. The position of VAL235, TYR393 and ARG42 acquired three conventional hydrogen bonds, two pi sigma bond ALA35 and ILE264 and THREE alkyl bond with PRO265, ILE14 and ALA439 position (Figs. 6B and 7B).
The protein–ligand complexes are able to interact in two dimensions. After molecular docking, the ligand makes contact with the MOAB protein, as shown in A ZINC12143050, B ZINC08301324, C ZINC16743012, and D ZINC64165826. Colors like blue, red, purple, light pink, deep pink, green and sky blue were used to denote various forms of bonds
For the compound ZINC16743012, the number of van der Waals interaction at position TYR60, PHE343, CYS397, GLY97, THR43, GLY40, ILE14, GLY13, ALA263, SER15, GLY425, GLY434, SER59 and TYR435 has found to be formed. Three conventional hydrogen bonds with GLN206, THR426 and MET436 position, one unfavorable positive-positive bond and one pi alkyl bond at ALA 439 have found to be formed (Figs. 6C and 7C). On the other hand, ZINC64165826, it has observed to formed six van der Waals carbon-hydrogen bonding in the position of PRO104, PHE99, VAL92, THR111, SER 160, and TRP107, three hydrogen bond with TYR97, ASN108 and HIS90, one pi-Alkyl bond with at position VAL106 (Fig. 6D).
Absorption, distribution, metabolism and excretion (ADME) and toxicity test Analysis of ADME properties
The drug is absorbed, distributed, metabolized, and excreted after being administered to an animal model or a human, resulting in active or passive transport to the target site. A pharmacological interaction with a biological macromolecule can be either positive or negative. The design of a drug is a step-by-step process, and failing to do so will lead to it being rejected, which is expensive for the company. The bioavailability of a drug is determined by its safety and efficacy, and lack of safety and efficacy are the primary reasons for drug failure, which are determined primarily by its ADME properties. SwissADME was utilized to assess the ADME qualities of the four compounds in terms of their pharmacokinetic parameters, including lipophilicity, water solubility, drug-likeness, and medicinal chemistry. The lipophilicity of a substance indicates that it may easily diffuse across the cell membrane; hence, an oral preparation is inappropriate. Moreover, since gastrointestinal absorption is limited, an injectable dose form may be an efficient way for achieving a quick beginning of action (Table 3). In addition, CoMFA stands for Comparative Molecular Field Analysis, an extensively employed 3D-QSAR method. Creating a three-dimensional grid around a molecule of interest and calculating the steric (shape) and electrostatic properties at each grid point are required (Fig. 8).
CoMFA result can provide valuable insights into the structure activity relationship of the studied molecules. This information can aid in the design of novel compounds with the desired activity profiles by assisting researchers in gaining a better understanding of the factors that influence the biological activity of the molecules. The spatial arrangement of the four finest compounds results in steric effects. Four compounds, including ZINC12143050, ZINC8301324, ZINC16743012, and ZINC64165826, yielded experimental activity values of 5.22184875, 5.21184875, 5.21184875, and 5.21184875, respectively (Additional file 3: Table S3).
Pharmacophore features analysis
A pharmacophore is a collection of either steric determinants or electronic properties that confirms optimal supramolecular interactions when conducting virtual screenings on large databases of molecules. Molecular docking is a powerful and more efficient method of finding molecules against specific targets that can induce or inhibit macromolecular activity. The compound with similar or relevant properties should exhibit the same or better activity as the query compound. Based on the docking score analysis, the pharmacophore features of ZINC12143050, ZINC08301324, ZINC16743012, and ZINC64165826, along with the antagonist CHEMBL3938629, were analyzed and compared. All the compounds have better pharmacophore properties than the antagonist CID: 56,961,657, so they are expected to be effective against our target protein. The pharmacophore feature found of the four compounds has shown in Fig. 9.
The evaluation of in-silico toxicity is an essential step that must be completed before clinical trials can begin for the purpose of selecting more effective lead compounds. Because of their precision, speed, and accessibility, computer-based toxicity measures have gained a lot of popularity in recent years. These qualities allow them to offer information on any molecule, whether it be natural or manufactured. Both the no-cost TEST tool and the ProTox II server were put to use as part of our investigation into the potential dangers posed by the four substances that were chosen. A number of toxicological parameters, including as acute toxicity, hepatotoxicity, cytotoxicity, carcinogenicity, mutagenicity, and immunotoxicity, were examined by the various software packages. Based on these evaluations, a median lethal dosage (LD50) in mg/kg was derived. According to the ProTox-II service, compounds ZINC12143050, ZINC08301324, and ZINC64165826 were classified as belonging to class 4, and LD50 ranges have also been compiled (Table 4).
Molecular dynamics (MD) simulation
MD simulations are used to investigate the binding stability of protein–ligand docking complexes. As an added benefit, MD simulations reveal details regarding intermolecular interactions over a given time scale. Here we use MD simulation methods to examine the docking contacts of four natural compounds and one reference antagonist with the MAOB protein, checking the stability of the protein-molecule complex and the strength of its intermolecular connections in less than 100 ns. The Maestro Desmond interface was used for MD trajectory extraction with SID, and the RMSD, RMSF, and Protein–Ligand (P-L) interaction mapping statistics were used to display the simulation results.
RMSD analysis
In MD simulation, the root mean square deviation (RMSD) is used to calculate the average distance caused by atom displacement for a given time frame in comparison with a reference time frame. The RMSD value of specific protein structure such as Cα, backbone, sidechain and heavy atoms have been estimated. The RMSD of the protein fit ligand captured from all the time frames during the reference time (in our case 100 ns). The RMSD value calculated from the X frame. It can be established whether the simulation has equilibrated or not based on the RMSD result. Within a reference protein structure, fluctuations of 1–3 Å are perfectly acceptable, but much greater values are not. A substantial conformational alteration in the protein indicates that the system is unstable. Except in the ZINC12143050 combination, the Cα atoms of MAOB displayed acceptable fluctuations in our four protein–ligand docking complexes. During a 100 ns simulation experiment, the compound ZINC12143050 showed an extended variation of 5.1 Å and a maximum fluctuation of 8.81 (between 26 and 28 ns) (Fig. 10). According to the data, MAOB undergoes protein conformation changes as a result of ZINC12143050 binding. Furthermore, at the end of the 100 ns simulation interval, measurement of RMSD using data acquired from protein fit ligands revealed minimal fluctuations (4.84 Å).
RMSF analysis
The local conformational change in the protein chain and the ligand molecules must be identified and quantified using the Root Mean Square Fluctuation (RMSF). When the MAOB protein was in contact with natural chemicals, the alterations caused by the residue index C were utilized to compute the local structural fluctuations. It's interesting to note that, with the exception of the N-terminal minimum 1.80 to maximum 9.90, all protein residues exhibit low RMSF values. With the exception of compound ZINC12143050, the combination of probable drugs against MOAB protein was validated by looking at the RMSF and RMSD values for all protein–ligand complexes. Hence, except for compound ZINC1070004335, the combined screened potential compounds were supported by examination of RMSF and RMSD values for all protein–ligand complexes. The apoprotein showed the most variation between residue positions 105 aa, with a fluctuation of 2.0 at PRO105. A minimal amount of variation was also visible in the apo structure at residue position THR241. The molecule Zinc 12,143,050 in combination with the protein was then compared to the apo structure, and a significant variation at residue position PRO333 was discovered (Fig. 11). In comparison with the apoprotein structure, zinc 08301324 appears to have the lowest average RMSF range between 1.0 and 1.3, and the variation of ASP153 and ALA355 was similarly minimal. The RMSF graph, on the other hand, revealed that the MOAB protein had lower average low and significant values in association with Zinc 16,743,012 (0.99 to 1.03) and Zinc 64,165,826 (1.3) than the reference apo structure. As previously mentioned, a low RMSF value denotes better protein stability, whereas the RMSF values discovered in this study for each protein–ligand system were lower than those for apoprotein. As a result, it is anticipated that the chemicals will maintain a stable interaction with the protein without changing its structure.
Protein–ligand interaction analysis
Hydrogen bonding, ionic bonding, water bridges, and hydrophobic bonding all play a role in turning a molecule into an effective drug. The MAOB protein had protein–ligand contact, and four natural chemicals were chosen from the MD trajectories and studied using the Desmond module's default parameters. All of the natural chemicals tested, ZINC12143050, ZINC08301324, ZINC16743012, and ZINC64165826, had tangible contact with the majority of the protein residues (Fig. 12). Furthermore, the four compounds evaluated using different filtering methods showed significant intermolecular interaction.
Solvent accessible surface area
Biological macromolecule structure and function are both influenced by their solvent-accessible surface area (SASA). Protein surface amino acid residues operate as active sites and bind ligands, providing insight into the solvent-like behavior (hydrophilic or hydrophobic) of molecules and protein–ligand complexes. Therefore, Fig. 13 displays the computed SASA value of the protein in association with the chemicals (A) ZINC12143050, (B) ZINC08301324, (C) ZINC16743012, and (D) ZINC64165826. For complicated systems, an average SASA value of 0–40 A was found, which indicates a significant amount of exposure of an amino acid residue to the molecule of interest.
Discussion
In current drug design, computer aided drug design (CADD) ushers in a new medicinal era by facilitating cost-effective processes, saving time, and lowering labor costs, all of which make drug discovery more feasible [32]. It is a necessary component as well as a tool for medication development. Therefore, the acceleration of CADD has provided scientists and researchers with the framework of biological and synthetic study. Thus, molecular docking, ADME, and molecular dynamic simulation procedures are employed to determine which drug candidates are most biologically effective. The severity of a disease can be mitigated by studying its mechanism, identifying the linked protein, and designing a ligand-binding method for the protein [33]. CADD is able to assist in the identification of specific target molecules by using information on their behavior and the way in which they bind ligands. Molecular docking, on the other hand, outlines the most prevalent binding modalities found within a ligand and protein, whereas MD simulation shows the complex method by which protein and ligand interact with one another. Because of this, small molecule candidates can be found that might be able to help treat a certain illness [34].
The compressive drug design method was used to look through a library of natural compounds to see if any could treat prostate cancer. Based on their molecular docking score, the four best compounds were chosen from the library of compounds with the strongest binding affinity. Also, compounds ZINC12143050, ZINC08301324, ZINC16743012, and ZINC64165826 have been shown to have stronger bonds with scores of − 11.7, − 11.4, − 11.2, and − 11.1 kcal/mol, respectively. The kinetics of metabolites in small molecular candidates have been studied using ADME analyses. The ADME mostly affects the drug in terms of its pharmacokinetic properties, which are also hard to evaluate quickly. Typically, the output of a CoMFA analysis includes contour maps that depict the favorable and unfavorable regions in the three-dimensional space around the molecule. These maps can highlight areas where modifications to the molecule's structure could lead to improved activity or selectivity. Overall, the results from a 3D-QSAR analysis like CoMFA contribute to the rational design of new drugs by providing a deeper understanding of how molecular structure influences activity, which can aid in the optimization and refinement of drug candidates. In traditional ways of making drugs, parts of rats or other animals are needed. Because a promising drug candidate needs to pass a standard clinical trial, the PK parameters should be optimized before the drug design process begins. This property affects the ability of small molecules to pass through biological systems. It also affects the weight of molecules and the topology of their polar surfaces (TPSA) [35]. Drug candidates with a large molecular weight may be less permeable, whereas TPSA improves the permeability of smaller molecules. LogP is used to determine whether polar and nonpolar solvents are suitable for dissolving a given chemical molecule. Both the inorganic logarithm and the coefficients of the relevant molecules were necessary for the aqueous phase partitioning. As a result, it affects how well different medication molecules are absorbed in the body, with a greater logP being associated with a naturally slower absorption rate. Nonetheless, LogS takes a pessimistic stance and influences the soluble state of candidate compounds with low values. The ability of a drug molecule to traverse a bilayer membrane is determined by the number of hydrocarbon bonds between the molecule's hydrogen bond donors and acceptors [36]. Rotatable bonds are prevalent in oral bioavailability because of their strong rational barriers. All four compounds have been tested for their PK properties, and needed evaluations were performed that found good value for each compound.
Further, toxicity tests are designed to provide information about adverse effects that can harm or damage an organism. About 20% of drug development failed because of a positive range of toxicity. Testing for toxicity is a crucial step before the experimentation of a drug that is very expensive and time-consuming to apply to an animal. Since there is no need for animal tests and is even time-consuming and reasonable, in-silico alternatives are chosen before drug development. This study identified four compounds that had a low toxic effect and optimal PK properties [37].
As a result, MD simulation has become an effective tool in the CADD process, as it determines how a compound moves within a macromolecular environment. It enables the analysis of the stability of a drug candidate against a targeted macromolecule [38]. To observe the RMSD, RMSF, and ligand–protein interaction of the complex system, MD simulation has been employed, which found the optimum RMSD and RMSF values for all four compounds with good protein–ligand contact [39]. In this way, the selected three compounds can be used to design additional classes of antiviral drugs for prostate cancer.
Conclusions
In studies assessing the ability of novel natural compounds to induce apoptosis in cells, four compounds were discovered, ZINC12143050, ZINC08301324, ZINC16743012, and ZINC64165826. The selected compounds have higher binding affinities of − 11.7, − 11.4, − 11.2, and − 11.1 kcal/mol with desired MAOB protein. Based on the in silico toxicity test, they have found a lower toxicity, and ADME analysis showed a readily soluble fat extract that is readily absorbed into the tissues. A structure-based model was initially developed, followed by molecular docking, ADMET analysis, and MD simulation. The top four natural compounds identified in the A-to-Z virtual screening process could serve as lead molecules in the fight against prostate cancer. Finally, the research findings are based on computational analysis, and further experimental validation is required to confirm the compound's efficacy.
Availability of data and materials
All data generated or analyzed during the study are included in this manuscript and supplementary files. Any further data required are available from the corresponding author on reasonable request.
Abbreviations
- ADME:
-
Absorption, distribution, metabolism and excretion
- MD:
-
Molecular dynamic simulation
- RMSD:
-
Root-mean-square deviation
- RMSF:
-
Root mean square fluctuation
- CADD:
-
Computer-aided drug design
- SASA:
-
Solvent accessible surface area
- MAOB:
-
Monoamine oxidase B
- EF:
-
Enrichment factors
- ROC:
-
Receiver operating curve
- PDB:
-
Protein data bank
- GH:
-
Güner-Henry Score
- ADT:
-
AutoDockTools
- UFF:
-
Universal force field
- LGA:
-
Lamarckian genetic algorithm
- QSAR:
-
Quantitative structure–activity relationship
- FEP:
-
Free energy perturbation
- EOS:
-
Estimate the equation of state
References
Whelan P (2014) The long perspective: Prostate cancer as a chronic disease. Prostate cancer diagnosis. Clin Manag. https://doi.org/10.1002/9781118347379.CH16
Siegel RL, Miller KD, Fuchs HE, Jemal A (2021) Cancer statistics, 2021. CA Cancer J Clin 71:7–33. https://doi.org/10.3322/CAAC.21654
Lodi A, Saha A, Lu X, Wang B, Sentandreu E, Collins M et al (2017) Combinatorial treatment with natural compouds in prostate cancer inhibits prostate tumor growth and leads to key modulations of cancer cell metabolism. Npj Precis Oncol 11:1–12. https://doi.org/10.1038/s41698-017-0024-z
Ostadkarampour M, Putnins EE (2021) Monoamine oxidase inhibitors: A review of their anti-inflammatory therapeutic potential and mechanisms of action. Front Pharmacol 12:889. https://doi.org/10.3389/FPHAR.2021.676239/BIBTEX
Binda C, Mattevi A, Edmondson DE (2011) Structural properties of human monoamine oxidases A and B. Int Rev Neurobiol 100:1–11. https://doi.org/10.1016/B978-0-12-386467-3.00001-7
Li M, Binda C, Mattevi A, Edmondson DE (2006) Functional role of the “aromatic cage” in human monoamine oxidase B: structures and catalytic properties of Tyr435 mutant proteins. Biochemistry 45:4775–4784. https://doi.org/10.1021/BI051847G/SUPPL_FILE/BI051847GSI20060301_101542.PDF
Tong J, Rathitharan G, Meyer JH, Furukawa Y, Ang LC, Boileau I et al (2017) Brain monoamine oxidase B and A in human parkinsonian dopamine deficiency disorders. Brain 140:2460–2474. https://doi.org/10.1093/BRAIN/AWX172
Aljanabi R, Alsous L, Sabbah DA, Gul HI, Gul M, Bardaweel SK (2021) Monoamine oxidase (MAO) as a potential target for anticancer drug design and development. Molecules. https://doi.org/10.3390/MOLECULES26196019
Chu GC-Y, Chung LWK, Gururajan M, Hsieh C-L, Josson S, Nandana S, et al. Regulatory signaling network in the tumor microenvironment of prostate cancer bone and visceral organ metastases and the development of novel therapeutics. Asian J Urol 2019; 6:65
Tainjie Pu et al. AACR 2022 Proceedings: Part B April 11–13 - American Association for Cancer Research - Google Books n.d. https://books.google.com.sa/books?id=_Z5vEAAAQBAJ&pg=PT1184&lpg=PT1184&dq=.+Following+the+analysis+of+three+independent+clinical+cohorts,+increased+stromal+MAOB+levels+were+shown+to+be+correlated+with+higher+Gleason+scores,+castration+resistance,+survival (accessed January 13, 2023).
Yin L, Liao C, Jason BW (2018) Monoamine oxidase deficiency causes prostate atrophy and reduces prostate progenitor cell activity. Stem Cells. https://doi.org/10.1002/stem.2831
Opo FADM, Rahman MM, Ahammad F, Ahmed I, Bhuiyan MA, Asiri AM (2021) Structure based pharmacophore modeling, virtual screening, molecular docking and ADMET approaches for identification of natural anti-cancer agents targeting XIAP protein. Sci Rep 11:4049. https://doi.org/10.1038/s41598-021-83626-x
Aljahdali MO, Molla MHR, Ahammad F (2021) Compounds identified from marine mangrove plant (Avicennia alba) as potential antiviral drug candidates against WDSV, an in-silico approach. Mar Drugs 19:253. https://doi.org/10.3390/MD19050253
Islam MR, Awal MA, Khames A, Abourehab MAS, Samad A, Hassan WMI et al (2022) Computational identification of druggable bioactive compounds from Catharanthus Roseus and Avicennia marina against colorectal cancer by targeting thymidylate synthase. Mol 27:2089. https://doi.org/10.3390/MOLECULES27072089
Aljahdali MO, Habibur M, Molla R, Ahammad F (2022) Immunoinformatics and computer-aided drug design as new approaches against emerging and re-emerging infectious diseases. Antivir Drugs. https://doi.org/10.5772/INTECHOPEN.101367
Makhouri FR, Ghasemi JB (2018) In silico studies in drug research against neurodegenerative diseases. Curr Neuropharmacol 16:664. https://doi.org/10.2174/1570159X15666170823095628
Arannilewa1 AJ, Alakanse OS, Adesola AO, Malachi OI, Obaidu IM, Oluwafemi EE, et al. Molecular docking analysis of Cianidanol fromGinkgo biloba with HER2+ breast cancer target. Bioinformation 2018;14:482. https://doi.org/10.6026/97320630014482.
Alamri MA, Altharawi A, Alabbas AB, Alossaimi MA, Alqahtani SM (2020) Structure-based virtual screening and molecular dynamics of phytochemicals derived from Saudi medicinal plants to identify potential COVID-19 therapeutics. Arab J Chem 13:7224–7234. https://doi.org/10.1016/J.ARABJC.2020.08.004
Bernal FA, Coy-Barrera E (2015) Molecular docking and multivariate analysis of xanthones as antimicrobial and antiviral agents. Molecules 20:13165–13204. https://doi.org/10.3390/MOLECULES200713165
Haider S, Barakat A, Ul-Haq Z (2020) Discovery of potential chemical probe as inhibitors of CXCL12 using ligand-based virtual screening and molecular dynamic simulation. Mol 25:4829. https://doi.org/10.3390/MOLECULES25204829
Pal S, Kumar V, Kundu B, Bhattacharya D, Preethy N, Reddy MP et al (2019) Ligand-based Pharmacophore modeling, virtual screening and molecular docking studies for discovery of potential topoisomerase i inhibitors. Comput Struct Biotechnol J 17:291. https://doi.org/10.1016/J.CSBJ.2019.02.006
Sangande F, Julianti E, Tjahjono DH (2020) Ligand-based pharmacophore modeling, molecular docking, and molecular dynamic studies of dual tyrosine kinase inhibitor of EGFR and VEGFR2. Int J Mol Sci 21:7779. https://doi.org/10.3390/IJMS21207779
Valasani KR, Vangavaragu JR, Day VW, Yan SS (2014) Structure based design, synthesis, pharmacophore modeling, virtual screening, and molecular docking studies for identification of novel cyclophilin D inhibitors. J Chem Inf Model 54:902–912. https://doi.org/10.1021/CI5000196/SUPPL_FILE/CI5000196_SI_001.PDF
Xie L, Li J, Xie L, Bourne PE (2009) Drug discovery using chemical systems biology: identification of the protein-ligand binding network to explain the side effects of CETP inhibitors. PLoS Comput Biol. https://doi.org/10.1371/JOURNAL.PCBI.1000387
Molla MHR, Aljahdali MO (2022) Identification of phytochemical compounds to inhibit the matrix-like linker protein VP26 to block the assembles of white spot syndrome virus (WSSV) envelope and nucleocapsid protein of marine shrimp: In silico approach. J King Saud Univ - Sci 34:102346. https://doi.org/10.1016/J.JKSUS.2022.102346
Forli S, Huey R, Pique ME, Sanner M, Goodsell DS, Olson AJ (2016) Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat Protoc 11:905. https://doi.org/10.1038/NPROT.2016.051
Krüger A, Maltarollo VG, Wrenger C, Kronenberger T (2019) ADME profiling in drug discovery and a new path paved on silica. Drug Discov Dev - New Adv. https://doi.org/10.5772/INTECHOPEN.86174
Hasan MR, Alsaiari AA, Fakhurji BZ, Molla MHR, Asseri AH, Sumon MAA et al (2022) Application of mathematical modeling and computational tools in the modern drug design and development process. Molecules 27:4169. https://doi.org/10.3390/molecules27134169
Islam MR, Awal MA, Khames A, Abourehab MAS, Samad A, Hassan WMI et al (2022) computational identification of druggable bioactive compounds from Catharanthus Roseus and Avicennia marina against colorectal cancer by targeting thymidylate synthase. Molecules. https://doi.org/10.3390/MOLECULES27072089
Fan T, Sun G, Zhao L, Cui X, Zhong R (2018) QSAR and classification study on prediction of acute oral toxicity of N-nitroso compounds. Int J Mol Sci 19:3015. https://doi.org/10.3390/IJMS19103015
Ivanova L, Tammiku-Taul J, García-Sosa AT, Sidorova Y, Saarma M, Karelson M (2018) Molecular dynamics simulations of the interactions between glial cell line-derived neurotrophic factor family receptor GFRα1 and small-molecule ligands. ACS Omega 3:11407–11414. https://doi.org/10.1021/ACSOMEGA.8B01524/ASSET/IMAGES/LARGE/AO-2018-01524B_0008.JPEG
Kapetanovic IM (2008) Computer-aided drug discovery and development (CADDD): in silico-chemico-biological approach. Chem Biol Interact 171:165. https://doi.org/10.1016/J.CBI.2006.12.006
De Vivo M, Masetti M, Bottegoni G, Cavalli A (2016) Role of molecular dynamics and related methods in drug discovery. J Med Chem 59:4035–4061. https://doi.org/10.1021/ACS.JMEDCHEM.5B01684/ASSET/IMAGES/LARGE/JM-2015-016843_0006.JPEG
Pinzi L, Rastelli G (2019) Molecular docking: shifting paradigms in drug discovery. Int J Mol Sci. https://doi.org/10.3390/IJMS20184331
Tibbitts J, Canter D, Graff R, Smith A, Khawli LA (2016) Key factors influencing ADME properties of therapeutic proteins: A need for ADME characterization in drug discovery and development. MAbs 8:229. https://doi.org/10.1080/19420862.2015.1115937
Bennion BJ, Be NA, McNerney MW, Lao V, Carlson EM, Valdez CA et al (2017) Predicting a drug’s membrane permeability: a computational model validated with in vitro permeability assay data. J Phys Chem B 121:5228–5237. https://doi.org/10.1021/ACS.JPCB.7B02914/ASSET/IMAGES/LARGE/JP-2017-02914T_0004.JPEG
Krewski D, Acosta D, Andersen M, Anderson H, Bailar JC, Boekelheide K et al (2010) Toxicity testing in the 21st century: a vision and a strategy. J Toxicol Environ Health B Crit Rev 13:51. https://doi.org/10.1080/10937404.2010.483176
Molla MHR, Aljahdali MO, Sumon MAA, Asseri AH, Altayb HN, Islam MS et al (2023) Integrative ligand-based pharmacophore modeling, virtual screening, and molecular docking simulation approaches identified potential lead compounds against pancreatic cancer by targeting FAK1. Pharm 16:120. https://doi.org/10.3390/PH16010120
Al-Karmalawy AA, Dahab MA, Metwaly AM, Elhady SS, Elkaeed EB, Eissa IH et al (2021) Molecular docking and dynamics simulation revealed the potential inhibitory activity of ACEIs against SARS-CoV-2 targeting the hACE2 receptor. Front Chem 9:227. https://doi.org/10.3389/FCHEM.2021.661230/BIBTEX
Acknowledgements
The authors also gratefully acknowledge technical and financial support provided by the Ministry of Education and King Abdulaziz University. DSR, Jeddah, Saudi Arabia.
Funding
This research work was funded by Institutional Fund Projects under grant no. (IFPIP: 1349–130-1443). The authors gratefully acknowledge technical and financial support provided by the Ministry of Education and King Abdulaziz University. DSR, Jeddah, Saudi Arabia.
Author information
Authors and Affiliations
Contributions
Conceptualization and data collection were contributed by MHRM, AHA and MSI. Data analysis and interpretation were contributed by MHRM and AHA. Writing and finalizing the manuscript were contributed by MHRM, AHA and MSI. All authors read and approved the final version of the study.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Molla, M.H.R., Asseri, A.H. & Islam, M.S. Integrated structure model-based virtual screening approaches identified anti-cancer agents against prostate cancer by targeting MAOB protein. Egypt J Med Hum Genet 24, 51 (2023). https://doi.org/10.1186/s43042-023-00431-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43042-023-00431-z