Abstract
Thaumatin-like proteins (TLPs) play potential roles in plant resistance to various diseases. Identifying TLPs is necessary to determine their function and apply them to plant disease resistance. However, limited information is available about TLP-family genes in wheat, especially regarding their responses to Fusarium species, which cause Fusarium head blight in wheat. In this study, we conducted a comprehensive genome-wide survey of TLP genes in wheat and identified 129 TLP genes in the wheat genome, which were unevenly distributed on 21 wheat chromosomes, with 5A containing the highest number. Phylogenetic analysis showed that these 129 wheat TLP genes together with 24 Arabidopsis TLPs were classified into 7 groups based on the protein sequences. We systematically analyzed the genes in terms of their sequence characterization, chromosomal locations, exon–intron distribution, duplication (tandem and segmental) events and expression profiles in response to Fusarium infection. Furthermore, we analyzed differentially expressed TLP genes based on publicly available RNA-seq data obtained from a resistant near isogenic wheat line at different time points after Fusarium graminearum inoculation. Then, the expression of 9 differentially expressed TLP genes was confirmed by real-time PCR, and these 9 genes were all upregulated in the resistant Sumai 3 variety, which was generally consistent with the RNA-seq data. Our results provide a basis for selecting candidate wheat TLP genes for further studies to determine the biological functions of the TLP genes in wheat.
Graphical Abstract

Similar content being viewed by others
Introduction
Fusarium head blight (FHB), which is mainly caused by Fusarium graminearum and F. venaceum, is one of the most serious and harmful wheat diseases worldwide and directly causes severe losses of quality and production (Bai & Shaner 2004). In recent years, due to changes in the climate and wheat farming systems, the occurrence of wheat FHB in China has become increasingly serious and begun to spread northward from the middle and lower reaches of the Yangtze River (Zhang et al. 2018). Because sources of wheat FHB resistance are very scarce, scientific researchers have tried many methods to improve the resistance to FHB (Bai & Shaner 2004; Fernando et al. 2021; Gorash et al. 2021; Steiner et al. 2017).
When plants are invaded by pathogens, they produce a series of defense responses, with resistance and defense genes being two major types of genes involved in disease resistance responses. Among them, pathogenesis-related (PR genes) proteins are the products of defense responses and are closely related to the hypersensitive reaction (HR) and systemic acquired resistance (SAR) (Edreva 2005; Liu & Ekramoddoullah 2006). According to structural characteristics, genetic relationships and biological activities, PR proteins are mainly divided into 17 families termed PR-1 to PR-17 (Van Loon et al. 2006). PR proteins are widely distributed in flowering plants and are an important part of plant defense systems (Liu & Ekramoddoullah 2006; Van Loon et al. 2006). One family of PR proteins (named the PR-5 family) has high sequence identity with the intensely sweet-tasing protein thaumatin first isolated from the fruit of the West African monocotyledon shrub Thaumatococcus daniellii (Benn.) Benth., and these proteins are therefore called thaumatin-like proteins (TLPs) (Cao et al. 2016). Purified TLPs from several plant species have shown in vitro anti-fungal activity, making them good research objects for disease resistance (Anžlovar & Dermastia 2003; Vigers et al. 1992). Most typical TLP proteins possess 16 conserved cysteine residues that form 8 disulfide bonds, and the molecular masses of these proteins range between 20 and 26 kDa (Liu, Zamani, & Ekramoddoullah 2010). In addition, researchers have also identified some small TLPs from monocots and conifers, which are characterized by smaller molecular weights (approximately 17 kDa) and only 10 conserved cysteine residues that form five disulfide bonds (Fierens et al. 2008; Liu, Sturrock, & Ekramoddoullah 2010).
Several experimental studies have indicated that the overexpression of TLP genes leads to increased disease resistance in plants (Chen et al. 1999; Datta et al. 1999; Mahdavi et al. 2012; Rout et al. 2016; Subramanyam et al. 2012; Van Loon et al. 2006; Yan et al. 2017), suggesting that TLPs play crucial roles in plant resistance to pathogen stress. TLPs can inhibit hyphal growth and spore germination by a membrane-permeabilizing mechanism (Abad et al. 1996) or by cell wall degradation (Osmond et al. 2001; Zareie et al. 2002). Some TLPs can function as β-1,3-glucanases, which mediate interactions with fungal cell walls or function as xylanase inhibitors (Fierens et al. 2007; Grenier et al. 2010). Some TLPs increase the permeability of fungal and oomycete plasma membranes (Kitajima & Sato 1999; Yun et al. 1998).
Considering the critical role of TLP proteins in host defenses during pathogen infection, investigations of the TLP gene family have been accomplished in several plant species, such as Cucumis melo (Liu et al. 2020), Gossypium barbadense (Zhang et al. 2021), Vitis vinifera L. (Yan et al. 2017), Brassica napus (Wang et al. 2020), Hordeum vulgare (Iqbal et al. 2020), etc. However, limited information on TLP proteins in bread wheat, especially in relation to disease resistance, has been reported. Thus, in this study, we aimed to identify the TLP genes in wheat at the whole-genome scale and analyze candidate TLP genes closely related to Fusarium head blight disease resistance. This research will lay a foundation for improving wheat FHB disease resistance by means of TLP genes.
Materials and methods
Identification of TLP-family genes in the wheat genome
Wheat genomic sequences were downloaded from the Triticum aestivum genome database (IWGSC RefSeq v1.1). A hidden Markov model (HMM) profile of the TLP DNA-binding domain (PF00314) was downloaded from the Pfam database (http://pfam.xfam.org/) (Finn et al. 2016) and used to query wheat TLP genes in the wheat genome sequence using HMMER software (version 3.0 from http://hmmer.janelia.org/). The obtained gene sequences were further confirmed by using SMART (http://smart.embl-heidelberg.de/smart/batch.pl) (Letunic et al. 2021) on the NCBI webserver. The molecular weights and theoretical isoelectric points (PIs) were calculated using the ExPASy Bioinformatics Resource Portal (http://web.expasy.org/compute_pi/) (Gasteiger et al. 2005).
Chromosomal localization and gene duplication
TaTLP genes were mapped onto wheat chromosomes according to their physical positions using TBtools (Chen et al. 2020). Duplicated genes were determined on the basis of multiple sequence alignments using ClustalW with the following criteria: shorter sequences covering > 75% of longer sequences after alignment and a similarity of aligned regions > 75%.
Phylogenetic tree, gene structure and conserved motif analysis
A phylogenetic tree including TaTLP genes identified in wheat as well as Arabidopsis was constructed with MEGA7 software (Molecular Evolutionary Genetics Analysis) (Kumar et al. 2015) using the neighbor-joining method based on the full-length protein sequences with default parameters. Bootstrap analysis was performed using 1000 replicates. The gene exon/intron structures of the wheat TLP genes were graphically visualized using the Gene Structure Display Server (GSDS) program (http://gsds.cbi.pku.edu.ch) (Hu et al. 2014). Conserved motifs in the TLP protein sequences were analyzed using the MEME program with the following parameters: number of repetitions, any; maximum number of motifs, 10; minimum motif width, 6; and maximum motif width, 100.
Differential expression analysis
RNA-seq expression data from two near isogenic wheat lines, which differed by the presence of both (NIL38) or none (NIL51) of the FHB-resistance QTLs Fhb1 and Qfhs.ifa-5A, under disease pressure (3, 6, 12, 24, 36, 48 hpi) as well as mock inoculation were subjected to analysis (Schweiger et al. 2013). Normalized expression values expressed as TPM (transcripts per kilobase of exon model per million mapped reads) from the gene expression profile dataset ERP013829 containing 298,774 wheat genes and 72 samples were downloaded from the Wheat Expression Browser (www.wheat-expression.com) (Ramírez-González et al. 2018). Differentially expressed genes (DEGs) between different comparison groups were detected using the R package ‘edgeR’. DEGs with a log fold change above 1 or below − 1 (false discovery rate < 0.01, P < 0.01) were used as the screening criteria. TBtools software was used to draw the heatmap.
Plant materials and FHB inoculation
The FHB-resistant wheat variety Sumai 3 was used in this study. Seeds of this variety were planted in the field during October 2017 at Jiangsu Academy of Agricultural Sciences Experiment Station, Nanjing, China (108° E, 34°15′ N). F. graminearum spores were sporulated on mung bean liquid medium at 25 °C and suspended in sterile water at a concentration of 1.0 × 106 conidia ml− 1. Approximately 10 μL of F. graminearum suspension was injected into the central spikelets at early anthesis. Sterile water was injected as a control at the same time points. Samples were collected at 0, 24, and 48 hours post-inoculation (hpi), immediately flash-frozen with liquid nitrogen and stored in a − 80 °C freezer until use, with three biological replications conducted for each sample.
RNA extraction and quantitative real-time PCR (qRT–PCR) analysis
Total RNA in the collected anthers was isolated using the RNAprep Pure Plant Kit (Tiangen Biotech, Beijing, China) following the manufacturer’s instructions. The total RNA concentrations were measured with a NanoDrop 2000 system (Thermo Fisher Scientific), and then the integrity was determined using an RNA Nano 6000 Assay Kit with an Agilent Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA). cDNA synthesis was performed using SuperScript II reverse transcriptase and oligo (dT) 12–18 primers (Invitrogen) according to the manufacturer’s protocol. The resulting cDNA was diluted 10-fold, and 1 μL of diluted cDNA was used as a template for PCR amplification in a 30-μL reaction using Power SYBR Green PCR master mix (Applied Biosystems). Gene-specific primers for each TaTLP gene were designed using Primer Premier 5.0 (Primer, Palo Alto, CA, USA). qRT–PCR analysis was conducted using the SYBR Premix Ex Taq Kit (TaKaRa) and the QuantStudio™ 7 Flex Real Time PCR System (Applied Biosystems) with a final volume of 20 μL per reaction. Each reaction mixture consisted of 10.0 μL SYBR Premix Ex Taq II, 1.0 μL cDNA, 0.8 μL each primer (10 μM), and 7.4 μL sterile H2O. The cycling parameters were 95 °C for 30 s, followed by 40 cycles at 95 °C for 5 s and 60 °C for 30 s. Melt-curve analyses were performed using a program of holding at 95 °C for 15 s, followed by a constant increase from 60 °C to 95 °C. Relative gene expression levels were calculated by the comparative Ct method according to the fold change (2–ΔΔCt) (Schmittgen & Livak 2008), and the relative expression level of each gene is presented as the mean ± standard deviation.
Results
Analysis of TLP-family genes in the wheat genome
A total of 129 TLP genes were identified in the wheat genome. The genes were named according to their physical positions on chromosomes. Detailed information about the genes, including gene IDs, chromosomal positions, and lengths of coding sequences, is presented in Table S1. As shown in Table S1, the predicted protein lengths of the TLPs ranged from 101 (TaTLP43) to 663 (TaTLP108) amino acids. The calculated MWs of the TLPs ranged from 10.6 kDa (TaTLP43) to 73.5 kDa (TaTLP12), and the calculated theoretical pIs of TLPs ranged from 4.23 (TaTLP56) to 12.13 (TaTLP78).
Chromosomal distribution and gene duplication of TaTLP genes
A total of 126 TLP genes (three identified TLPs had no known physical positions) were unevenly distributed on 21 chromosomes (Fig. 1). Chromosome 5A contained the highest number (22 genes) of TLP genes. The chromosome with the second-highest number of TLP genes was 5B, which included 17 TLPs. Interestingly, the fifth homologous group of chromosome D contained only 8 genes. Two chromosomes, 1D and 6A, each contained only one TLP gene. The remaining chromosomes contained less than 10 TLP genes, with totals ranging from 2 to 9.
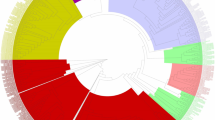
Phylogenetic relationships among TLP proteins from wheat and A. thaliana. Genes from wheat and Arabidopsis on branch ends are denoted by turquoise solid circles and red stars, respectively. Different arcs indicate different groups of TLP proteins. Differently colored gene names indicate different groups. Different subgroup classifications are marked on branch nodes
There were 255 duplication pairs of TLP genes, including 54 tandem duplication pairs and 201 segmental duplication pairs, suggesting that segmental duplication events played a role in the expansion of TLP genes (Fig. 1, Table S2).
Phylogenetic analysis, gene structure, and motif composition of TaTLP genes
A phylogenetic tree was constructed from the protein sequences of 129 wheat TLP genes and 24 Arabidopsis TLP genes. According to Fig. 2, all 153 TLP genes were divided into seven groups. Group 1 and Group 3 contained 6 and 13 genes, respectively, which were all from wheat. Group 2 included only one member from Arabidopsis. Group 4 contained 5 members—one from Arabidopsis and 4 from wheat. Group 5 contained 4 members, which were all from Arabidopsis. Group 6 and Group 7 contained the highest (62) number of genes among all groups but showed difference in the number of included Arabidopsis genes, with 16 and 2 Arabidopsis genes in Group 6 and Group 7, respectively. Group 6 and Group 7 could be further divided into 6 (Subgroup 6–1 to Subgroup 6–6) and 3 (Subgroup 7–1 to Subgroup 7–3) subgroups, respectively.
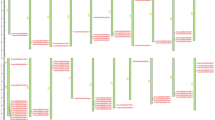
Gene structures and conserved motif compositions of wheat TLP proteins. a Motifs numbered 1–10 are displayed in differently colored boxes. The lengths of proteins can be estimated using the scale at the bottom. b Exon/intron organizations of 129 TLP genes. Solid green boxes, solid yellow boxes and black lines indicate exons, untranslated regions and introns, respectively
Combining the gene structure with the phylogenetic tree, we found that the TaTLP exon–intron distribution was related to the TaTLP classification. The numbers of TaTLP exons were discontinuously distributed from 1 through 4. The majority of TaTLP gene structures contained 1–2 exons (Fig. 3).
Gene motif analysis also indicated that the gene motifs and distribution patterns were closely related to the gene groups and subgroups. As shown in Fig. 3, motif 5, motif 6 and motif 7 are fundamental in TaTLP domains, as most TaTLP genes contained these motifs. The number of TaTLP motifs ranged from 1 to 8. TaTLP members of Group 3 contained motif 6 and motif 9, and three members (TaTLP40, TaTLP47 and TaTLP53) contained an additional motif 10. Most members (17 out of 23) of Subgroup 7–3 contained motif 2, motif 3 and motif 4, while others did not contain these three motifs. In general, members of the same group had similar exon/intron structures and motifs (Fig. 2b).
Identification of differentially expressed TLP genes
DEG analysis identified 10,363 DEGs (8367 upregulated and 1996 downregulated) in the near-isogenic wheat line containing both FHB-resistance QTLs Fhb1 and Qfhs.ifa-5A (NIL38) between the F. graminearum inoculation and mock inoculation scenarios at different time points (Table S3). We further analyzed the induction of TLP genes at different times after inoculation, and the results identified 16 TLP DEGs in NIL38 between the F. graminearum inoculation and mock inoculation scenarios at 36 hpi, followed by 48 hpi (9) and 24 hpi (7) (Fig. 4). Among the 16 differentially expressed genes in NIL38 between the F. graminearum inoculation and mock inoculation scenarios, 12 were differentially expressed in at least two comparisons, and 4 genes (TaTLP12, TaTLP17, TaTLP43, and TaTLP79) were differentially expressed in only one comparison. Among the 12 repeated differentially expressed genes, 4 (TaTLP42, TaTLP51, TaTLP58 and TaTLP61) appeared in all three comparisons, and the remaining 8 genes appeared in two of the comparisons, including 3 genes (TaTLP114, TaTLP120, and TaTLP126) between 24 hpi and 36 hpi and 5 genes (TaTLP13, TaTLP33, TaTLP50, TaTLP80, and TaTLP118) between 36 hpi and 48 hpi (Table S4) (Fig. 4).
Expression patterns of TLP members in response to F. graminearum infection
To verify whether these genes are involved in the wheat response to pathogen stress, according to the gene copy number and amplification specificity of the TLP DEGs, 9 of the 16 TLP DEGs were further selected to validate the roles of TLP genes in the disease-resistant line Sumai 3 in response to F. graminearum inoculation by using qRT–PCR analysis. The primer sequences used are shown in Table S5. The results showed that the expression of all 9 TLP genes was upregulated after inoculation, with an intitial increase at 24 hpi and a high expression level at 48 hpi (Fig. 5). The gene expression pattern results obtained by qRT–PCR were generally the same as the RNA-Seq results, suggesting the potential roles of these genes in resistance to F. graminearum infection.
qRT–PCR was used to analyze the expression profiles of 9 selected TLP genes in response to F. graminearum infection at different time points. The data were normalized to the expression level of the tublin gene. The error bars represent the standard deviation (SD) of three biological replicates. Values of P < 0.05 were considered statistically significant
Discussion
Wheat FHB is a very important disease. Although resistance QTLs, including Fhb1, have played very important roles in wheat FHB resistance breeding, the currently achieved resistance level is still not satisfactory. Other resources for FHB resistance are also limited; therefore, more methods to improve FHB resistance are urgently needed. Plant thaumatin-like proteins are widely distributed in a variety of plants, animals and microorganisms (Liu, Sturrock, & Ekramoddoullah 2010; Shatters et al. 2006). It has been proven that they mainly exert antifungal activity and that their expression can be induced when plants are under stress. Studies have shown that TLPs can not only induce programmed cell death in fungi but also inhibit fungal infection, activate signal pathways in plant defense responses, and enhance the ability of plants to resist adversity (Liu & Ekramoddoullah 2010; Abad et al. 1996; Liu et al. 2012; Niu et al. 2011; Van Loon et al. 2006; Ward 1991). Due to their antifungal activities and crucial roles in plant defense responses against biotic and abiotic stresses, it is necessary to study TLPs.
In this study, based on the high abundance of TLP genes obtained from the transcriptome dataset of wheat inoculated with Fusarium, 129 wheat TLP sequences were obtained. Through homologous evolutionary tree analysis with Arabidopsis TLP genes, the wheat TLP proteins and Arabidopsis TLP proteins were divided into seven cluster groups. The clustering results also showed that TLP proteins are strongly conserved. For example, four Arabidopsis TLP genes (AtTLP4, AtTLP14, AtTLP15, and AtTLP23) gathered to form Group 5. Group 6 contained the most Arabidopsis TLP genes, and 5 Arabidopsis genes (AtTLP3, AtTLP9, AtTLP11, AtTLP18, and AtTLP20) in Subgroup 6–1, 3 Arabidopsis genes (AtTLP16, AtTLP17, and AtTLP19) in Subgroup 6–2, and 6 Arabidopsis genes (AtTLP1, AtTLP2, AtTLP5, AtTLP6, AtTLP7, and AtTLP8) in Subgroup 6–4 were closely clustered.
According to previous studies of Arabidopsis TLPs, Arabidopsis TLP genes are closely related to disease resistance. For example, Hu & Reddy (1997) reported that AtTLP1 (AT1G18250) showed antifungal activity against several fungal pathogens and that its expression can be induced by pathogen infection and salicylic acid. It is known as a marker gene of systemic acquired resistance induced by the salicylic acid-dependent pathway (Li & Fan 2000), which suggests that the 3 wheat TLP genes (TaTLP112, TaTLP117, and TaTLP123) closely clustered to this gene may also be related to SA-dependent plant disease resistance.
Three wheat TLP genes (TaTLP36, TaTLP49, and TaTLP56) were clustered with four Arabidopsis TLP genes, and among these four Arabidopsis genes, two were reported to be related to disease resistance. For example, AtTLP6 (AT1G75030) showed antifungal activity against several fungal pathogens, suggesting that AT1G75030 may be involved in plant defenses against fungal pathogens. The expression of PR5 (AT1G75040 and AtTLP7) was strongly induced with a 15-fold increase in the susceptible accession Col-0 during the infection time course, whereas it was not induced in the nonsusceptible accession (Lemarié et al. 2015), which was in agreement with the RNA sequencing results obtained by Schweiger et al. (2016). Zhu et al. (2021) also reported that the expression of PR5 (AT1G75040) was also increased in overexpression lines following Botrytis cinerea infection. The AT4G11650 (AtTLP13) gene was clustered with 13 wheat genes, and previous analyses of the AT4G11650 gene showed that it was closely related to defense responses and was also a marker gene of pathways related to defense responses (Berrocal-Lobo & Molina 2008; Guo & Stotz 2007; Mukherjee et al. 2010), which provided important information about the functions of the 13 wheat genes. AT4G24180 (AtTLP16) and the remaining two Arabidopsis genes AT4G36000 (AtTLP17) and AT4G38660 (AtTLP19) were clustered with 12 wheat TLP genes, also suggesting that these 12 wheat genes are closely related to disease resistance, as AT4G24180 (AtTLP16) was reported to be induced by colonization with nonpathogenic fluorescent Pseudomonas spp. and to activate systemic resistance (Léon-Kloosterziel et al. 2005).
The above information only facilitates speculation that wheat TLP genes may be related to resistance based on the clustering relationships between wheat TLP genes and Arabidpsis TLP genes. In addition, a large number of studies have also shown that the overexpression of TLP genes can improve the antifungal ability of transgenic plants. For example, the overexpression of a rice TLP gene in banana plants enhanced the resistance to Fusarium wilt caused by Fusarium oxysporum sp. cubensec (race 4) (Mahdavi et al. 2012). Zamani et al. (2012) reported that the overexpression of a TLP gene isolated from rye (Secale cereal L.) enhanced the resistance to canola (Brassica napus L.) stem rot caused by Sclerotinia sclerotiorum. Acharya et al. (2013) reported that overexpressing a TLP gene of Camellia sinensis (CsTLP) in potato plants enhanced the resistance to two important fungal pathogens, Macrophomina phaseolina (necrotrophic) and Phytophthora infestans (hemibiotrophic).
Recent advances in high-throughput technologies, including whole-genome de novo sequencing and RNA-seq in wheat (Appels et al. 2018; Borrill et al. 2016; Pearce et al. 2015; Stark et al. 2019), provide novel insights into the molecular identification and understanding of disease resistance genes. Using transcriptome data to identify upregulated and downregulated genes related to disease resistance responses will provide important information for selecting candidate genes. In the present study, the FHB-related dataset ERP013829, which encompasses the RNA-seq data of 72 samples, was downloaded from the ENA database. Differential gene expression analysis was used to compare different inoculation time points in resistant materials and susceptible materials with the control. In total, 32 TLP genes were detected, including 16 TLP genes that were differentially expressed between the F. graminearum inoculation and mock inoculation scenarios in the resistant near isogenic wheat line (NIL38) containing both FHB-resistance QTLs, Fhb1 and Qfhs.ifa-5A, among which 15 were upregulated and only one was downregulated. The qRT–PCR results were also consistent with the RNA-seq data, but the roles of these genes in disease resistance will be the focus of follow-up work. The present study provides useful and important information about wheat TLP genes to allow breeding programs to increase the resistance of wheat to Fusarium infections.
Conclusion
In the present study, 129 members of the TLP family in the wheat genome were identified. Sequence characterization, chromosomal locations, exon–intron distribution, duplication (tandem and segmental) events and expression profiles in response to Fusarium infection were analyzed. Phylogenetic analysis showed that these 129 wheat TLP genes together with 24 Arabidopsis TLPs were classified into 7 groups based on the protein sequences. The expression patters of the selected 9 TLP genes were confirmed by real-time PCR, and these 9 genes were all upregulated in the resistant Sumai 3 variety after Fusarium graminearum inoculation, which was generally consistent with the RNA-seq data. Our results provide a basis for selecting candidate wheat TLP genes for further studies to determine the biological function for Fusarium head blight resistance.
Availability of data and materials
Plant materials and the datasets supporting the results of this article are included within the article and its additional files.
References
Abad, L. R., D’Urzo, M. P., Liu, D., Narasimhan, M. L., Reuveni, M., Zhu, J. K., Bressan, R. A. (1996). Antifungal activity of tobacco osmotin has specificity and involves plasma membrane permeabilization. Plant Science, 118(1), 11–23.
Acharya, K., Pal, A. K., Gulati, A., Kumar, S., Singh, A. K., & Ahuja, P. S. (2013). Overexpression of Camellia sinensis thaumatin-like protein, CsTLP in potato confers enhanced resistance to Macrophomina phaseolina and phytophthora infestans infection. Molecular Biotechnology, 54(2), 609–622.
Anžlovar, S., & Dermastia, M. (2003). The comparative analysis of osmotins and osmotin-like PR-5 proteins. Plant Biology, 5(2), 116–124.
Appels, R., Eversole, K., Stein, N., Feuillet, C., Keller, B., Rogers, J., Wang, L. (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science, 361(6403), eaar7191.
Bai, G., & Shaner, G. (2004). Management and resistance in wheat and barley to Fusarium head blight. Annual Review of Phytopathology, 42, 135–161.
Berrocal-Lobo, M., & Molina, A. (2008). Arabidopsis defense response against Fusarium oxysporum. Trends in Plant Science, 13(3), 145–150.
Borrill, P., Ramirez-Gonzalez, R., & Uauy, C. (2016). expVIP: A customizable RNA-seq data analysis and visualization platform. Plant Physiology, 170(4), 2172–2186.
Cao, J., Lv, Y., Hou, Z., Li, X., & Ding, L. (2016). Expansion and evolution of thaumatin-like protein (TLP) gene family in six plants. Plant Growth Regulation, 79(3), 299–307.
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., & Xia, R. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 13(8), 1194–1202.
Chen, W., Chen, P., Liu, D., Kynast, R., Friebe, B., Velazhahan, R., Gill, B. S. (1999). Development of wheat scab symptoms is delayed in transgenic wheat plants that constitutively express a rice thaumatin-like protein gene. Theoretical and Applied Genetics, 99(5), 755–760.
Datta, K., Velazhahan, R., Oliva, N., Ona, I., Mew, T., Khush, G. S., Datta, S. K. (1999). Over-expression of the cloned rice thaumatin-like protein (PR-5) gene in transgenic rice plants enhances environmental friendly resistance to Rhizoctonia solani causing sheath blight disease. Theoretical and Applied Genetics, 98, 1138–1145.
Edreva, A. (2005). Pathogenesis-related proteins: Research progress in the last 15 years. General and Applied Plant Physiology, 31(1–2), 105–124.
Fernando, W. G. D., Oghenekaro, A. O., Tucker, J. R., & Badea, A. (2021). Building on a foundation: Advances in epidemiology, resistance breeding, and forecasting research for reducing the impact of Fusarium head blight in wheat and barley. Canadian Journal of Plant Pathology, 43(4), 495–526.
Fierens, E., Gebruers, K., Courtin, C. M., & Delcour, J. A. (2008). Xylanase inhibitors bind to nonstarch polysaccharides. Journal of Agricultural and Food Chemistry, 56(2), 564–570.
Fierens, E., Rombouts, S., Gebruers, K., Goesaert, H., & Delcour, J. A. (2007). TLXI, a novel type of xylanase inhibitor from wheat (Triticum aestivum) belonging to the thaumatin family. Biochemical Journal, 403(3), 583–591.
Finn, R. D., Penelope, C., Eberhardt, R. Y., Eddy, S. R., Jaina, M., Mitchell, A. L., Amaia, S. V. (2016). The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Research, 44(D1), D279–D285.
Gasteiger, E., Hoogland, C., Gattiker, A., Wilkins, M. R., Appel, R. D., & Bairoch, A. (2005). Protein identification and analysis tools on the ExPASy server. In The proteomics protocols handbook, (pp. 571–607).
Gorash, A., Armoniene, R., & Kazan, K. (2021). Can effectoromics and loss-of-susceptibility be exploited for improving Fusarium head blight resistance in wheat? Crop Journal, 9(1), 1–16.
Grenier, J., Potvin, C., Trudel, J., & Asselin, A. (2010). Some thaumatin-like proteins hydrolyse polymeric beta-1,3-glucans. Plant Journal, 19(4), 473–480.
Guo, X., & Stotz, H. U. (2007). Defense against Sclerotinia sclerotiorum in Arabidopsis is dependent on jasmonic acid, salicylic acid, and ethylene signaling. Molecular Plant-Microbe Interactions, 20(11), 1384.
Hu, B., Jin, J., Guo, A., Zhang, H., Luo, J., & Gao, G. (2014). GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics, 31(8), 1296–1297.
Hu, X., & Reddy, A. S. N. (1997). Cloning and expression of a PR5-like protein from Arabidopsis: Inhibition of fungal growth by bacterially expressed protein. Plant Molecular Biology, 34(6), 949–959.
Iqbal, I., Tripathi, R. K., Wilkins, O., & Singh, J. (2020). Thaumatin-like protein (TLP) gene family in barley: Genome-wide exploration and expression analysis during germination. Genes, 11(9), 1180.
Kitajima, S., & Sato, F. (1999). Plant pathogenesis-related proteins: Molecular mechanisms of gene expression and protein function. Journal of Biochemistry, 125(1), 1–8.
Kumar, S., Stecher, G., & Tamura, K. (2015). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33, 1870–1874.
Lemarié, S., Robert-Seilaniantz, A., Lariagon, C., Lemoine, J., Marnet, N., Jubault, M., Gravot, A. (2015). Both the jasmonic acid and the salicylic acid pathways contribute to resistance to the biotrophic clubroot agent Plasmodiophora brassicae in Arabidopsis. Plant and Cell Physiology, 56(11), 2158–2168.
Léon-Kloosterziel, K. M., Verhagen, B. W. M., Keurentjes, J. J. B., Van Pelt, J. A., Rep, M., Van Loon, L. C., & Pieterse, C. M. J. (2005). Colonization of the Arabidopsis rhizosphere by fluorescent Pseudomonas spp. activates a root-specific, ethylene-responsive PR-5 gene in the vascular bundle. Plant Molecular Biology, 57(5), 731–748.
Letunic, I., Khedkar, S., & Bork, P. (2021). SMART: Recent updates, new developments and status in 2020. Nucleic Acids Research, 49(D1), D458–D460.
Li, R., & Fan, Y. (2000). Advances on genetic engineering of plant resistance against fungal diseases. Progress in Biotechnology, 20(2), 9–13.
Liu, D., He, X., Li, W., Chen, C., & Ge, F. (2012). Molecular cloning of a thaumatin-like protein gene from Pyrus pyrifolia and overexpression of this gene in tobacco increased resistance to pathogenic fungi. Plant Cell, Tissue and Organ Culture, 111(1), 29–39.
Liu, J., & Ekramoddoullah, A. (2006). The family 10 of plant pathogenesis-related proteins: Their structure, regulation, and function in response to biotic and abiotic stresses. Physiological and Molecular Plant Pathology, 68, 3–13.
Liu, J., Sturrock, R., & Ekramoddoullah, A. (2010). The superfamily of thaumatin-like proteins: Its origin, evolution, and expression towards biological function. Plant Cell Reports, 29(5), 419–436.
Liu, J., Zamani, A., & Ekramoddoullah, A. (2010). Expression profiling of a complex thaumatin-like protein family in western white pine. Planta, 231(3), 637–651.
Liu, Y., Cui, J., Zhou, X., Luan, Y., & Luan, F. (2020). Genome-wide identification, characterization and expression analysis of the TLP gene family in melon (Cucumis melo L.). Genomics, 112(3), 2499–2509.
Mahdavi, F., Sariah, M., & Maziah, M. (2012). Expression of rice thaumatin-like protein gene in transgenic banana plants enhances resistance to Fusarium wilt. Applied Biochemistry and Biotechnology, 166(4), 1008–1019.
Mukherjee, A. K., Carp, M. J., Zuchman, R., Ziv, T., Horwitz, B. A., & Gepstein, S. (2010). Proteomics of the response of Arabidopsis thaliana to infection with Alternaria brassicicola. Journal of Proteomics, 73(4), 709–720.
Niu, D., Liu, H., Jiang, C., Wang, Y., Wang, Q., Jin, H., & Guo, J. (2011). The plant growth-promoting rhizobacterium Bacillus cereus AR156 induces systemic resistance in Arabidopsis thaliana by simultaneously activating salicylate- and jasmonate/ethylene-dependent signaling pathways. Molecular Plant-Microbe Interactions, 24(5), 533–542.
Osmond, R. I. W., Hrmova, M., Fontaine, F., Imberty, A., & Fincher, G. B. (2001). Binding interactions between barley thaumatin-like proteins and (1,3)-beta-D-glucans. Kinetics, specificity, structural analysis and biological implications. European Journal of Biochemistry, 268(15), 4190–4199.
Pearce, S., Vazquez-Gross, H., Herin, S. Y., Hane, D., Wang, Y., Gu, Y., & Dubcovsky, J. (2015). WheatExp: An RNA-seq expression database for polyploid wheat. BMC Plant Biology, 15(1), 1–8.
Ramírez-González, R. H., Borrill, P., Lang, D., Harrington, S. A., Brinton, J., Venturini, L., Uauy, C. (2018). The transcriptional landscape of polyploid wheat. Science, 361(6403), eaar6089.
Rout, E., Nanda, S., & Joshi, R. K. (2016). Molecular characterization and heterologous expression of a pathogen induced PR5 gene from garlic (Allium sativum L.) conferring enhanced resistance to necrotrophic fungi. European Journal of Plant Pathology, 144(2), 345–360.
Schmittgen, T. D., & Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nature Protocols, 3(6), 1101–1108.
Schweiger, W., Steiner, B., Ametz, C., Siegwart, G., Wiesenberger, G., Berthiller, F., Buerstmayr, H. (2013). Transcriptomic characterization of two major Fusarium resistance quantitative trait loci (QTLs), Fhb1 and Qfhs.Ifa-5A, identifies novel candidate genes. Molecular Plant Pathology, 14(8), 772–785.
Shatters, R. G., Boykin, L. M., Lapointe, S. L., Hunter, W. B., & Iii, A. (2006). Phylogenetic and structural relationships of the PR5 gene family reveal an ancient multigene family conserved in plants and select animal taxa. Journal of Molecular Evolution, 63(1), 12–29.
Stark, R., Grzelak, M., & Hadfield, J. (2019). RNA sequencing: The teenage years. Nature Reviews Genetics, 20(11), 631–656.
Steiner, B., Buerstmayr, M., Michel, S., Schweiger, W., Lemmens, M., & Buerstmayr, H. (2017). Breeding strategies and advances in line selection for Fusarium head blight resistance in wheat. Tropical Plant Pathology, 42(3), 165–174.
Subramanyam, K., Arun, M., Mariashibu, T. S., Theboral, J., Rajesh, M., Singh, N. K., Ganapathi, A. (2012). Overexpression of tobacco osmotin (Tbosm) in soybean conferred resistance to salinity stress and fungal infections. Planta, 236(6), 1909–1925.
Van Loon, L. C., Rep, M., & Pieterse, C. M. J. (2006). Significance of inducible defense-related proteins in infected plants. Annual Review of Phytopathology, 44(1), 135–162.
Vigers, A. J., Wiedemann, S., Roberts, W. K., Legrand, M., Selitrennikoff, C. P., & Fritig, B. (1992). Thaumatin-like pathogenesis-related proteins are antifungal. Plant Science, 83(2), 155–161.
Wang, T., Hu, J., Ma, X., Li, C., & Song, X. (2020). Identification, evolution and expression analyses of whole genome-wide TLP gene family in Brassica napus. BMC Genomics, 21(1), 1–14.
Ward, E. R. (1991). Coordinate gene activity in response to agents that induce systemic acquired resistance. The Plant Cell, 3(10), 1085–1094.
Yan, X., Qiao, H., Zhang, X., Guo, C., Wang, M., Wang, Y., & Wang, X. (2017). Analysis of the grape (Vitis vinifera L.) thaumatin-like protein (TLP) gene family and demonstration that TLP29 contributes to disease resistance. Scientific Reports, 7(1), 4269.
Yun, D., Ibeas, J. I., Lee, H., Coca, M. A., Narasimhan, M. L., Uesono, Y., Bressan, R. A. (1998). Osmotin, a plant antifungal protein, subverts signal transduction to enhance fungal cell susceptibility. Molecular Cell, 1(6), 807.
Zamani, A., Motallebi, M., Jonoubi, P., Ghafarian-Nia, N. S., & Zamani, M. R. (2012). Heterologous expression of the Secale cereal thaumatinlike protein in transgenic canola plants enhances resistance to stem rot disease. Iranian Journal of Biotechnology, 10(2), 87–95.
Zareie, R., Melanson, D. L., & Murphy, P. J. (2002). Isolation of fungal cell wall degrading proteins from barley (Hordeum vulgare L.) leaves infected with Rhynchosporium secalis. Molecular Plant-Microbe Interactions, 15(10), 1031–1039.
Zhang, A., Yang, W., Li, X., & Sun, J. (2018). Current status and perspective on research against Fusarium head blight in wheat. Hereditas, 40(10), 858–873.
Zhang, Y., Chen, W., Sang, X., Wang, T., Gong, H., Zhao, Y., Wang, H. (2021). Genome-wide identification of the thaumatin-like protein family genes in Gossypium barbadense and analysis of their responses to Verticillium dahliae infection. Plants, 10(12), 2647.
Zhu, Y., Guo, M., Song, J., Zhang, S., Guo, R., Hou, D., Huang, X. (2021). Roles of endogenous melatonin in resistance to botrytis cinerea infection in an Arabidopsis model. Frontiers in Plant Science, 12, 683228.
Acknowledgements
Not applicable.
Funding
This study is based upon work supported partially by the National Key Project for the Research and Development of China (2017YFE0126700), Jiangsu Agricultural Science and Technology Innovation Fund (CX(21)3109), Jiangsu seed Industry Revitalization Project (JBGS(2021)052).
Author information
Authors and Affiliations
Contributions
RR, XLZ and PZ conceived and designed the research. RR, XZ, XL, and YH collected the data and completed the bioinformatics analyses. RR and YH performed the experiments. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1
. Characteristic features of the 129 TLP-family genes identified in this Study. Table S2. Duplicated TLP gene pairs in the present study. Table S3. RNAseq data on differentially expressed genes between the F. graminearum inoculation and mock inoculation scenarios at different time points in the resistant near-isogenic wheat line NIL38. Table S4. RNAseq data on differentially expressed TLP genes between the F. graminearum inoculation and mock inoculation scenarios at different time points in the resistant near-isogenic wheat line NIL38. Table S5. Primers used for qRT–PCR of 9 TLP genes.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ren, R., Zhou, X., Zhang, X. et al. Genome-wide identification and characterization of thaumatin-like protein family genes in wheat and analysis of their responses to Fusarium head blight infection. Food Prod Process and Nutr 4, 24 (2022). https://doi.org/10.1186/s43014-022-00105-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43014-022-00105-0