Abstract
Stony corals generate their calcium carbonate exoskeleton in a highly controlled biomineralization process mediated by a variety of macromolecules including proteins. Fully identifying and classifying these proteins is crucial to understanding their role in exoskeleton formation, yet no optimal method to purify and characterize the full suite of extracted coral skeletal proteins has been established and hence their complete composition remains obscure. Here, we tested four skeletal protein purification protocols using acetone precipitation and ultrafiltration dialysis filters to present a comprehensive scleractinian coral skeletal proteome. We identified a total of 60 proteins in the coral skeleton, 44 of which were not present in previously published stony coral skeletal proteomes. Extracted protein purification protocols carried out in this study revealed that no one method captures all proteins and each protocol revealed a unique set of method-exclusive proteins. To better understand the general mechanism of skeletal protein transportation, we further examined the proteins’ gene ontology, transmembrane domains, and signal peptides. We found that transmembrane domain proteins and signal peptide secretion pathways, by themselves, could not explain the transportation of proteins to the skeleton. We therefore propose that some proteins are transported to the skeleton via non-traditional secretion pathways.
Similar content being viewed by others
Introduction
Scleractinian corals, or stony corals, are the most prolific biomineralizers in phylum Cnidaria [1]. They are a key component of shallow-water tropical reefs, often forming massive structures that serve as the foundation of an ecosystem which hosts some of the more biodiverse communities on the planet [2, 3]. They are amongst the oldest biomineralizing metazoans, producing calcium carbonate (CaCO3) exoskeletons in the form of aragonite through biologically-directed mechanisms [4, 5], which makes up ≥ 95% of the entire skeletal mass, the remainder of which is the skeletal organic matrix (SOM) [6,7,8].
Biomineralization refers to the ability of a living organism to selectively exploit elements from its surrounding environment to build a biologically functioning crystalline structure [9, 10]. Biomineralizers can be found throughout all kingdoms of life, from bacteria [e.x., 11], to algae [e.x., 12], mollusks [e.x., 13], corals [e.x., 10] and mammals [e.x., 14]. The minerals formed through this process differ in structure from their non-biological counterparts and their formation is mediated by a variety of organic molecules (the SOM), which have been intensively studied since the 1960′s in diverse organisms [reviewed by [10]. The SOM consists of proteins, lipids, and polysaccharides, which are not present in the abiotic mineral form [reviewed by [15]. SOM proteins (SOMPs) embedded within the mineral are hypothesized to serve as a framework for crystal nucleation [reviewed by [16]. The proteins involved in the process of skeletal biomineralization have been described and studied extensively in echinoderms [e.x., 17, 18] [and reviewed by [19, 20], mollusks [e.x., 21,22,23,24] and mammals [e.x., 25,26,27], among others [28]. At present, the best described SOMP complex is of mammalian bone and teeth [15].
In stony corals, the most current knowledge of SOMPs is limited and based on intraskeletal protein extraction [29,30,31,32,33]. It has been suggested that coral SOMPs aid in the molecular processes of crystallization as well as in the development and strengthening of the minerals ([34,35,36,37] among others). The constant advancement of mass spectrometry technology has broadened our capability to identify many proteins in skeletal extracts, even those proteins in low abundance [38]. However, this technology is sensitive to contamination by organic matter remnants from soft tissue and cell debris from the study organism, and the little-addressed issue remains of contamination by researchers during the protein extraction, preparation, and sequencing steps [39,40,41]. Upon extraction, the SOMPs are usually divided into two fractions: soluble and insoluble matrix proteins (SSOM and ISOM respectively), based on their solubility in the acid of choice or in water [22, 31,32,33, 42]. While some past attention has been directed towards the soluble fraction [30, 43, 44], Pereira-Mouriès et al. [45] showed that, in the bivalve Pinctada maxima, the classification of SSOM and ISOM is misleading and that both fractions share common features. Furthermore, Goffredo et al. [42] found in the stony coral Balanophyllia europaea that both fractions consist of the same macromolecules; they associated the degree of solubility to the difference in cross-linking. They also showed that each solubility fraction has a different influence on calcium carbonate crystal morphology, aggregation, and polymorphism in vitro. In contrast, Ramos-Silva et al. [32] observed a different SOMP composition between solubility fractions in the scleractinian coral Acropora millepora,. Out of 36 SOMPs, only two were found exclusively in the soluble fraction and twelve were exclusive to the insoluble fraction. These examples demonstrate the attempts to attribute different properties to the two fractions, but the data remain inconclusive.
To date, three major coral skeletal proteomes have been published [31,32,33] with each proteome consisting of 30–40 proteins. Of the 30 proteins sequenced from A. digitifera skeleton, 26 were also detected in A. millepora skeleton [32, 33]. They consist mostly of either transmembrane (TM) domain proteins or secretory proteins [33]. However, only 12 of the proteins identified in A. millepora skeleton matched those found in S. pistillata skeleton [31, 32]. In A. millepora, 11 TM domain-containing proteins were identified, as well as two proteases that were not detected in S. pistillata [32]. The authors suggested that the proteases’ role is in cleaving the extracellular domain of TM proteins and incorporating them into the skeleton.
The coral skeletal proteomes published to date reveal an overlap of several detected proteins, but at least 10 proteins from each species appear to be unique. It is currently unknown if this is truly due to species-specific gene expression and protein localization or to methods in extracting, purifying, and sequencing the proteins. In this study we analyzed several methods for extracted protein purification to increase the detection of the full suite of SOMPs from cleaned coral skeleton powder. We show that the use of acetone precipitation versus centrifugal filter washing, and the degree to which each purification method is performed, affects the numbers and types of proteins that can be sequenced by mass spectrometry. Further, we suggest that there is no one ‘best’ method for coral skeletal protein purification to capture all SOMPs such that future research projects may need to utilize several preparation methods to detect the full breadth of proteins embedded in coral skeleton.
Methods
Sample collection and preparation for protein extraction
The hermatypic coral Stylophora pistillata (Esper, 1797) was collected under a special permit from the Israeli Natural Parks Authority in the waters in front of the H. Steinitz Marine Biology Laboratory, Eilat, Israel, Red Sea (29° 30 N, 34° 56 E), using SCUBA diving.
We fragmented one S. pistillata colony into small pieces, approximately 2 × 2 cm, with a diamond band saw. Coral fragments were transferred to 50-ml Falcon brand conical vials (Falcon tubes) and oxidized with 20 mL 1:1 of 30% H2O2: 3% NaClO solution for 1 h, during which 1.5 mL of 3% NaClO solution were gently added to the tubes every 20 min and continued the incubation overnight at room temperature following modified methods of Stoll et al. [46]. Fragments were washed five times with ultra-pure water for one minute each time and dried at 60 C˚ overnight. We crushed the cleaned fragments to ≤ 63 µm diameter with a mortar and pestle. Skeleton powder, in sterile Falcon tubes, was then oxidized and washed in ultra-pure water three more times (i.e., four complete rounds of oxidative cleaning) to ensure that no organic residue remained on the skeletal grains. In each cycle, the removal of the oxidizing or wash solution was performed by centrifugation at 5000 × g for 3 min at 4 C˚. Cleaned skeletal powder was then dried overnight at 60 C˚. We carried out all the described processes in a laminar flow biological hood (apart from oven drying) with all preparation tools and surfaces bleached to avoid contamination.
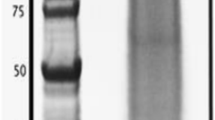
To monitor the removal of proteins from the skeletal powder, we checked the cleaning efficiency under SEM after the fourth oxidative cleaning. Samples were sputter-coated with 4 nm gold prior to examination using a ZEISS Sigma TM scanning electron microscope with in-lens detector (5 kV, WD = 5–7 mm) (Additional file 1: Figure 1a,b) [17, 47]. In addition, we sonicated the cleaned powder at 4 °C in filter-sterilized phosphate buffered saline (PBS, pH 7.4) for 30 min, pelleted the powder at 5000 x g for 3 min at 4 °C, concentrated the supernatant on a 3-kDa cutoff centrifugal filter unit (Amicon) and loaded samples of supernatant on a 8–16% SDS-PAGE TGX Stain-Free gels (Bio-Rad) (Additional file 1: Figure 1c).
Extraction and purification of skeletal proteins
We used four samples of approximately 1.3 g cleaned skeleton powder each to test four protein purification protocols as described in Fig. 1. All samples were decalcified in 0.5 M acetic acid (30 ml acid/g cleaned skeleton powder) in Falcon tubes while rotating the tubes at room temperature for 3 h. Samples were then centrifuged at 5000 x g for 5 min at 4 °C and supernatant was transferred to a new tube and stored at 4 °C. We continued the decalcification of the undissolved pellets with a second volume of 0.5 M acetic acid and allowed decalcification to proceed to completion, at which point the pH of the solution was measured at 5.5–6.5. We then combined both decalcification rounds (70 ml total) for each sample containing both the acid-soluble and -insoluble fractions, froze the total volumes at -80 °C, and dried them by overnight lyophilization. The dried pellets were stored at -80 °C until further processing. Because washes of the extracted proteins can reduce the representation of proteins, in this study we examined two protein concentrating and cleaning methods similar to those performed for previously published coral skeletal proteomes; (i) centrifugation ultrafiltration (CF methods [32]) and (ii) acetone precipitation (ACT methods, [31]) (Fig. 1; Additional file 1: Table 1).
To desalt and concentrate the extracted SOM, the lyophilized pellets of all samples were re-suspended in 12 ml MilliQ water and the proteins were concentrated on 3 kDa cutoff Amicon® Ultra 15 centrifugal filter units (Merck-Millipore) 5000 x g at 4 °C to reach a final volume of 0.5 ml. This process was repeated once. At this stage, we observed a water-insoluble pellet (ISOM) in all samples, which for samples CF2, ACT1, and ACT3 was pelleted from the water-soluble fraction (SSOM) by centrifugation at 5000 x g for 5 min at 4 °C. This initial wash, concentration, and solubility fractionation protocol was the terminal step for centrifugal filter sample CF2. Sample CF4 was resuspended one more time in MilliQ and concentrated before separation of SSOM and ISOM as above. For acetone precipitation samples ACT1 and ACT3, the two rounds of desalting and concentration were followed upon with successive washes of the SSOM and ISOM in acetone. To sample ACT1 SSOM was added 2 ml 100% ice cold acetone. The sample was vortexed for 10 s, incubated at − 20 °C for 30 min, and centrifuged at 4300×g for 30 min at 4 °C. The resulting pellet was washed three more times with 2 mL of ice cold 80% acetone. The ISOM fraction was washed four times with 80% acetone. Both solubility fractions of sample ACT3 were treated as in ACT1 but with one less washing step of each fraction. All fractions across all purification treatments were stored at −80 °C. An aliquot of each fraction was analyzed by SDS-PAGE on a 8–16% SDS-PAGE TGX Stain-Free gels with subsequent silver staining and displays the commonly-observed smearing of extracted biomineral proteins with minimal observable banding (Additional file 1: Figure 2).
LC MS\MS
Stylophora pistillata skeletal protein samples were dissolved in 5% SDS and digested with trypsin using the S-trap method overnight at room temperature. We analyzed the resulting peptides using a nanoflow ultra-performance liquid chromatograph (nanoAcquity) coupled to a high resolution, high mass accuracy mass spectrometer (Fusion Lumos). The sample was trapped on a Symmetry C18 0.18*20 mm trap column (Waters, Inc) and separated on a HSS T3 0.075*250 mm column (Waters, Inc.) using a gradient of 4–28% (80% acetonitrile, 0.1% Formic acid) for 150 min. Spray voltage was set to +2 kV. The data were acquired in the Fusion Lumos using a Top Speed Data-Dependent Acquisition method using a cycle time of 3 s. An MS1 scan was performed in the Orbitrap at 120,000 resolution with a maximum injection time of 60 ms. The data were scanned between 300 and 1800 m/z. MS2 was selected using a monoisotopic precursor selection set to peptides, peptide charge states set to +2 to +8 and dynamic exclusion set to 30 s. MS2 was performed using HCD fragmentation scanned in the Orbitrap, with the first mass set to 130 m/z at a resolution of 15,000. Maximum injection time was set to 60 ms with automatic gain control of 5 × 10−4 ions as a fill target. The resulting data were searched against the NCBI Stylophora pistillata protein database using the Byonic search engine (Protein Metrics Inc.)—the first search was carried out without any false discovery rate (FDR) filtering, to generate a focused database for a second search. The second search was set to 1% FDR, allowing fixed carbamidomethylation on C and variable oxidation on MW, deamidation on NQ and protein N-terminal acetylation. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the Pride partner repository [48], under the dataset identifier PXD017891.
Data sorting
We used the S. pistillata genome database as a reference peptide database for the mass spectrometry analysis [49] (NCBI BioProjects PRJNA281535 and PRJNA415215) appended with known NCBI S. pistillata skeletal proteins[e.x., 31, 50] S. pistillata carbonic anhydrase, ACE95141.1). We also included a common contaminants database. Despite this inclusion of a contaminants database, several proteins likely of human origin were sequenced and attributed to S. pistillata. To filter out these potential contaminants from our final list of coral-specific proteins, we BLASTed all sequences against the ‘Primates’ database in NCBI using Blast2GO. We then examined NCBI-generated sequence alignments of coral versus Homo sapiens proteins with e-values lower than e−50 and percent mean similarity greater than 50%, all sequences with e-values lower than e−100, and all sequences with percent similarity greater than 80%, and removed from our final list of coral proteins any sequences with three or more peptides each of seven or more amino acids in length that were identical between S. pistillata and humans.
All remaining coral-specific proteins identified by the LC MS\MS analysis were filtered to those with at least two significant peptides or at least one significant peptide with at least 10 spectra and an identification score of 250 or greater. Skeletal proteins were first sorted by fractions and methods (i.e., SSOM and ISOM for each purification method). Next, we sorted all skeletal proteins by their gene ontology (GO) terms [51, 52]. Finally, we grouped all by terms of interest: protein modification, transmembrane, and ECM; membrane processing; metal binding and vesicular.
Proteins detected by LC–MS/MS were annotated using the Trinotate pipeline which relies on both Pfam and UniProt data [53]. GO terms and Pfam annotations were assigned to Stylophora pistillata predicted proteins and transcripts using Trinotate 3.0.1 (https://github.com/Trinotate). Transmembrane regions were predicted using the TMHMM server v2.0 [54]. Signal peptides on the N-termini of proteins were predicted using SignalP 5.0 [55, 56]. Attachment of proteins to the exterior of the cell membrane by glycosylphosphatidylinositol (GPI) anchors was predicted using PredGPI [57]. These computational analyses used program default settings and cutoffs. Completeness of protein sequences was determined by comparing all returned coral proteins to the Acropora digitifera genome [58]; NCBI RefSeq assembly GCF_000222465.1). Detection of various proteins across solubility and protocol fractions was visualized in Venny 2.1 online software.
To examine the conserved coral biomineralization proteins, we determined orthologous biomineralization genes, known from skeletal proteomic analysis, across coral taxa. First, we estimated orthology relationships between all non-redundant genes of selected metazoa species using OrthoFinder. We included all genes of all Cnidaria species with known genome-based annotations. OrthoFinder generates orthology groups (Orthogroups) based on normalized reciprocal best BLAST hits’ bit scores [59], and then estimates orthologues genes pairs within Orthogroups [60]. We then selected all pairs of Acropora spp. orthologs to S. pistillata (1:1, 1:many, many:many relationships) ([31,32,33], this study). From these pairs we further selected S. pistillata spectra-based identified proteins, or skeletal proteins known in the literature. Since not all skeletal protein annotations from the literature were included in our reference OrthoFinder proteome datasets, we further found their best matches in the reference OrthoFinder proteome using BLASTP.
Results
After extensive cleaning of the powdered skeleton and acid-extraction of embedded organic matter we identified in total 60 coral-specific proteins meeting our criteria in S. pistillata skeleton as predicted by the species genome [49] (Table 1, Additional file 2: Table 2). Trinotate [53, 61] returned GO annotations [51] for all of these proteins, although many remain uncharacterized (Additional file 3: Table 3).
In order to evaluate the efficacy and improve current methods for stony coral skeletal protein purification, we examined four different protocols; two centrifugation ultrafiltration filters (CF) and two further acetone precipitation (ACT) protocols. Proteomes of the four methodologies differed in composition and variety (Fig. 2). Combining results of all CF fractions identified 52 coral-specific proteins while combined ACT protocols yielded 13 such proteins (Fig. 2a). Moreover, redundancy between methodologies was low. Only 8.3% of the proteins overlapped between methods while 78.3% and 13.3% of the proteins were exclusive to combined CF and combined ACT fraction data, respectively.
Distribution of proteins numbers according of the purification methods. Distribution of all proteins (SOM and ISOM combined) by purification methods (a). SOM and ISOM distribution by acetone precipitation methods (ACT; b). SOM and ISOM distribution by centrifugation and ultrafiltration method (CF; c). Note that CF methods yielded more proteins in total. ISOM Insoluble Skeletal Organic Matrix; SOM Skeletal Organic Matrix
To evaluate the purification efficiency of each protocol, we first compared the number of proteins detected in each method (e,x,, ACT1 vs ACT 3 and CF2 vs CF4). Of the 8 proteins found only in samples from the acetone wash protocols (ACT 1 and ACT 3), one was observed only in ACT1 while five were observed only ACT3 (Fig. 2, Additional file 2: Table 2); both ACT1 solubility fractions had one more wash step than did those in ACT3. In contrast, of the 47 proteins observed only in CF samples, 12 were found only in CF2, which went through two filter centrifugation steps, while 16 were found only in CF4, which went through a third filter centrifugation step; the remaining 19 were found using both CF protocols. Further, only two proteins were observed in all four purification fractions in at least one solubility form: CARP4/SAARP1 and synapsin 2-like. Two additional proteins were observed in three of the four protocols: STPCA2 and an uncharacterized protein.
We next examined the difference in protein composition obtained from the solubility fractions SSOM versus ISOM. We identified different distributions of proteins in the SSOM versus ISOM in both purification methods (Fig. 2b, c). Notably, when combining all methods, more proteins were identified in the ISOM compared to the SSOM. A total of 57 coral proteins were identified in the ISOM fraction compared to a total of 8 in the SSOM fraction (Additional file 2: Table 2), of which 5 (8.3%) were identify both in SSOM and ISOM.
Our data compared to other coral skeletal proteomes
Since our analysis yielded a large amount of new skeletal proteins, we also compared our results with the three previously published proteomes of S. pistillata, A. digitifera and A. millepora [31,32,33]. Out of our entire identified skeletal proteome containing 60 proteins, using OrthoFinder and BLASTP, only 16 were found to be similar to proteins identified in these studies. Yet, this proportion of overlap (16 out of 60) is significantly greater than the expected proportion by chance, since the proportion of known skeletal matrix proteins in the reference coral proteomes is extremely small (less than ~ 0.2%). Seven proteins were found to overlap all four proteomes: a coadhesin-like protein, an EGF and laminin G domain-containing protein, a hypothetical protein, a MAM and LDL-receptor class A domain-containing protein, a mucin, aspartic acid-rich protein 2-like, and a ZP domain-containing protein (Table 2).
Skeletal proteome characterization
We interrogated the mechanisms by which proteins may be exported from or attached to the cell (Additional file 2: Table 2). Seventeen coral skeletal proteins with likely complete N-terminus predictions possess signal peptides as a potential mechanism for export from the cell. Eight proteins contain at least one transmembrane span suggesting that they are embedded in the cell membrane. Further, 10 proteins likely interact with the exterior of the cell membrane by GPI anchors. In total, 25 of the 60 sequenced coral skeletal proteins exhibit documented characteristics for localization in the ECM.
Because the majority of the proteins sequenced from the S. pistillata skeleton do not possess features for signaling their export from the cell, we queried the data set for further suggestions of positioning the proteins in the membrane or that the proteins may be exported by vesicles such as those that may be involved in calcium concentration. To do this, we examined the skeletal proteome annotations and GO classifications toward finding common features to allow grouping of proteins. Out of the entire skeletal proteome sequenced in this study (60 proteins), 39 genes were returned with GO terms that allowed their classification into five groups of interest based on their cellular component, biological process, and molecular function to suggest likely cellular locations pertinent to the calcification mechanism, which may therefore be indicative of their function in this process: lipid\phosphate\glycan related proteins (i.e., membrane processing); ECM-related, transmembrane, and protein modification proteins; and metal binding proteins vesicular/secretion related proteins (Additional file 3: Table 3).
Of the proteins with GO terms, 10 proteins are suggested to be involved with processing of the cell membrane (Fig. 3), A much larger number are related to vesicles/secretion as well as binding metal, with 21 and 25 assigned to each category, respectively. We combined these two categories in our proposed cellular location Fig. 3, as some of the skeletal proteins proposed to be found intracellularly in vesicles are also known to bind calcium [34, 62]. Finally, 19 and 12 are potential ECM proteins or are involved in protein modification, respectively. Of the proteins in the vesicles/secretion and ECM categories, several appear to be completely predicted, based on comparison to orthologs in the A. digitifera genome, yet lack a signal peptide (Additional file 2: Table 2). It should be noted that many proteins are assigned to multiple categories.
Suggested cellular and extracellular locations of S. pistillata skeletal protein based on GO terms. Large yellow and red circles represent calicoblastic cells with red cell membrane facing the skeleton. Small green circles represent vesicles. Purple squiggles represent transmembrane proteins that may or may not span the width of the calicoblastic space plus ECM proteins
Discussion
In this study we show the importance of using complementary post-extraction methods to purify and concentrate coral skeletal proteins for sequencing the full breadth of the skeletal proteome. Our results show a clear and marked difference in detected proteins between protein purification methods, with only two proteins observed in all methods. Centrifugal Filter (CF) methods yielded a much greater abundance and diversity of proteins than did acetone precipitation (ACT) (Fig. 2). Moreover, we found that protein detection is largely method-exclusive (Fig. 2). We hypothesize that the protein yield differences between purification methods is based on the different properties of each method. CF is a mechanical filtration based on size and has a bias toward hydrophobic proteins, whereas ACT is based mainly on the chemical interactions of proteins resulting in increased precipitation of hydrophilic proteins [63]. We speculate that the CF methods yielded more total proteins in our study as acetone precipitation does not recover all proteins [64] and those proteins which are recovered in a pellet may be difficult to resolubilize due to molecular interactions formed within the aggregate by protein denaturation [65], potentially leading to the loss of many proteins in the pellet which are not transferred to the trypsin digestion step.
A major challenge in working with intra-skeletal proteins is isolating the true skeletal proteins from soft tissue contamination [40, 41]. In the present study, as in previous work on S. pistillata skeletal proteins [40] we carried out an intensive oxidative cleaning step on the skeletal powder, in addition to cleaning the skeletal fragments, to avoid contamination. We did not observe any organic residues on cleaned powders examined by SEM (Additional file 1: Figure 1a,b) or in PBS soaked on the powders and concentrated (Additional file 1: Figure 1c), and we are therefore confident that all sequenced proteins are endogenous to the skeleton and are not coral cellular contamination.
In this study, we examined the protein composition of the SSOM and ISOM proteins separately. Although we see variations due to the different purification techniques (Fig. 2), our results indicate that, broadly, the ISOM is distinguished in composition from the SSOM. Across all purification techniques, the ISOM fractions yielded more total proteins and fraction-exclusive proteins than the SSOM fraction did (Fig. 2b, c). Since all extraction and purification experiments were carried out simultaneously, we can rule out batch effect and technical differences. This ISOM versus SSOM skeletome fractional composition is similar to previous studies carried out on other marine biomineralizers such as mollusks [45] as well as to the stony corals A. millepora [32] and S. pistillata [31]. However, the overlap between the two solubility fractions reported for A. millepora [32] was 61%, in contrast to our study which shows an overlap across all purification treatments combined of only 10% (Fig. 2). One plausible explanation for this difference is the use of improved LC–MS/MS performance over the past several years enabled us to detect more proteins and obtain a more comprehensive proteome. In previous studies of coral skeletal proteomes which detailed differences between solubility fractions, the instruments used were low resolution, low mass accuracy, which tend to result in a lower percent identification of the data. Additionally, most search engines and FDR-calculating algorithms struggle with very small datasets [66, 67]. Our use of Byonic helps to alleviate statistical limitations that can result in false negative results.
Further differences in proteomes, beyond species differences, are the differing reagents used in precipitation (compared to [31]) and our smaller centrifugal filter cutoffs (compared to [32]). This second difference is particularly important with respect to protein degradation. Even when embedded in biominerals so that amino acids and even short peptides persist, proteins may still succumb to degradation [68]. This result in small peptide fragments that may be lost from centrifugal filter units with 10 kDa pore sizes and larger. It is reasonable to assume that coral skeletal proteins go through the same process, and if so, the cutoff of the membrane directly affects the number of peptide spectra matches (PSM). SDS-PAGE analysis shows smearing of extracted skeletal proteins, which in addition to likely differential addition of glycans, sulfates, and phosphates to proteins [e.x., 30, 69, 70], may indicate protein degradation (Additional file 1: Figure 2). Using smaller cutoff filters in this study might have allowed us to capture some of these sheered peptides and led to higher PSMs.
Traditional protein trafficking by signal peptides and transmembrane domain does not explain the full extent of protein transportation to skeleton
Corals’ skeleton is external to the animal; therefore, proteins in the skeletal matrices must be transported outside the cells or span the membrane and have an extracellular portion to reach the skeletal crystallization front. Indeed, recent studies of anthozoans reveal a significant proportion of TM domain proteins (~ 35%) in the SOM [32, 33], similar to that found in the better-studied Echinoidia [71]. Based on these findings, we examined the hypothesis that many of the skeletal proteins originate in the plasma membrane.
Our analysis revealed that TM domain proteins are not the major component of the SOM protein complex; in the present proteome, TMHMM prediction suggests that eight are embedded in the membrane. Hence, we examined a cellular secretion option. Ramos-Silva et al. [32] reported 15 proteins (41%) with SPs that did not also possess a TM domain in the A. millepora proteome. In their study of the A. digitifera proteome, Takeuchi et al. [33] reported a similar proportion of proteins with SPs but no TM domains (40%). In the present S. pistillata proteome, 17 out of 60 (28%) were positive for SPs (Additional file 2: Table 2, [31]), likely a combination of incomplete gene prediction and other mechanisms for exporting the proteins to the calicoblastic space.
Hydrophobic regions of TM domains are similar to those of SPs, making SP prediction difficult [54, 72]. We therefore cannot say for sure, based on these two analyses (TMHMM and SignalP) alone, which is the more dominant pathway for protein transport for many of the SOMPs. Further, while SP by itself can help predict traditional secretory pathways [72], proteins that do not possess classical SPs or TM regions may still leave the cell via other means (reviewed in [72,73,74]). This includes membrane pores [72], ATP-binding cassette transporters [75] and autophagosome/endosome-based secretion [73, 76], as well as exosomes [77] which can transport proteins that lack signal peptides in extracellular vesicles [78]. Since TM and SP analysis did not fully explain the mechanism of protein transportation, we classified the likely functions and locations of the skeletal proteins according to their GO terms as lipid\phosphate\glycan related proteins, metal binding proteins, vesicular/secretion related proteins, ECM-related and transmembrane proteins, and protein modification proteins (Fig. 3, Additional file 3: Table 3). Looking at the proposed locations or functions of skeletal proteins (Fig. 3), we find comparable numbers of vesicular and metal binding proteins versus TM domain/ECM proteins. Further, several of these proteins appear to be completely predicted at their N-termini yet lack signal peptides (Additional file 2: Table 2). These results are in-line with the trend reported in previous coral skeletal proteomes [31,32,33]. These proteins may leave the calicoblastic cells through one of the non-classical mechanisms described above. While we do not exclude the role of SP and TM proteins in the skeletal deposition process, we suggest that the biological mechanism of SOM transportation is enhanced by exosomes and other non-traditional secretion pathways.
A further method by which proteins may be exported to the site of calcification is a vesicular pathway that differs from the conventional SP and TM pathway and remains to be fully characterized. Previous studies have shown Ca2+ rich granules in the calicoblastic epithelium (skeletogenic cells), but not in the other tissue layers, suggesting their role as a Ca2+ reservoirs in the cells. Vesicles were previously identified in corals [79, 80]; however, their origin and content was not detailed, and they are sometimes attributed to preservation byproducts. These intracellular ion-rich vesicles may endocytose sea water through macropinocytosis [81, 82], after which they are enriched in carbonate ions and then form hydrated ACC and anhydrous ACC precursors stabilized by acidic biomolecules including CARPs [83]. Using cell cultures, Mass et al. [62] suggest that the vesicles, which contain Asp rich proteins, then transport their contents to the ECM, releasing their content by exocytosis. The biomineral then further develops extracellularly, likely aided by other ECM proteins [30, 35, 50, 84] as well as other biomolecules [85,86,87,88]. At present the processes of calcium delivery to the skeleton and the roles of most of the proteins in coral skeletal deposition remain to be determined.
It is difficult to map and characterize proteins in non-model organisms such S. pistillata [89,90,91]. High quality proteomic mapping requires knowledge of phosphorylation, glycosylation, proteolytic cite activities and other modifications [31, 92,93,94], in order to create a more thorough database [89,90,91]. Further, many of the coral skeletal proteins reported to date remain uncharacterized. Uncharacterized proteins were reported in Acropora skeletons at a rate of approximately 25% [32, 33] while, they were previously reported at less than 10% in S. pistillata skeleton [31]. Our study revealed a greater proportion, of 28% uncharacterized proteins, in S. pistillata skeleton, in line with Acropora spp. While partially attributed to sample size, it is most likely due to quality of genomic data available, since stony corals are non-model organisms and their genomic libraries are far from complete, resulting in incomplete databases on which to map the proteome and many uncharacterized genes.
Conclusion
In this study we have considered the differential effects of coral skeletal protein extract preparation as well as the method by which these proteins, or parts thereof, are transported from intracellular to extracellular locations. When preparing coral skeletal proteomes, we propose that a multi-method approach to cleaning, demineralization, and protein purification should be used. Our results showed that each protein preparation protocol yielded exclusive sets of proteins with little overlap between ACT and CF fractions. While CF protocols yielded many more proteins than did ACT methods, use of a single protocol to clean and concentrate coral skeletal proteins results in a significant amount of data loss, and it is therefore of crucial importance to consider alternative and complementary methods to obtain a fully comprehensive skeletal proteome. We showed that while the role of TM domain proteins cannot be overlooked, many of the proteins detected in the S. pistillata skeletal proteomes as well as in that of other species point toward other secretory or vesicular pathways. Our categorization method, supported by data from other recent studies, also suggests that corals use an alternative secretory pathway, such as exosomes or non-classical secretion vesicles, and much work is required in order to determine the calcium deposition pathway and the proteins involved. Our study provides a large set of new uncharacterized coral skeletal proteins as well as others of purported function but that have not been observed before in the coral SOM. These data expand the current knowledge of the SOM in corals and will help, in future studies, to resolve corals’ calcium deposition mechanism and the various roles of the proteins involved.
Availability of data and materials
The datasets generated during the current study are available in the ProteomeXchange repository under file number PXD017891 (https://www.ebi.ac.uk/pride/archive/projects/PXD017891).
Abbreviations
- ACT:
-
acetone precipitation of proteins
- CF:
-
centrifugation ultrafiltration of proteins
- FDR:
-
false discovery rate
- GO:
-
gene ontology
- GPI:
-
glycosylphosphatidylinositol
- ISOM:
-
insoluble skeletal organic matrix
- PSM:
-
peptide spectra matches
- PBS:
-
phosphate buffered saline
- SOM:
-
skeletal organic matrix
- SOMP:
-
skeletal organic matrix proteins
- SSOM:
-
soluble skeletal organic matrix
- TM:
-
transmembrane
References
Cairns SD. Deep-water corals: An overview with special reference to diversity and distribution of deep-water scleractinian corals. Bull Mar Sci. 2007;81:311–22.
Reaka-Kudla ML. The global biodiversity of coral reefs: a comparison with rain forests. In: Biodiversity II: understanding and protecting our biological resources 1997, 2:551.
Knowlton N, Brainard RE, Fisher R, Moews M, Plaisance L, Caley MJ. Coral reef biodiversity. In Life in the world’s oceans: diversity distribution and abundance; 2010:65–74.
Erwin DH, Laflamme M, Tweedt SM, Sperling EA, Pisani D, Peterson KJ. The Cambrian conundrum: early divergence and later ecological success in the early history of animals. Science. 2011;334:1091–7.
Tambutté S, Tambutté E, Zoccola D, Allemand D. Organic matrix and biomineralization of scleractinian corals. In: Handbook of biomineralization: biological aspects and structure formation 2007:243–259.
Cuif J-P, Dauphin Y, Berthet P, Jegoudez J. Associated water and organic compounds in coral skeletons: quantitative thermogravimetry coupled to infrared absorption spectrometry. Geochem Geophys Geosyst. 2004;5:Q11011.
Wainwright SA. Skeletal organization in the coral, Pocillopora damicornis. Quarterly Journal of Microscopical Science. 1963;s3-104:169–83.
Von Euw S, Zhang Q, Manichev V, Murali N, Gross J, Feldman LC, Gustafsson T, Flach C, Mendelsohn R, Falkowski PG. Biological control of aragonite formation in stony corals. Science. 2017;356:933–8.
Mann S. Biomineralization: principles and concepts in bioinorganic materials chemistry. New York: Oxford University Press; 2001.
Weiner S, Dove PM. An overview of biomineralization processes and the problem of the vital effect. Rev Miner Geochem. 2003;54:1–29.
Schüler D, Frankel RB. Bacterial magnetosomes: microbiology, biomineralization and biotechnological applications. Appl Microbiol Biotechnol. 1999;52:464–73.
Westbroek P, Young J, Linschooten K. Coccolith production (biomineralization) in the marine alga Emiliania huxleyi. J Protozool. 1989;36:368–73.
Addadi L, Joester D, Nudelman F, Weiner S. Mollusk shell formation: a source of new concepts for understanding biomineralization process. Chem Eur J. 2006;12:980–7.
Levine P, Glimcher M. The isolation and amino acid composition of the organic matrix and neutral soluble proteins of developing rodent enamel. Arch Oral Biol. 1965;10:753-IN751.
Beniash E. Biominerals—hierarchical nanocomposites: the example of bone. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2011;3:47–69.
Lowenstam HA, Weiner S. On biomineralization. New York: Oxford University Press; 1989.
Karakostis K, Zanella-Cléon I, Immel F, Guichard N, Dru P, Lepage T, Plasseraud L, Matranga V, Marin F. A minimal molecular toolkit for mineral deposition? Biochemistry and proteomics of the test matrix of adult specimens of the sea urchin Paracentrotus lividus. J Proteom. 2016;136:133–44.
Shashikant T, Khor JM, Ettensohn CA. From genome to anatomy: the architecture and evolution of the skeletogenic gene regulatory network of sea urchins and other echinoderms. Genesis. 2018;56:e23253.
Nebelsick JH, Dynowski JF, Grossmann JN, Tötzke C: Echinoderms: hierarchically organized light weight skeletons. In: Evolution of lightweight structures. Springer; 2015: 141–155.
Killian CE, Wilt FH. Molecular aspects of biomineralization of the echinoderm endoskeleton. Chem Rev. 2008;108:4463–74.
Weiner S, Hood L. Soluble protein of the organic matrix of mollusk shells: a potential template for shell formation. Science. 1975;190:987–9.
Mann K, Edsinger-Gonzales E, Mann M. In-depth proteomic analysis of a mollusc shell: acid-soluble and acid-insoluble matrix of the limpet Lottia gigantea. Proteome Sci. 2012;10:1–18.
Politi Y, Mahamid J, Goldberg H, Weiner S, Addadi L. Asprich mollusk shell protein: in vitro experiments aimed at elucidating function in CaCO3 crystallization. CrystEngComm. 2007;9:1171–7.
Bouyoucef M, Rakic R, Gómez-Leduc T, Latire T, Marin F, Leclercq S, Carreiras F, Serpentini A, Lebel J-M, Galéra P. Regulation of extracellular matrix synthesis by shell extracts from the marine bivalve Pecten maximus in human articular chondrocytes—application for cartilage engineering. Mar Biotechnol. 2018;20:436–50.
Weiner S, Wagner HD. The material bone: structure-mechanical function relations. Annu Rev Mater Sci. 1998;28:271–98.
Reid DG, Duer MJ, Murray RC, Wise ER. The organic–mineral interface in teeth is like that in bone and dominated by polysaccharides: universal mediators of normal calcium phosphate niomineralization in vertebrates? Chem Mater. 2008;20:3549–50.
Sharma V, Srinivasan A, Roychoudhury A, Rani K, Tyagi M, Dev K, Nikolajeff F, Kumar S. Characterization of protein extracts from different types of human teeth and insight in biomineralization. Sci Rep. 2019;9:9314.
Kamel BS, Voolstra CR, Medina M. BioMine-DB: A database for metazoan biomineralization proteins. PeerJ Preprints. 1983;2016:e1983v1982.
Fukuda I, Ooki S, Fujita T, Murayama E, Nagasawa H, Isa Y, Watanabe T. Molecular cloning of a cDNA encoding a soluble protein in the coral exoskeleton. Biochem Biophys Res Commun. 2003;304:11–7.
Puverel S, Tambutté E, Pereira-Mouries L, Zoccola D, Allemand D, Tambutté S. Soluble organic matrix of two Scleractinian corals: partial and comparative analysis. Compar Biochem Physiol Part B. 2005;141:480–7.
Drake JL, Mass T, Haramaty L, Zelzion E, Bhattacharya D, Falkowski PG. Proteomic analysis of skeletal organic matrix from the stony coral Stylophora pistillata. Proc Natl Acad Sci. 2013;110:3788–93.
Ramos-Silva P, Kaandorp J, Huisman L, Marie B, Zanella-Cleon I, Guichard N, Miller DJ, Marin F. The skeletal proteome of the coral Acropora millepora: the evolution of calcification by cooption and domain shuffling. Mol Biol Evol. 2013;30:2099–112.
Takeuchi T, Yamada L, Shinzato C, Sawada H, Satoh N. Stepwise evolution of coral biomineralization revealed with genome-wide proteomics and transcriptomics. PLoS ONE. 2016;11:e0156424.
Mass T, Drake JL, Haramaty L, Kim JD, Zelzion E, Bhattacharya D, Falkowski PG. Cloning and characterization of four novel coral acid-rich proteins that precipitate carbonates in vitro. Curr Biol. 2013;23:1126–31.
Puverel S, Tambutté E, Zoccola D, Dumart-Coulon I, Bouchot A, Lotto S, Allemand D, Tambutté S. Antibodies against the organic matrix in scleractinians: a new tool to study coral biomineralization. Coral Reefs. 2005;24:149–56.
Clode PL, Marshall AT. Calcium associated with a fibrillar organic matrix in the scleractinian coral Galaxea fascicularis. Protoplasma. 2003;220:153–61.
Watanabe T. Molecular analyses of protein components of the organic matrix in the exoskeleton of two scleractinian coral species. Comp Biochem Phys B. 2003;136:767–74.
Scheltema RA, Hauschild J-P, Lange O, Hornburg D, Denisov E, Damoc E, Kuehn A, Makarov A, Mann M. The Q Exactive HF, a Benchtop mass spectrometer with a pre-filter, high-performance quadrupole and an ultra-high-field Orbitrap analyzer. Mol Cell Proteom. 2014;13:3698–708.
Drake JL, Mass T, Falkowski PG. The evolution and future of carbonate precipitation in marine invertebrates: witnessing extinction or documenting resilience in the Anthropocene? Elem Sci Anthropocene. 2014;2:000026.
Drake JL, Mass T, Haramaty L, Zelzion E, Bhattacharya D, Falkowski PG. Reply to Ramos-Silva et al.: Regarding coral skeletal proteome. Proc Natl Acad Sci. 2013;110:E2147–8.
Ramos-Silva P, Marin F, Kaandorp J, Marie B. LETTER: Biomineralization toolkit: the importance of sample cleaning prior to the characterization of biomineral proteomes. Proc Natl Acad Sci. 2013;110:E2144–6.
Goffredo S, Vergni P, Reggi M, Caroselli E, Sparla F, Levy O, Dubinsky Z, Falini G. The skeletal organic matrix from Mediterranean coral Balanophyllia europaea influences calcium carbonate precipitation. PLoS ONE. 2011;6:e22338.
Dauphin Y. Comparative studies of skeletal soluble matrices from some scleractinian corals and molluscs. Int J Biol Macromol. 2001;28:293–304.
Weiner S. Aspartic acid-rich proteins: major components of the soluble organic matrix of mollusk shells. Calcif Tissue Int. 1979;29:163–7.
Pereira-Mouriès L, Almeida MJ, Ribeiro C, Peduzzi J, Barthélemy M, Milet C, Lopez E. Soluble silk-like organic matrix in the nacreous layer of the bivalve Pinctada maxima: a new insight in the biomineralization field. Eur J Biochem. 2002;269:4994–5003.
Stoll HM, Ruiz Encinar J, Ignacio Garcia Alonso J, Rosenthal Y, Probert I, Klaas C. A first look at paleotemperature prospects from Mg in coccolith carbonate: cleaning techniques and culture measurements. Geochem Geophys Geosyst. 2001;. https://doi.org/10.1029/2000GC000144.
Nothdurft LD, Webb GE. Earliest diagenesis in scleractinian coral skeletons: implications for palaeoclimate-sensitive geochemical archives. Facies. 2009;55:161–201.
Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–50.
Voolstra CR, Li Y, Liew YJ, Baumgarten S, Zoccola D, Flot J-F, Tambutté S, Allemand D, Aranda M. Comparative analysis of the genomes of Stylophora pistillata and Acropora digitifera provides evidence for extensive differences between species of corals. Sci Rep. 2017;7:17583.
Bertucci A, Tambutté S, Supuran C, Allemand D, Zoccola D. A new coral carbonic anhydrase in Stylophora pistillata. Mar Biotechnol. 2011;13:992–1002.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–9.
Consortium GO. Expansion of the gene ontology knowledgebase and resources. Nucleic Acids Res. 2017;45:D331–8.
Bryant DM, Johnson K, DiTommaso T, Tickle T, Couger MB, Payzin-Dogru D, Lee TJ, Leigh ND, Kuo T-H, Davis FG. A tissue-mapped axolotl de novo transcriptome enables identification of limb regeneration factors. Cell Rep. 2017;18:762–76.
Krogh A, Larsson B. Heijne Gv, Sonnhammer EL: predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–80.
Armenteros JJA, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol. 2019;37:420.
Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6.
Poisson G, Chauve C, Chen X, Bergeron A. FragAnchor: a large-scale predictor of glycosylphosphatidylinositol anchors in eukaryote protein sequences by qualitative scoring. Genom Proteom Bioinform. 2007;5:121–30.
Shinzato C, Shoguchi E, Kawashima T, Hamada M, Hisata K, Tanaka M, Fujie M, Fujiwara M, Koyanagi R, Ikuta T, et al. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature. 2011;476:320–3.
Emms DM, Kelly S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015;16:157.
Emms DM, Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20:1–14.
Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talón M, Dopazo J, Conesa A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420–35.
Mass T, Drake JL, Heddleston JM, Falkowski PG. Nanoscale visualization of biomineral formation in coral proto-polyps. Curr Biol. 2017;27(3191–3196):e3193.
Thongboonkerd V, Mcleish KR, Arthur JM, Klein JB. Proteomic analysis of normal human urinary proteins isolated by acetone precipitation or ultracentrifugation. Kidney Int. 2002;62:1461–9.
Fic E, Kedracka-Krok S, Jankowska U, Pirog A, Dziedzicka-Wasylewska M. Comparison of protein precipitation methods for various rat brain structures prior to proteomic analysis. Electrophoresis. 2010;31:3573–9.
Fields GB, Alonso DO, Stigter D, Dill KA. Theory for the aggregation of proteins and copolymers. J Phys Chem. 1992;96:3974–81.
Bern MW, Kil YJ. Two-dimensional target decoy strategy for shotgun proteomics. J Proteome Res. 2011;10:5296–301.
Fondrie WE, Noble WS. A machine learning strategy that leverages large datasets to boost statistical power in small-scale experiments. BioRxiv 2019:849331.
Sykes GA, Collins MJ, Walton DI. The significance of a geochemically isolated intracrystalline organic fraction within biominerals. Org Geochem. 1995;23:1059–65.
Goldberg WM. Acid polysaccharides in the skeletal matrix and calicoblastic epithelium of the stony coral Mycetophyllia reesi. Tissue Cell. 2001;33:376–87.
Cuif J-P, Dauphin Y, Doucet J, Salome M, Susini J. XANES mapping of organic sulfate in three scleractinian coral skeletons. Geochim Cosmochim Acta. 2003;67:75–83.
Mann K, Wilt FH, Poustka AJ. Proteomic analysis of sea urchin (Strongylocentrotus purpuratus) spicule matrix. Proteome Sci. 2010;8:33.
Von Heijne G, Abrahmsèn L. Species-specific variation in signal peptide design Implications for protein secretion in foreign hosts. FEBS Lett. 1989;244:439–46.
Rabouille C. Pathways of unconventional protein secretion. Trends Cell Biol. 2017;27:230–40.
Rabouille C, Malhotra V, Nickel W. Diversity in unconventional protein secretion. J Cell Sci. 2012;125:5251–5.
McGrath JP, Varshavsky A. The yeast STE6 gene encodes a homologue of the mammalian multidrug resistance P-glycoprotein. Nature. 1989;340:400–4.
Nickel W, Rabouille C. Mechanisms of regulated unconventional protein secretion. Nat Rev Mol Cell Biol. 2009;10:148–55.
Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–89.
Whitham M, Parker BL, Friedrichsen M, Hingst JR, Hjorth M, Hughes WE, Egan CL, Cron L, Watt KI, Kuchel RP. Extracellular vesicles provide a means for tissue crosstalk during exercise. Cell Metab. 2018;27(237–251):e234.
Clode PL, Marshall AT. Kalisomes in corals: a novel KCl concentrating organelle? Tissue Cell. 2002;34:199–209.
Marshall AT, Clode PL, Russell R, Prince K, Stern R. Electron and ion microprobe analysis of calcium distribution and transport in coral tissues. J Exp Biol. 2007;210:2453–63.
Mass T, Giuffre AJ, Sun C-Y, Stifler CA, Frazier MJ, Neder M, Tamura N, Stan CV, Marcus MA, Gilbert PUPA. Amorphous calcium carbonate particles form coral skeletons. Proc Natl Acad Sci. 2017;114:E7670–8.
Ganot P, Tambutté E, Caminiti-Segonds N, Toullec G, Allemand D, Tambutté S. Ubiquitous macropinocytosis in anthozoans. Elife. 2020;9:e50022.
Neder M, Laissue PP, Akiva A, Akkaynak D, Albéric M, Spaeker O, Politi Y, Pinkas I, Mass T. Mineral formation in the primary polyps of pocilloporoid corals. Acta Biomater. 2019;96:631–45.
Mass T, Drake JL, Peters EC, Jiang W, Falkowski PG. Immunolocalization of skeletal matrix proteins in tissue and mineral of the coral Stylophora pistillata. Proc Natl Acad Sci. 2014;111:12728–33.
Takeuchi T, Plasseraud L, Ziegler-Devin I, Brosse N, Shinzato C, Satoh N, Marin F. Biochemical characterization of the skeletal matrix of the massive coral, Porites australiensis—the saccharide moieties and their localization. J Struct Biol. 2018;203:219–29.
Puverel S, Houlbreque F, Tambutté E, Zoccola D, Payan P, Caminiti N, Tambutté S, Allemand D. Evidence of low molecular weight components in the organic matrix of the reef building coral, Stylophora pistillata. Compar Biochem Physiol A Mol Integr Physiol. 2007;147:850–6.
Farre B, Cuif J-P, Dauphin Y. Occurrence and diversity of lipids in modern coral skeletons. Zoology. 2010;113:250–7.
Naggi A, Torri G, Iacomini M, Colombo Castelli G, Reggi M, Fermani S, Dubinsky Z, Goffredo S, Falini G. Structure and function of stony coral intraskeletal polysaccharides. ACS Omega. 2018;3:2895–901.
Carpentier SC, Panis B, Vertommen A, Swennen R, Sergeant K, Renaut J, Laukens K, Witters E, Samyn B, Devreese B. Proteome analysis of non-model plants: a challenging but powerful approach. Mass Spectrom Rev. 2008;27:354–77.
Dowd WW. Challenges for biological interpretation of environmental proteomics data in non-model organisms. Integr Comp Biol. 2012;52:705–20.
Forné I, Abian J, Cerdà J. Fish proteome analysis: model organisms and non-sequenced species. Proteomics. 2010;10:858–72.
Butler WT, Brunn JC, Qin C. Dentin extracellular matrix (ECM) proteins: comparison to bone ECM and contribution to dynamics of dentinogenesis. Connect Tissue Res. 2003;44:171–8.
Qin C, Baba O, Butler WT. Post-translational modifications of SIBLING proteins and their roles in osteogenesis and dentinogenesis. Crit Rev Oral Biol Med. 2004;15:126–36.
Sarashina I, Endo K. Skeletal matrix proteins of invertebrate animals: comparative analysis of their amino acid sequences. Paleontol Res. 2006;10:311–36.
Acknowledgements
We thank staff at the Bioinformatics Core Unit, University of Haifa, and The Crown Genomics Institute of the Nancy and Stephen Grand Israel National Center for Personalized Medicine, Weizmann Institute of Science. We also thank Shahar Mass for production of Figure 3.
Funding
This project has received funding from the Israel Science Foundation (Grant 312/15) and the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No 755876). JLD was supported by the Zuckerman STEM Leadership Program. Funding bodies played no role in the design of the study; data collection, analysis or interpretation; or in writing the manuscript.
Author information
Authors and Affiliations
Contributions
YP, JD, and TM designed the study. YP and RA prepared the samples. YP, RA, AM, ML, DM, and JD analyzed the data. All authors wrote the manuscript and approve this submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1. Table 1
. Adapted from [Marie, 2013] and edited. A brief summary of different cnidaria; cleaning, demineralization and extraction protocols for skeletal matrix proteins. (ASM, Acid soluble matrix; AIM, Acid insoluble matrix; R.T, room temperature). Figure 1. Scanning electron micrographs (a, b) showing that S. pistillata ground to 63 μm and cleaned as described are free of visible exogenous contaminating organic material. Soaking the cleaned ground skeleton in PBS while sonicating for an hour also did not reveal any protein banding on TGX Stain-free gels (c). Lanes in (c) are: 1a = Precision Plus Unstained ladder at auto-exposure (~5 seconds); 1b = the same lane as (1a) but exposed for 20 seconds; 1c = concentrated PBS soak exposed at 20 seconds; gels were activated under UV light for five minutes. No protein smearing or banding was observed in the concentrated PBS soak of the cleaned skeleton powder. Figure 2. Protein gel of organic matrix proteins extracted from cleaned S. pistillata skeleton powders following UV activation for five minutes (A) and then silver staining (B).
Additional file 2.
Proteins sequenced from S. pistillata skeleton grouped by gene ontology cellular location and function. A given protein may be assigned to more than one group.
Additional file 3.
Byonic output of S. pistillata skeletal proteins sequenced by LC-MS/MS across all solubility groups and purification methods.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Peled, Y., Drake, J.L., Malik, A. et al. Optimization of skeletal protein preparation for LC–MS/MS sequencing yields additional coral skeletal proteins in Stylophora pistillata. BMC Mat 2, 8 (2020). https://doi.org/10.1186/s42833-020-00014-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s42833-020-00014-x