Abstract
Targeted genome editing technology is becoming one of the most important genetic tools and widely employed in the plant pathology community. In recent years, CRISPR (Clustered regularly interspaced short palindromic repeats) and CRISPR-associated proteins discovered in the adaptive immune system in prokaryotes have been successfully reprogrammed into various genome editing tools and have caught the attention of the scientific community due to its simplicity, high efficiency, versatility. Here, we provide an overview of various CRISPR/Cas systems, the derived tools and their applications in plant pathology. This review highlights the advantages of knocking-out techniques to target major susceptibility genes and negative regulators of host defense pathways for gaining resistance to bacterial, fungal and viral pathogens in model and crop plants through utilizing the CRISPR/Cas-based tools. Besides, we discuss the possible strategies of employing the CRISPR-based tools for both fundamental studies on plant-pathogen interactions and molecular crop breeding towards the improvement of resistance in the future.
Similar content being viewed by others
Background
The world’s population will reach at least 9.8 billion by 2050, more and more food is needed to provide sufficient nutrients for the rising populations. Crops are susceptible to a larger set of pathogens (fungi, bacteria, oomycetes, viruses, etc.) causing severe economic loss. Thus, the development of plant resistance plays a key role in adjusting crop production to meet the global population requirements (Nejat et al. 2017; Dong and Ronald 2019). Understanding the molecular mechanisms of host-pathogen interactions has been a significant area of investigation in plant pathology for many years.
It’s well known that plant and pathogen are locked in a battle of recognition and evasion, in which a multilayered defense system including both pathogen-associated molecular patterns (PAMPs) -triggered immunity (PTI) and effector-triggered immunity (ETI) has evolved in plants to fight against invading pathogens for survival. Generally speaking, PTI is rapidly activated through the recognition of PAMPs by pattern recognition receptors (PRRs). This basal resistance response, which includes reactive oxygen species (ROS) production, callose deposition and transcriptional reprogramming, usually prevents the invasion of non-adapted pathogens. To counteract this, pathogens secrete effectors to interrupt PTI and modulate host cell physiology, resulting in effector-triggered susceptibility (ETS). However, in resistant plants effectors or its byproducts can be recognized by intracellular immune receptors and induce ETI (a robust resistance response) which is usually associated with localized plant cell death leading to pathogen arrest. Some virulent pathogens can overcome the host’s ETI response via loss and/or modification of ETI-eliciting effectors as well as meta-effector interactions (Chisholm et al. 2006; Laflamme et al. 2020). Thus, the pathogen and the host play an endless arms race game between them. Despite decades of research, we still have only a limited understanding of the molecular mechanisms of host-pathogen interactions. Therefore, further extensive investigations of host-pathogen interactions, especially the identification of key targets related to defense responses in plants would provide a great opportunity to engineer broad-spectrum and durable resistance in many crops.
Gene loss-of-function mutants and gain-of-function germplasms are important resources for gene function studies and crop genetic improvements (Borisjuk et al. 2019). In recent years, genome-editing (GE) technologies with homing endonucleases (Meganucleases), zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) and newly-emerged CRISPR/Cas systems, which enable precise DNA modification of the genome, have greatly transformed the researches on plants (Gaj et al. 2016). The CRISPR/Cas9 system was initially identified as an RNA-mediated adaptive immune system against viral invasion in bacteria and archaea (Rath et al. 2015). Currently, CRISPR/Cas9 has been adapted for genome engineering in diverse eukaryotes due to the simplicity and high efficiency, becoming an alternative to ZFNs or TALENs. To date, CRISPR-based tools have been employed in a wide range of plant-pathogen interaction studies, including host responses to bacteria, fungi, oomycetes, viruses, etc.
CRISPR technology has provided new opportunities to identify important defense-related genes in plants and improve crop resistance to pathogens to ensure food safety and sustainable agricultural development in the future. In this review, we summarize the main features of various CRISPR-based genome editing techniques and address their recent applications in a large number of plant-pathogen interactions. Also, we discuss potential applications of CRISPR/Cas9 tools in defense-related gene dissection and crop improvement.
An overview of CRISPR-based tools
Based on phylogenetic, structural, and functional characteristics of Cas proteins, CRISPR/Cas systems are categorized into types I, III and IV (Class 1), and types II, V, and VI (Class 2) with distinct mechanisms of guide RNA biogenesis and target interference (Makarova et al. 2015, 2018; Garcia-Doval and Jinek 2017), in which the Class 2 CRISPR-associated nucleases have been extensively adapted and utilized for nucleic acid manipulation (Garcia-Doval and Jinek 2017; Ji et al. 2019) (Table 1 and Fig. 1).
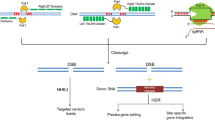
Diagrammatic representation of the cleavage site architecture of different CRISPR system. sgRNA consists of a spacer (light blue), crRNA (purple) and tracrRNA (dark orange). Optimal PAMs (orange) and PFSs (red) are important for recognizing target of these corresponding systems. Cas12a, Cas12b, and Cas14 are validated subtypes in type V and each shows a distinct architecture. Only crRNA is required for Cas12a, while the other subtypes require an additional tracrRNA. Cas14 cleaves ssDNA without PAM specificity. Cas13a and Cas13b belong to subtype VI with recognition of the PFS sequence, the crRNA-Cas13 complex induces ssRNA cleavage activity. Cas9, CRISPR associated protein 9; crRNA, CRISPR RNA; sgRNA, single guide RNA; tracrRNA, transactivating CRISPR RNA; PAM, protospacer adjacent motif; PFS, protospacer flanking sites
The type II CRISPR/SpCas9 system is the best characterized and widely used for plant targeted genome targeting. It is derived from S. pyogenes and originally consists of three components: the SpCas9 nuclease, a trans-activating small RNA (tracrRNA), and a small mature CRISPR RNA (crRNA) (Bhaya et al. 2011). To date, the optimized system only contains two components, a SpCas9 protein and an easily-engineered guide RNA (gRNA), an artificial fusion RNA with both the custom-designed crRNA and the scaffold tracrRNA (Jinek et al. 2012; Cong et al. 2013). SpCas9 is guided by the gRNA to the specific DNA site which contains the sequence complementary to the first 17–20 nucleotides of the gRNA and is followed by an NGG PAM (protospacer adjacent motif). Upon binding, the HNH and RuvC-like nuclease domains of SpCas9 cleaves both strands of the DNA (complementary and non-complementary to the guide RNA) at exactly -3 position upstream of PAM, resulting in a blunt-ended DNA double-strand break (DSB) (Jinek et al. 2012; Hsu et al. 2014). One of the most significant limitations of the CRISPR/SpCas9 application is the requirement of the PAM sequence for DNA target cleavage. Therefore, many Cas9 variants and orthologues with different PAM specificities, such as ScCas9, SaCas9, SpCas9-NG, xCas9, SpCas9(VQR), SpCas9-NRRH, SpCas9-NRCH, etc., have been isolated, greatly expanding the targeting scope of genome-editing tools in plants (Hu et al. 2016, 2018; Ren et al. 2017, 2019; Chatterjee et al. 2018; Hua et al. 2018, 2019; Wang et al. 2020). Other than that, FnCas9 from F. novicida, which is unlike ScCas9 and recognizes a YG PAM, was implicated in RNA targeting (Price et al. 2015).
CRISPR/Cas12 is the second most well-documented CRISPR system. It is classified as type V and has distinct evolutionary origins and structural architecture from Cas9. To date, some RuvC-like domain-containing Cas12 (i.e. Cas12a, b, c, d, e, g, h, i, k, etc.) have been defined with diverse functions, and both CRISPR/Cas12a (Cpf1) and CRISPR/Cas12b (C2c1) have been successfully applied for genome editing in plants (Zetsche et al. 2015; Zaidi et al. 2017; Ming et al. 2020). Cas12a is guided by a single mature crRNA in DNA targeting (Zetsche et al. 2015), whereas Cas12b requires the presence of both tracrRNA and crRNA to function (Teng et al. 2018; Strecker et al. 2019). Both Cas12a and Cas12b recognize a T-rich PAM and generate 4–5 nt long staggered ends distal to the PAM (Yang et al. 2016; Garcia-Doval and Jinek 2017; Teng et al. 2018; Strecker et al. 2019). Cas14, another type V protein isolated from non-culturable archaea, has been identified and reported to cleave single-stranded DNA (ssDNA) without restrictive sequence requirements. Thus, Cas14 is an ideal tool for engineering resistance against ssDNA plant viruses and high-fidelity SNP genotyping due to its sequence-independent and unrestricted cleavage (Harrington et al. 2018; Khan et al. 2019).
The type VI CRISPR/Cas13 systems function as a ribonuclease and have been divided into four subtypes, including type VI-A (Cas13a), VI-B (Cas13b), VI-C (Cas13c) and VI-D (Cas13d) (Abudayyeh et al. 2016, 2017; Cox et al. 2017; Shmakov et al. 2017; Konermann et al. 2018; Zhang et al. 2018a, 2018b). Cas13 proteins target RNA rather than DNA molecules and are characterized by two distinct HEPN RNase domains and assembled with single crRNA to form a crRNA-guided protein complex for RNA targeting. Similar to PAM for Cas9 and Cas12, 5′- and/or 3′-protospacer-flanking site (PFS) are needed for redirecting the Cas13/crRNA complex to the target site. Cas13 also showed the promiscuous ability of the RNase to cleave collateral RNAs once activated in the presence of template targets. Cas13 proteins are being used to localize, detect, and track RNA molecules of different types.
Single nucleotide polymorphisms (SNPs) are the genetic basis of many important agronomic traits variations (i.e. pathogen resistance, yield) of crop plants. Therefore, a series of base editing tools, including cytidine base editor (CBE) and adenine base editor (ABE), have been developed to achieve specific base substitution in plants (Ren et al. 2017; Yan et al. 2018). It has been engineered by fusing various cytidine deaminase and/or adenine deaminase to impaired Cas9 (Cas9 nickase or dead Cas9), which induces cytosine-to-thymine (C > T) and adenine-to-guanine (A > G) substitutions in the target region in genome, respectively (Komor et al. 2016; Gaudelli et al. 2017). Besides, prime editors, constructed with Moloney murine leukemia virus reverse transcriptase (M-MLV RT) and Cas9 nickase, have been further developed, enabling all 12 base-to-base conversions as well as indels and insertions (Anzalone et al. 2019; Lin et al. 2020).
Genome editing in plant pathogens
The development of efficient genome-editing techniques, particularly CRISPR/SpCas9, has emerged a broad array of probable uses that could be explored in plant pathogens. The ability to modify plant pathogen genomes offers the possibility to confer desirable phenotypes for numerous purposes (Zhang et al. 2018). Compared to traditional methods for genetic manipulation of the microbial genome which are usually associated with inefficient homologous recombination, the CRISPR/SpCas9 tools are highly efficient and much simpler in some cases. Besides, they provide a high-throughput experimental platform to dissect gene function at the whole-genome level in plant pathogens.
After the first example of genome editing with high efficiency in Escherichia coli was reported (Jiang et al. 2013), the application of CRISPR technology has been greatly expanded into other bacterial species such as Pseudomonas, Yersina, Bacillus, Streptomyces and Corynebacterium (Liu et al. 2019). For example, CRISPR/SpCas9 and CRISPR/FnCas12a systems have been established in P. putida KT2440. Both systems are successful in achieving gene deletions, gene insertions, and gene replacements with efficiency as high as 100% (Aparicio et al. 2018; Sun et al. 2018). Furthermore, a base editing system pnCasPA-BEC was developed by engineering cytidine deaminase APOBEC1 to the Cas9 nickase, which allowed a highly-efficient C > T substitution in P. aeruginosa, P. putida, P. fluorescens, Staphylococcus aureus and P. syringae (Chen et al. 2018; Gu et al. 2018). Besides, dCas9-based transcription inhibition system (CRISPRi) was recently applied for target gene repression in Pseudomonas spp. using a deactivated SpCas9 (Bikard et al. 2013; Tan et al. 2018).
The CRISPR/SpCas9-mediated genome editing technology has been successfully established in a wide range of fungal species, including Magnaporthe oryzae (Arazoe et al. 2015; Foster et al. 2018), Alternaria alternata (Wenderoth et al. 2017), Leptosphaeria maculans (Idnurm et al. 2017), Fusarium oxysporum (Wang et al. 2018; Wang and Coleman 2019), Fusarium graminearum (Gardiner and Kazan 2018), Fusarium fujikuroi (Shi et al. 2019), Fusarium proliferatum (Ferrara et al. 2019), Sclerotinia sclerotiorum (Li et al. 2018), Colletotrichum sansevieriae (Nakamura et al. 2019), B. cinerea (Leisen et al. 2020), Sporisorium scitamineum (Lu et al. 2017), Ustilaginoidea virens (Liang et al. 2018), Ustilago maydis (Schuster et al. 2016) and Ustilago tricophora (Huck et al. 2019). In fungi, both SpCas9 and the sgRNA can be stably or transiently expressed by polyethylene glycol (PEG)-mediated transformation, Agrobacterium-mediated transformation, electroporation, and biolistic transformation (Schuster and Kahmann 2019). Alternatively, the SpCas9/sgRNA ribonucleoprotein (RNP) complex can be assembled in vitro and directly applied in M. oryzae and F. oxysporum (Foster et al. 2018; Wang et al. 2018).
CRISPR/SpCas9 tools have also been successfully established and applied for genome editing in oomycetes, such as P. sojae, P. capsici, P. palmivora and P. litchii. For example, a CRISPR/SpCas9-mediated genome editing method with or without donor DNA has been established for genetic manipulation of P. sojae through PEG-mediated protoplast transformation. In these studies, disruption of the RXLR effector Avr4/6 proteins prevented its recognition by the corresponding soybean R protein Rps4 and Rps6 (Dou et al. 2010; Barakate and Stephens 2016; Fang and Tyler 2016). Furthermore, CRISPR/SpCas9 was also used to knockout another RXLR effector gene PcAvh1 in P. capsici. In this study, inoculation of PcAvh1 mutants on N. benthamiana showed that PcAvh1 was required for the full virulence of P. capsici (Chen et al. 2019). CRISPR/SpCas9 system has also been used in P. palmivora to generate homozygous PpalEPIC8 mutants via Agrobacterium-mediated transformation. In their experiment, mutations in PpalEPIC8 decreased the pathogenicity of P. palmivora in papaya fruits, probably by inhibiting the action of papain (Gumtow et al. 2018). In oomycetes (i.e. P. sojae, P. capsici and P. litchii), the CRISPR/SpCas9-mediated genome editing system has been optimized by using the oxathiapiprolin-resistance gene PcMuORP1 as the selection marker. This has enabled a much higher efficiency for screening of transformants as compared to the conventional selection marker NptII (Kong et al. 2019; Schuster and Kahmann 2019; Wang et al. 2019).
Genome editing for plant disease resistance against bacterial, fungal and oomycete pathogens
During the infection process of bacteria, type III effectors are secreted into the plant cell (Büttner and He 2009). These effectors primarily interrupt the host’s defense pathways and/or activate the S genes for disease development (Yang et al. 2006; Zaidi et al. 2018). Therefore, both S genes and negative regulators of plant innate immune response are good target sites for CRISPR/Cas9-mediated gene editing to improve plant resistance. For example, knockout of the endogenous OsSWEET11 gene which is the target of TALEs in rice resulted in significantly enhanced resistance to X. oryzae pv. oryzae (Xoo) without altering pollen development (Kim et al. 2019). TALE-binding elements (EBEs) in the promoter region of both OsSWEET11 and OsSWEET14 were also targeted, the resulting rice lines carrying indels conferred robust resistance to most Xoo strains (Xu et al. 2019). Moreover, recent studies showed that multiplex genome editing of the promoter regions of OsSWEET11, OsSWEET13, and OsSWEET14 in three rice varieties confered broad-spectrum resistance to Xoo (Oliva et al. 2019). CRISPR/SpCas9 and CRISPR/Cas12 have also been used to generate plant resistance to other pathogens. For example, genome editing of the promoter region of the S gene CsLOB1 in citrus, which is the target of TALE from X. citri pv. citri (Xcc), confered a high degree of resistance to the citrus canker disease (Jia et al. 2016, 2017; Peng et al. 2017). DMR6 is essential for resistance to downy mildew in A. thaliana, knockout of its homolog SIDMR6–1 in tomato by CRISPR/SpCas9 rendered plant resistant to different bacterial pathogens, including P. syringae, P. capsici and Xanthomonas spp. (Thomazella et al. 2016). Besides, the SlJAZ2Δjas tomato germplasm generated by CRISPR/SpCas9 provided resistance to P. syringae pv. tomato DC3000 without altering its defense response to the necrotrophic fungal pathogen Botrytis cinerea (Ortigosa et al. 2019)
Host colonization by fungal pathogens is a complex process. To date, much attention has been paid to S genes and negative regulators involved in the defense pathway. For example, the mildew resistance locus O (MLO) is a well-known S gene loci originally identified in barley (Jørgensen 1992). CRISPR/SpCas9-mediated knockout of its homologs in wheat (TaMLOs) resulted in resistance against the powdery mildew fungal pathogen Blumeria graminis f.sp. tritici (Wang et al. 2014). Furthermore, knockout of its homologs in tomato (SlMLO1) confered resistance to the powdery mildew fungus Oidium neolycopersici (Nekrasov et al. 2017). Likewise, given that EDR1 plays a negative role in the defense response against powdery mildew in A. thaliana, CRISPR/SpCas9 was used to simultaneously knock out three TaEDR copies, developing powdery mildew resistance in wheat (Frye et al. 2001; Zhang et al. 2017). Another example is OsERF922, which encodes a transcription factor belonging to the APETALA2/ethylene response factor (AP2/ERF) superfamily in rice, and has been reported to act as both positive and negative regulators in plant defense against different pathogens (Langner et al. 2018). Wang and colleagues employed CRISPR/SpCas9 to edit OsERF922, and the resulting rice lines carrying different frameshift indels showed enhanced resistance to the rice blast fungus M. oryzae without affecting plant development and other agronomic traits tested (Wang et al. 2016). Recently, CRISPR/SpCas9 has also been used to knock out the BSR-K1 gene in rice, resulting in resistance to both M. oryzae and Xoo (Zhou et al. 2018).
Besides the knockout strategy mentioned above, defective R gene correction through CRISPR/Cas9-mediated precise base editing is another efficient and time-saving way to improve crop disease resistance. For example, an SNP at position 441 in the recessive allele of Pi-d2 has been reported to be associated with the resistance to M. oryzae. The recessive allele pi-d2(M441) in rice results in loss of resistance to the Chinese blast isolate ZB15. On the other hand, incorporation of the dominant R gene Pi-d2(I441) into the rice variety Digu confered gene-for-gene resistance to ZB15 (Chen et al. 2006). Based on these findings, Ren and colleagues employed a cytidine base editor rBE5 to rapidly correct the recessive pi-d2 gene in rice variety Kitaake by introducing a G > A substitution (M411I) into the genome (Ren et al. 2018). This fast and precise genome engineering method can greatly speed up the resistance breeding program as compared to traditional hybrid breeding and marker-assisted selection methods.
As fungal-like eukaryotes, Oomycetes belong to the kingdom Chromista. The most representative pathogenic oomycetes are Phytophthora spp., Pythium spp., and Peronospora spp. CRISPR/Cas tools have been successfully established and applied for controlling plant oomycete disease, which mainly focuses on editing the effectors of Oomycetes, and editing the plants to improve the resistance to oomycetes is limited. The only example came from Fister and coworker who developed a CRISPR/Cas system to target TcNPR3 by transient expression in Theobroma cacao leaves, gene editing in 27% of TcNPR3 copies was achieved and the gene-edited leaves showed enhanced resistance to the pathogen P. tropicalis, and stably genome-edited somatic embryos were obtained via Agrobacterium-mediated transformation (Fister et al. 2018). Given that NPR3 and NPR4 serve as redundant transcriptional co-repressors for salicylic acid-responsive defense genes in A. thaliana, it’s worth editing their homologs in other crops to develop novel resistant germplasms.
Genome editing in the study of plant-virus interactions
The CRISPR/Cas-mediated gene-editing technology has been developed rapidly and has been used to explore effective ways for engineering virus resistance. Virus resistance can be accomplished by (1) targeting host factors that are involved in the replications of the virus or (2) targeting and destroying the viral genome itself and thus preventing the replication of the virus.
The eukaryotic translation initiation factor 4E (eIF4E), also known as a cap-binding protein, has been proven as a susceptibility factor in plant-virus interactions. Disruption of eIF4E gene results in innate immunity against many potyviruses (virus belonging to the family of Potyviridae) in various plant species. For example, CRISPR/SpCas9-mediated eIF4E-edited cucumber plants exhibited resistance to zucchini yellow mosaic virus (ZYMV), papaya ringspot mosaic virus-W (PRSMV-W) and cucumber vein yellowing virus (CVYV) (Chandrasekaran et al. 2016). Moreover, mutagenesis of eIF4E alleles in cassava and A. thaliana by CRISPR/SpCas9 system reduced cassava brown streak disease symptom and turnip mosaic virus (TuMV) infection, respectively (Pyott et al. 2016; Gomez et al. 2019). Similarly, CRISPR/SpCas9 has also been used to mutate eIF4G in rice and generated resistance to rice tungro spherical virus (RTSV) (Macovei et al. 2018). Recently, a C > G conversion (N176K) was introduced into the wild-type eIF4E1 in A. thaliana by cytidine base editor, conferring resistance to clover yellow vein virus (ClYVV) (Bastet et al. 2019).
Geminiviruses, a group of circular single-stranded DNA viruses, cause acute damage to economically important crops, like tomato, sugar beet, and pepper (Langner et al. 2018). Many studies have been carried out to directly target geminiviral genomic DNA with CRISPR/SpCas9 (Kalinina et al. 2020). Constructs containing SpCas9 and sgRNAs which targeted the Rep (replication-associated protein) gene and the intergenic region (IR) in viral genomes of beet severe curly top virus (BSCTV) and bean yellow dwarf virus (BeYDV) were transformed into N. benthamiana and A. thaliana, respectively. The resulting plants exhibited high levels of resistance to the targeted virus (Baltes et al. 2015; Ji et al. 2015). Similarly, N. benthamiana and tomato plants expressing SpCas9 and sgRNA specific for CP (coat protein) or Rep sequences of tomato yellow leaf curl virus (TYLCV) exhibited significant virus resistance (Tashkandi et al. 2018). More studies reported that the CRISPR/SpCas9 system targeting MP, CP and any other conserved regions of viral genomes established wheat dwarf virus (WDV) resistance in barley and banana streak virus (BSV) resistance in banana, respectively (Kis et al. 2019; Tripathi et al. 2019).
RNA viruses can be also directly targeted by Cas protein (i.e. Cas13a and FnCas9) which target RNA molecules rather than DNA. For example, Aman and colleagues employed CRISPR/Cas13a to target and degrade the viral RNA of TuMV in N. benthamiana (Aman et al. 2018). Furthermore, Zhang and colleagues used CRISPR/FnCas9 to degrade the viral genome of cucumber mosaic virus (CMV) and tobacco mosaic virus (TMV) in transgenic N. benthamiana and A. thaliana plants, respectively (Zhang et al. 2018c). Similar approaches have been successfully applied to produce resistance to potato virus Y (PVY) in tobacco and resistance to rice stripe mosaic virus (RSMV) and southern rice black-streaked draft virus (SRBSDV) in rice (Zhang et al. 2019). Although CRISPR/Cas has been successfully employed to inhibit the viral growth in transgenic plants, the potential risks of viruses escaping the CRISPR/Cas9 cleavage and leading to loss of resistance caused by the fast-evolving virus are under concern. Mehta and colleagues reported that between 33 and 48% of edited virus genomes evolved a conserved single nucleotide mutation which conferred resistance to CRISPR/Cas9 cleavage, resulting in failed resistance to the geminivirus during glasshouse inoculations (Mehta et al. 2019).
Conclusions and future directions
Today, the CRISPR/Cas system is the most extensively used technology for targeted genomic editing compared to other GE technologies, and has been developed and applied in a large number of host plants and plant pathogens for dissecting the molecular mechanisms underlying the plant-pathogen interactions and for improving host resistance against bacteria, fungi, oomycetes, DNA viruses, and RNA viruses. The CRISPR/Cas system is a valuable tool for creating gene loss-of-function and gain-of-function mutants and for understanding plant-pathogen interactions as well as reducing the damage caused by the destructive pathogens in agricultural application.
CRISPR/Cas-based tools can be used for single and multiple gene knockouts. Moreover, CRISPR/Cas-based tools can also be used to create high-throughput mutant libraries, providing a powerful approach to accelerate gene function studies on plant resistance and pathogen pathogenesis. Therefore, the potential players involved in plant-pathogen interactions, such as resistance gene family, receptor-like kinase gene family, transcriptional factor gene family and differentially expressed transcripts corresponding to pathogen challenge, would be recommended to be targeted on a large scale by the highly-efficient CRISPR/Cas tools, and the crucial defense-related genes can be identified, characterized and further utilized in agricultural application.
SNPs and quantitative trait locus (QTL), abundant forms of genetic variation among individuals in crop species, are responsible for diverse pleiotropic phenotypes, including crop resistance. To date, crucial SNPs and SNP-typed QTLs have been identified in associated with a quite number of resistance and resistance-related genes in many crops, such as Pi-ta, Pi-d2, bsr-d1, bsr-k1, Xa4, Xa5, CsSGR, etc. CRISPR/Cas9-mediated base editors and prime editors can be employed to perform precise genome editing of SNPs and SNP-typed QTLs easily in cash crops, endowing multiple resistance to pathogens. Recently, the base-editing-mediated gene evolution (BEMGE) method was developed (Kuang et al. 2020). This novel crop breeding method can artificially evolve any endogenous gene in planta with a tiled sgRNA library corresponding to the target genomic locus. Thus, BEMGE is a promising method for the transformation of any functional gene related to plant defense response.
In conclusion, the CRISPR/Cas system and its derivatives provide a novel opportunity to explore the complex area of plant-pathogen interactions. Along with the continuous changes in agricultural production activities and plant disease systems, we expect that the CRISPR technologies will make a big contribution in deciphering the interaction between plant and pathogen and designing durable and broad-spectrum disease resistance plants in the future.
Availability of data and materials
Not applicable.
Abbreviations
- ABE:
-
Adenine base editor
- BEMGE:
-
Base-editing-mediated gene evolution;
- BeYDV:
-
Bean yellow dwarf virus
- BSCTV:
-
Beet severe curly top virus
- BSV:
-
Banana streak virus
- CBE:
-
Cytidine base editor
- CMV:
-
Cucumber mosaic virus
- ClYVV:
-
Clover yellow vein virus
- CVYV:
-
Cucumber vein yellowing virus
- CRISPR:
-
Clustered regularly interspaced short palindromic repeats;
- CRISPRi:
-
dCas9-based transcription inhibition
- crRNA:
-
CRISPR RNA
- CP:
-
Coat protein
- DSB:
-
DNA double-strand break
- EBEs:
-
TALE-binding elements
- ETI:
-
Effector-triggered immunity
- ETS:
-
Effector-triggered susceptibility
- GE:
-
Genome-editing
- gRNA:
-
Guide RNA
- IR:
-
Intergenic region
- MLO :
-
Mildew resistance locus O
- M-MLV RT:
-
Moloney murine leukemia virus reverse transcriptase
- PAM:
-
Protospacer adjacent motif
- PAMPs:
-
Pathogen-associated molecular patterns
- PEG:
-
Polyethylene glycol
- PFS:
-
Protospacer-flanking site
- PRRs:
-
Pattern recognition receptors
- PRSMV-W:
-
Papaya ring spot mosaic virus-W
- PTI:
-
PAMP-triggered immunity
- PVY:
-
Potato virus Y
- QTL:
-
Quantitative trait locus
- Rep:
-
Replication-associated protein
- RNP:
-
Ribonucleoprotein
- ROS:
-
Reactive oxygen species
- RSMV:
-
Rice stripe mosaic virus
- RTSV:
-
Rice tungro spherical virus
- SNP:
-
Single-nucleotide polymorphism
- SRBSDV:
-
Southern rice black-streaked draft virus
- ssDNA:
-
Single-stranded DNA
- TAL:
-
Transcription activator-like
- TALENs:
-
Transcription activator-like effector nucleases
- TMV:
-
Tobacco mosaic virus
- tracrRNA:
-
Trans-activating small RNA
- TYLCV:
-
Tomato yellow leaf curl virus
- TuMV:
-
Turnip mosaic virus
- WDV:
-
Wheat dwarf virus
- Xcc :
-
Xanthomonas citri subsp. citri
- ZFNs:
-
Zinc-finger nucleases;
- ZYMV:
-
Zucchini yellow mosaic virus
References
Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, et al. RNA targeting with CRISPRCas13. Nature. 2017;550:280–4.
Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DB, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:aaf5573.
Aman R, Ali Z, Butt H, Mahas A, Aljedaani F, Khan MZ, et al. RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol. 2018;19:1.
Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576:149–57.
Aparicio T, de Lorenzo V, Martinez-Garcia E. CRISPR/Cas9-based counterselection boosts recombineering efficiency in Pseudomonas putida. Biotechnol J. 2018;13:e1700161.
Arazoe T, Miyoshi K, Yamato T, Ogawa T, Ohsato S, Arie T, et al. Tailor-made CRISPR/Cas system for highly efficient targeted gene replacement in the rice blast fungus. Biotechnol Bioeng. 2015;112:2543–9.
Baltes NJ, Hummel AW, Konecna E, Cegan R, Bruns AN, Bisaro DM, et al. Conferring resistance to geminiviruses with the CRISPR-Cas prokaryotic immune system. Nat Plants. 2015;1:15145.
Barakate A, Stephens J. An overview of CRISPR-based tools and their improvements: new opportunities in understanding plant-pathogen interactions for better crop protection. Front Plant Sci. 2016;7:765.
Bastet A, Zafirov D, Giovinazzo N, Guyon-Debast A, Nogue F, Robaglia C, et al. Mimicking natural polymorphism in eIF4E by CRISPR-Cas9 base editing is associated with resistance to potyviruses. Plant Biotechnol J. 2019;17:1736–50.
Bhaya D, Davison M, Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu Rev Genet. 2011;45:273–97.
Bikard D, Jiang W, Samai P, Hochschild A, Zhang F, Marraffini LA. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013;41:7429–37.
Borisjuk N, Kishchenko O, Eliby S, Schramm C, Anderson P, Jatayev S, et al. Genetic modification for wheat improvement: from transgenesis to genome editing. Biomed Res Int. 2019;2019:6216304 https://doi.org/10.1155/2019/6216304.
Büttner D, He SY. Type III protein secretion in plant pathogenic bacteria. Plant Physiol. 2009;4:1656–64.
Chandrasekaran J, Brumin M, Wolf D, Leibman D, Klap C, Pearlsman M, et al. Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol Plant Pathol. 2016;17:1140–53.
Chatterjee P, Jakimo N, Jacobson JM. Minimal PAM specificity of a highly similar SpCas9 ortholog. Sci Adv. 2018;4:eaau0766.
Chen W, Zhang Y, Zhang Y, Pi Y, Gu T, Song L, et al. CRISPR/Cas9-based genome editing in Pseudomonas aeruginosa and cytidine deaminase-mediated base editing in Pseudomonas species. iScience. 2018;6:222–31 https://doi.org/10.1016/j.isci.2018.07.024.
Chen X, Shang J, Chen D, Lei C, Zou Y, Zhai W, et al. A B-lectin receptor kinase gene conferring rice blast resistance. Plant J. 2006;46:794–804.
Chen X-R, Zhang Y, Li H-Y, Zhang Z-H, Sheng G-L, Li Y-P, et al. The RXLR effector PcAvh1 is required for full virulence of Phytophthora capsici. Mol Plant-Microbe Interact. 2019;32:986–1000.
Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: shaping the evolution of the plant immune response. Cell. 2006;124:803–14.
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23.
Cox DBT, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J, et al. RNA editing with CRISPR-Cas13. Science. 2017;358:1019–27.
Dong OX, Ronald PC. Genetic engineering for disease resistance in plants: recent progress and future perspectives. Plant Physiol. 2019;180:26–38.
Dou D, Kale SD, Liu T, Tang Q, Wang X, Arredondo FD, et al. Different domains of Phytophthora sojae effector Avr4/6 are recognized by soybean resistance genes Rps4 and Rps6. Mol Plant-Microbe Interact. 2010;23:425–35.
Fang Y, Tyler BM. Efficient disruption and replacement of an effector gene in the oomycete Phytophthora sojae using CRISPR/Cas9. Mol Plant Pathol. 2016;17:127–39.
Ferrara M, Haidukowski M, Logrieco AF, Leslie JF, Mulèm G. A CRISPR-Cas9 system for genome editing of Fusarium proliferatum. Sci Rep. 2019;9:19836.
Fister AS, Landherr L, Maximova SN, Guiltinan MJ. Transient expression of CRISPR/Cas9 machinery targeting TcNPR3 enhances defense response in Theobroma cacao. Front Plant Sci. 2018;9:268.
Foster AJ, Martin-Urdiroz M, Yan X, Wright HS, Soanes DM, Talbot NJ. CRISPR-Cas9 ribonucleoprotein-mediated co-editing and counterselection in the rice blast fungus. Sci Rep. 2018;8:14355.
Frye CA, Tang D, Innes RW. Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc Natl Acad Sci U S A. 2001;98:373–8.
Gaj T, Sirk SJ, Shui SL, Liu J. Genome-editing technologies: principles and applications. Cold Spring Harb Perspect Biol. 2016;8(12):a023754.
Garcia-Doval C, Jinek M. Molecular architectures and mechanisms of class 2 CRISPR-associated nucleases. Curr Opin Struct Biol. 2017;47:157–66.
Gardiner DM, Kazan K. Selection is required for efficient Cas9-mediated genome editing in Fusarium graminearum. Fungal Biol. 2018;122:131–7.
Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature. 2017;551:464–71.
Gomez MA, Lin Z-JD, Moll T, Luebbert C, Chauhan RD, Hayden L, et al. Simultaneous CRISPR/Cas9-mediated editing of cassava eIF 4E isoforms nCBP-1 and nCBP-2 reduces cassava brown streak disease symptom severity and incidence. Plant Biotechnol J. 2019;17:421–34.
Gu T, Zhao S, Pi Y, Chen W, Chen C, Liu Q, et al. Highly efficient base editing in Staphylococcus aureus using an engineered CRISPR RNA-guided cytidine deaminase. Chem Sci. 2018;9:3248–53.
Gumtow R, Wu D, Uchida J, Tian M. A Phytophthora palmivora extracellular cystatin-like protease inhibitor targets papain to contribute to virulence on papaya. Mol Plant-Microbe Interact. 2018;31:363–73.
Harrington LB, Burstein D, Chen JS, Paez-Espino D, Ma E, Witte IP, et al. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science. 2018;362:839–42.
Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–78.
Hu JH, Miller SM, Geurts MH, Tang W, Chen L, Sun N, et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 2018;556:57–63.
Hu X, Wang C, Fu Y, Liu Q, Jiao X, Wang K. Expanding the range of CRISPR/Cas9 genome editing in rice. Mol Plant. 2016;9:943–5.
Hua K, Tao X, Han P, Wang R, Zhu J-K. Genome engineering in rice using Cas9 variants that recognize NG PAM sequences. Mol Plant. 2019;12:1003–14.
Hua K, Tao X, Yuan F, Wang D, Zhu J-K. Precise A•T to G•C base editing in the rice genome. Mol Plant. 2018;11:627–30.
Huck S, Bock J, Girardello J, Gauert M, Pul Ü. Marker-free genome editing in Ustilago trichophora with the CRISPR-Cas9 technology. RNA Biol. 2019;16:397–403.
Idnurm A, Urquhart AS, Vummadi DR, Chang S, Van de Wouw AP, López-Ruiz FJ. Spontaneous and CRISPR/Cas9-induced mutation of the osmosensor histidine kinase of the canola pathogen Leptosphaeria maculans. Fungal Biol Biotechnol. 2017;4:12.
Ji X, Wang D, Gao C. CRISPR editing-mediated antiviral immunity: a versatile source of resistance to combat plant virus infections. Sci China Life Sci. 2019;62:1246–9.
Ji X, Zhang H, Zhang Y, Wang Y, Gao C. Establishing a CRISPR-Cas-like immune system conferring DNA virus resistance in plants. Nat Plants. 2015;1:15144.
Jia H, Orbovic V, Jones JB, Wang N. Modification of the PthA4 effector binding elements in type I CsLOB1 promoter using Cas9/sgRNA to produce transgenic Duncan grapefruit alleviating Xcc ΔpthA4:dCsLOB1.3 infection. Plant Biotechnol J. 2016;14:1291–301.
Jia H, Zhang Y, Orbovic V, Xu J, White FF, Jones JB, et al. Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnol J. 2017;15:817–23.
Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–9.
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–21.
Jørgensen IH. Discovery, characterization and exploitation of Mlo powdery mildew resistance in barley. Euphytica. 1992;63:141–52.
Kalinina NO, Khromov A, Love AJ, Taliansky ME. CRISPR applications in plant virology: virus resistance and beyond. Phytopathology. 2020;110:18–28.
Khan MZ, Haider S, Mansoor S, Amin I. Targeting plant ssDNA viruses with engineered miniature CRISPR-Cas14a. Trends Biotechnol. 2019;37:800–4.
Kim YA, Moon H, Park CJ. CRISPR/Cas9-targeted mutagenesis of Os8N3 in rice to confer resistance to Xanthomonas oryzae pv. oryzae. Rice. 2019;12:67.
Kis A, Hamar É, Tholt G, Bán R, Havelda Z. Creating highly efficient resistance against wheat dwarf virus in barley by employing CRISPR/Cas9 system. Plant Biotechnol J. 2019;17:1004–6.
Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–4.
Konermann S, Lotfy P, Brideau NJ, Oki J, Shokhirev MN, Hsu PD. Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell. 2018;173:665–76.
Kong G, Wan L, Deng YZ, Yang W, Li W, Jiang L, et al. Pectin acetylesterase PAE5 is associated with the virulence of plant pathogenic oomycete Peronophythora litchii. Physiol Mol Plant Pathol. 2019;106:16–22.
Kuang Y, Li S, Ren B, Yan F, Spetz C, Li X, et al. Base-editing-mediated artificial evolution of OsALS1 in planta to develop novel herbicide-tolerant rice germplasms. Mol Plant. 2020;13:565–72.
Laflamme B, Dillon MM, Martel A, Almeida RND, Desveaux D, Guttman DS. The pan-genome effector-triggered immunity landscape of a host-pathogen interaction. Science. 2020;367:763–8.
Langner T, Kamoun S, Belhaj K. CRISPR crops: plant genome editing toward disease resistance. Annu Rev Phytopathol. 2018;56:479–512.
Leisen T, Bietz F, Werner J, Wegner A, Schaffrath U, Scheuring D, et al. CRISPR/Cas with ribonucleoprotein complexes and transiently selected telomere vectors allows highly efficient marker-free and multiple genome editing in Botrytis cinerea. bioRxiv. 2020; https://doi.org/10.1101/2020.01.20.912576.
Li J, Zhang Y, Zhang Y, Yu PL, Pan H, Rollins JA. Introduction of large sequence inserts by CRISPR-Cas9 to create pathogenicity mutants in the multinucleate filamentous pathogen Sclerotinia sclerotiorum. mBio. 2018;9:e00567–18.
Liang Y, Han Y, Wang C, Jiang C, Xu JR. Targeted deletion of the USTA and UvSLT2 genes efficiently in Ustilaginoidea virens with the CRISPR-Cas9 system. Front Plant Sci. 2018;9:699.
Lin Q, Zong Y, Xue C, Wang S, Jin S, Zhu Z, et al. Prime genome editing in rice and wheat. Nat Biotechnol. 2020;38:582–5.
Liu D, Huang C, Guo J, Zhang P, Chen T, Wang Z, et al. Development and characterization of a CRISPR/Cas9n based multiplex genome editing system for Bacillus subtilis. Biotechnol Biofuels. 2019;12:197.
Lu S, Shen X, Chen B. Development of an efficient vector system for gene knock-out and near in-cis gene complementation in the sugarcane smut fungus. Sci Rep. 2017;7:3113.
Macovei A, Sevilla NR, Cantos C, Jonson GB, Slamet-Loedin I, Cermak T, et al. Novel alleles of rice eIF4G generated by CRISPR/Cas9-targeted mutagenesis confer resistance to Rice tungro spherical virus. Plant Biotechnol J. 2018;16:1918–27.
Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, et al. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015;13:722–36.
Makarova KS, Wolf YI, Koonin EV. Classification and nomenclature of CRISPR-Cas systems: where from here? CRISPR J. 2018;1:325–36.
Mehta D, Sturchler A, Anjanappa RB, Zaidi SS, Hirsch-Hoffmann M, Gruissem W, et al. Linking CRISPR-Cas9 interference in cassava to the evolution of editing-resistant geminiviruses. Genome Biol. 2019;20:80.
Ming M, Ren Q, Pan C, He Y, Zhang Y, Liu S, et al. CRISPRCas12b enables efficient plant genome engineering. Nat Plants. 2020;6:202–8.
Nakamura M, Okamura Y, Iwai H. Plasmid-based and-free methods using CRISPR/Cas9 system for replacement of targeted genes in Colletotrichum sansevieriae. Sci Rep. 2019;9:18947.
Nejat N, Rookes J, Mantri NL, Cahill DM. Plant-pathogen interactions: toward development of next-generation disease-resistant plants. Crit Rev Biotechnol. 2017;37:229–37.
Nekrasov V, Wang C, Win J, Lanz C, Weigel D, Kamoun S. Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci Rep. 2017;7:482.
Oliva R, Ji C, Atienza-Grande G, Huguet-Tapia JC, Perez-Quintero A, Li T, et al. Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat Biotechnol. 2019;37:1344–50.
Ortigosa A, Gimenez-Ibanez S, Leonhardt N, Solano R. Design of a bacterial speck resistant tomato by CRISPR/Cas9-mediated editing of SlJAZ2. Plant Biotechnol J. 2019;17:665–73.
Peng A, Chen S, Lei T, Xu L, He Y, Wu L, et al. Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol J. 2017;15:1509–19.
Price AA, Sampson TR, Ratner HK, Grakoui A, Weiss DS. Cas9-mediated targeting of viral RNA in eukaryotic cells. Proc Natl Acad Sci U S A. 2015;112:6164–9.
Pyott DE, Sheehan E, Molnar A. Engineering of CRISPR/Cas9-mediated potyvirus resistance in transgene-free Arabidopsis plants. Mol Plant Pathol. 2016;17:1276–88.
Rath D, Amlinger L, Rath A, Lundgren M. The CRISPR-Cas immune system: biology, mechanisms, and applications. Biochimie. 2015;117:119–28.
Ren B, Liu L, Li S, Kuang Y, Wang J, Zhang D, et al. Cas9-NG greatly expands the targeting scope of the genome-editing toolkit by recognizing NG and other atypical PAMs in rice. Mol Plant. 2019;12:1015–26.
Ren B, Yan F, Kuang Y, Li N, Zhang D, Lin H, et al. A CRISPR/Cas9 toolkit for efficient targeted base editing to induce genetic variations in rice. Sci China Life Sci. 2017;60:516–9.
Ren B, Yan F, Kuang Y, Li N, Zhang D, Zhou X, et al. Improved base editor for efficiently inducing genetic variations in rice with CRISPR/Cas9-guided hyperactive hAID mutant. Mol Plant. 2018;11:623–6.
Schuster M, Kahmann R. CRISPR-Cas9 genome editing approaches in filamentous fungi and oomycetes. Fungal Genet Biol. 2019;130:43–53.
Schuster M, Schweizer G, Reissmann S, Kahmann R. Genome editing in Ustilago maydis using the CRISPR-Cas system. Fungal Genet Biol. 2016;89:3–9.
Shi T-Q, Gao J, Wang W-J, Wang K-F, Xu G-Q, Huang H, et al. CRISPR/Cas9-based genome editing in the filamentous fungus Fusarium fujikuroi and its application in strain engineering for gibberellic acid production. ACS Synth Biol. 2019;8:445–54.
Shmakov S, Smargon A, Scott D, Cox D, Pyzocha N, Yan W, et al. Diversity and evolution of class 2 CRISPR-Cas systems. Nat Rev Microbiol. 2017;15:169–82.
Strecker J, Jones S, Koopal B, Schmid-Burg J, Zetsche B, Gao L, et al. Engineering of CRISPR-Cas12b for human genome editing. Nat Commun. 2019;10:212.
Sun J, Wang Q, Jiang Y, Wen Z, Yang L, Wu J, et al. Genome editing and transcriptional repression in Pseudomonas putida KT2440 via the type II CRISPR system. Microb Cell Factories. 2018;17:41.
Tan SZ, Reisch CR, Prather KLJ. A robust CRISPR interference gene repression system in Pseudomonas. J Bacteriol. 2018;200:e00575–17.
Tashkandi M, Ali Z, Aljedaani F, Shami A, Mahfouz MM. Engineering resistance against tomato yellow leaf curl virus via the CRISPR/Cas9 system in tomato. Plant Signal Behav. 2018;13:e1525996.
Teng F, Cui T, Fen G, Guo L, Xu K, Gao Q, et al. Repurposing CRISPR-Cas12b for mammalian genome engineering. Cell Discov. 2018;4:63.
Thomazella DP, Brail Q, Dahlbeck D, Staskawicz B. CRISPR-Cas9 mediated mutagenesis of a DMR6 ortholog in tomato confers broad-spectrum disease resistance. bioRxiv. 2016; https://doi.org/10.1101/064824.
Tripathi JN, Ntui VO, Ron M, Muiruri SK, Britt A, Tripathi L. CRISPR/Cas9 editing of endogenous banana streak virus in the B genome of Musa spp. overcomes a major challenge in banana breeding. Commun Biol. 2019;2:46.
Wang F, Wang C, Liu P, Lei C, Hao W, Gao Y, et al. Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PLoS One. 2016;11:e0154027.
Wang M, Xu Z, Gosavi G, Ren B, Cao Y, Kuang Y, et al. Targeted base editing in rice with CRISPR/ScCas9 system. Plant Biotechnol J. 2020; https://doi.org/10.1111/pbi.13330.
Wang Q, Cobine PA, Coleman JJ. Efficient genome editing in Fusarium oxysporum based on CRISPR/Cas9 ribonucleoprotein complexes. Fungal Genet Biol. 2018;117:21–9.
Wang Q, Coleman JJ. CRISPR/Cas9-mediated endogenous gene tagging in Fusarium oxysporum. Fungal Genet Biol. 2019;126:17–24.
Wang W, Xue Z, Miao J, Cai M, Zhang C, Li T, et al. PcMuORP1, an oxathiapiprolin-resistance gene, functions as a novel selection marker for Phytophthora transformation and CRISPR/Cas9 mediated genome editing. Front Microbiol. 2019;10:2402.
Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, et al. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol. 2014;32:947–51.
Wenderoth M, Pinecker C, Voß B, Fischer R. Establishment of CRISPR/Cas9 in Alternaria alternata. Fungal Genet Biol. 2017;101:55–60.
Xu Z, Xu X, Gong Q, Li Z, Li Y, Wang S, et al. Engineering broad-spectrum bacterial blight resistance by simultaneously disrupting variable TALE-binding elements of multiple susceptibility genes in rice. Mol Plant. 2019;12:1434–46.
Yan F, Kuang Y, Ren B, Wang J, Zhang D, Lin H, et al. Highly efficient A·• T to G·• C base editing by Cas9n-guided tRNA adenosine deaminase in rice. Mol Plant. 2018;11:631–4.
Yang B, Sugio A, White FF. Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc Natl Acad Sci U S A. 2006;103:10503–8.
Yang H, Gao P, Rajashankar KR, Patel DJ. PAM-dependent target DNA recognition and cleavage by C2c1 CRISPR-Cas endonuclease. Cell. 2016;167:1814 28.e12.
Zaidi SS, Mahfouz MM, Mansoor S. CRISPR-Cpf1: a new tool for plant genome editing. Trends Plant Sci. 2017;22:550–3.
Zaidi SS, Mukhtar MS, Mansoor S. Genome editing: targeting susceptibility genes for plant disease resistance. Trends Biotechnol. 2018;36(9):898–906.
Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–71.
Zhang B, Ye W, Ye Y, Zhou H, Saeed AFUH, Chen J, et al. Structural insights into Cas13b-guided CRISPR RNA maturation and recognition. Cell Res. 2018a;28:1198–201.
Zhang C, Konermann S, Brideau NJ, Lotfy P, Wu X, Novick SJ, et al. Structural basis for the RNA-guided ribonuclease activity of CRISPR-Cas13d. Cell. 2018b;175:212–23.
Zhang T, Zhao Y, Ye J, Cao X, Xu C, Chen B, et al. Establishing CRISPR/Cas13a immune system conferring RNA virus resistance in both dicot and monocot plants. Plant Biotechnol J. 2019;17:1185–7.
Zhang T, Zheng Q, Yi X, An H, Zhao Y, Ma S, et al. Establishing RNA virus resistance in plants by harnessing CRISPR immune system. Plant Biotechnol J. 2018c;16:1415–23.
Zhang Y, Bai Y, Wu G, Zou S, Chen Y, Gao C, et al. Simultaneous modification of three homoeologs of TaEDR1 by genome editing enhances powdery mildew resistance in wheat. Plant J. 2017;91:714–24.
Zhang ZT, Jiménez-Bonilla P, Seo SO, Lu T, Jin YS, Blaschek HP, et al. Bacterial genome editing with CRISPR-Cas9: taking Clostridium beijerinckii as an example. In: Braman JC, editor. Synthetic biology. Methods in molecular biology, vol. 1772. New York: Humana Press; 2018. p. 297–325.
Zhou X, Liao H, Chern M, Yin J, Chen Y, Wang J, et al. Loss of function of a rice TPR-domain RNA-binding protein confers broad-spectrum disease resistance. Proc Natl Acad Sci U S A. 2018;115:3174–9.
Acknowledgements
We thank Dr. Fangfang Li at Institute of Plant Protection, Chinese Academy of Agriculture Sciences for her helpful advice, and Dr. Carl Spetz at Norwegian Institute of Bioeconomy Research and other members (Meixia Wang, Jingwen Wang, Lang Liu, Zhenwan Lu, Ziyan Xu and Guigen Ma) of our laboratory for proofreading of the manuscript. We apologize to our colleagues whose work could not be cited and discussed due to space limitations.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31871948), and the National Youth Talent Support Program and the Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences to HZ.
Author information
Authors and Affiliations
Contributions
GG, FY, BR, KY, DY, XZ and HZ wrote the manuscript. GG drew the figures. KY finished the Tables. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gosavi, G., Yan, F., Ren, B. et al. Applications of CRISPR technology in studying plant-pathogen interactions: overview and perspective. Phytopathol Res 2, 21 (2020). https://doi.org/10.1186/s42483-020-00060-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s42483-020-00060-z