Abstract
Background
By resolving complicated spectra from drug combinations, chemometric techniques are valuable for multi-component investigation. The capacity to properly estimate combinations of components without separating drugs from their mixture is one of the benefits of chemometric analysis approaches over traditional analytical methods. These approaches are easy to use and sensitive even to the lowest concentrations. They are also practical, affordable, and cost-effective. In the current study, the chemometric aided spectrophotometric approach was used to evaluate the two drugs naringin and verapamil. The approach is multidimensional and based on chemometrics, which includes an orthogonal partial least square method that is a new refinement of the partial least squares regression analysis method. With this technique, no conversions are made to the spectrum that overlaps the two drugs. The tools UV-PROBE, SIMCA version 17, and excel were used to process the chemometric data.
Results
According to results from an orthogonal partial least square model, the mean percent recovery and relative standard deviation for the combination of verapamil with naringin were 100.80/1.19 and 100.836/1.35, respectively.The calibration model was used to predict known synthetic mixtures.This method shows good consistency in recovery ranging between 98.92 and 103.59% for VER and from 96.21 to 101.84% NAR. As saying the synthetic mixture revealed that it had a high percentage of purity.
Conclusions
The proposed chemometric method can estimate the quantitative amount of pharmaceuticals based on their dosage forms. This approach meets the requirements for the international conference on harmonization's (ICH) analytical criteria, such as precision and accuracy.
Graphical Abstract

Similar content being viewed by others
Background
Chemometric methods are useful for analyzing multi-components by solving their complex spectrums from mixtures (Dinc et al. 2002). Compared to conventional analysis methods, chemometrics can accurately determine the combination of medications without isolating them from the mixture. These techniques have many other advantages, including being easy to use, sensitive even to the lowest concentrations and practical. They are also cost-effective compared with the alternative techniques used for simultaneous identification. These procedures do not affect the analysis or calibration of pharmaceuticals due to the concentration of the other components. It is also faster to identify each component in the combination (Prachi et al. 2013). Chemometric methods for measuring naringin and verapamilin synthetic combinations were refined and verified. These methods are easy to use, precise, accurate, and economical.
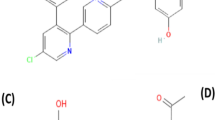
Citrus flavonoid naringin (NAR), one of the most promising substances in the human diet, is readily available. Citrus fruits contain the abundant flavone glycoside naringin (4,5,7-trihydroxyflavanone-7-rhamnoglucoside). Chemical structure of naringin is shown in (Fig. 1), which has intriguing biological and pharmacological effects (Opinion et al. 2011). The bitterness of naringin is due to its glucose moiety. Its contents are naringenin (an aglycone) and neohesperidose, linked with the C-7 hydroxyl. Naringin can now be added in foods without restriction because it has been classified as a flavoring compound (Sharma et al. 2011).
Naringin contain a flavanone-7-o-glycoside which has a number of pharmacological actions, including antioxidant, anti-carcinogenic, and anti-diabetic properties. Additionally, it blocks some cytochrome P450 enzymes, such as CYP3A4 and CYP1A2, which can lead to a variety of medication interactions in-vitro (Tulasi et al. 2019, Kim et al. 2023).
Verapamil is chemically known as 2-(3,4-dimethoxyphenyl)-5-[2-(3,4-dimethoxyphenyl) ethyl-methylamino)-2-propan-2-ylpentanenitrile. Verapamil's chemical structure is illustrated in Fig. 2. Verapamil is a P-glycoprotein Inhibitor, cytochrome P450 3A4 and cytochrome P4503A inhibitor. Verapamil increases the size of the coronary arteries and reduces the contractility of the myocardium by blocking transmembrane calcium ion input into smooth muscle and myocardial cells. This substance also blocks drug efflux pumps P-glycoprotein, which are overexpressed by some tumors resistant to many drugs and can enhance the effect of certain antineoplastic agents (National Center for Biotechnology Information 2023).
Verapamil has been analyzed by various methods like UV (Trivedi et al. 2017), High-performance liquid chromatography (HPLC) (Auti et al. 2019), Ultra performance liquid chromatography (UPLC) (Vijayabaskar et al. 2017), thin layer chromatography (Bhushan et al. 2005), LC–MS/MS (Ravi et al. 2019), chiral capillary electrophoresis (Cârcu-Dobrin et al. 2021). Similarly, Naringin also has been estimated by various methods like Chromatography-Tandem Mass Spectrometry (Xiong et al. 2014), Capillary electrophoresis (Akiyama et al. 2000), HPLC (Ishii et al. 1997), HPTLC (Alam et al. 2014), UPLC-MS/MS (Wen et al. 2012), thin layer chromatography and fourier transform infrared spectroscopy (FTIR) (Hakim et al. 2019). Each method has yet to be developed to estimate naringin together with verapamil. In the present work, chemometric techniques have been effectively used to estimate verapamil and naringin simultaneously by OPLS.
Methods
Materials
Verapamil and naringin (C27H38N2O4 and C27H32O14) with purity levels of ≥ 99% were purchased from Yerrow Chem products, Mumbai. Methanol (AR Grade) was purchased from S.D fine Chemicals Ltd, Mumbai, water (Milli Q. All other reagents and materials were of AR grade and commercially available). Verapamil and Naringin concentrations were determined using a UV–visible spectrophotometer (Shimadzu-Kyoto, Japan Model 1800 double beam with matching 1 cm path-length quartz cells) using methanol as the solvent at wavelengths between 200 and 400 nm. The drug's solubility, melting point, and molar absorptivity were its distinguishing qualities. The drug was analyzed using the UV–visible technique using software UV Probe and SIMCA (Sartorius, version 17) for handling the orthogonal partial least square (OPLS) model, which satisfied the requirements set out by the International Conference on harmonization (ICH) for analytical criteria such as linearity, precision, and accuracy.
Preparation of standard solution
Weight precisely 10.0 mg each of NAR and VER. The stock solution was prepared by dissolving the VER in 10.0 mL methanol and then adding the required volume using the same solvent. VER or NAR stock solutions were prepared using concentrations ranging between 0.5–12 and 2–48 ug/mL. The zero-order spectra were recorded at wavelengths between 200 and 400 nm compared to a blank solvent (Mazumder et al. 2020).
One component calibration
The linear dynamic range of each drug was determined by filling a 10.0 mL volumetric container with different amounts of the stock solution. This was diluted in methanol at the correct concentration. The electronic absorption spectrum was recorded in the 200–400 nm wavelength range. Absorption values at 200 nm wavelengths and 400 nm were measured for various concentrations (Prasad et al. 2017). The light dependent resistor (LDR) was calculated by plotting the absorbance against concentration. The LDR for the two drugs was achieved between 0.5 and 50 ug/mL.
Standard calibration and prediction samples set
The multilevel, multi-factor architecture was used to develop the calibration and validation sets. A calibration set with four component mixtures has been designed to improve the prediction of OPLS. To prepare the calibration set, sixteen samples were used, as shown in Table 1, and nine samples in Table 2, and then the volume of the methanol was adjusted to match the NAR or VER concentrations. These concentrations ranged from 2–48 to 0.5–12 mg/mL, respectively, against blank methanol in a range between 200 and 400 nm at intervals of one nm. The recorded spectrum from UV Probe was transferred to SIMCA 17, using OPLS Toolbox, for data analysis. A calibration model was created (Zou et al. 2022).
Building an OPLS model
A multivariate calibration method like OPLS could be applied with a filter derivative to address the overlap between the medicines studied. As a part of the pre-processing, data from calibration samples was automatically scaled to be used in OPLS. The cross-validation method using one sample was left out and then followed. The ideal number of components were important for the OPLS method. If the components were too many, noise was added, but if they were too few, noise was reduced. The established models of the OPLS method could accurately characterize two latent variables (Abdallah et al. 2019). A calibration curve is created by comparing projected concentrations with actual concentrations.
Construction of calibration and prediction
To prepare the calibration and validation mixes, we combined working standard solutions from NAR and VER at different ratios based on their linearity concentration ranges. For the calibration set, the concentration was calculated using a general factorial system (2 factors with 4 levels each) and for the predictions set (2 factors with 3 levels each) (Sonawane et al. 2019). The calibration and validation mixtures totaled 16 in number and were prepared independently. All mixtures were recorded in the absorption spectra range of 200–400 nm, with 1 nm increments. Calculations were made for regression equations and statistical data related to the thesis model. The calculations were also done for recoveries and mean recoveries.
Preparation of sample solution
To perform the assay, a quantity of powder equal to 20.0 mg VER is weighed and placed in a calibrated 100.0 mL volumetric flask (Sonawane et al. 2019). Then it's dissolved in methanol. In a 25.0 mL volumetric flask calibrated to 0.5 mg/mL, a solution of 0.5 mg/mL was pipetted and then dissolved in the methanol. This solution was then diluted further with methanol to the desired concentration within the calibration range. The proposed chemometric method (OPLS), which was scanned in a spectrophotometer to determine sample solutions, was then used.
Assay of synthetic mixture
To perform the assay, a quantity of powder equal to 20.0 mg VER was accurately weighed and placed in a calibrated 100.0 ml volumetric flask. Then it was dissolved in Methanol. In a 25.0 ml volumetric flask calibrated to 0.5 mg/ml, a solution of 0.5 mg/ml was pipetted and then dissolved in the Methanol. The resulting solution is further diluted using Methanol at a concentration within the calibration range. After scanning with a spectrophotometer, the proposed chemometric method (opls) was used to determine sample solutions (Ríos-Reina et al. 2023). Table 6 shows the results of an assay.
Results
In methanol, the VER and NAR spectra overlapped with the binary combination of NAR and zero-order superimposed spectra.To overcome this problem, it is possible to use a chemometric technique to separate these two drugs from the mixture. Under conditions where the spectra overlap, chemometric techniques can be employed to estimate drugs (Biancolillo et al. 2018). Multivariate chemometrics was developed to offer a simple, reliable, selective and valid method of determining the drugs without interference.
Statistical parameters
The regression model can be defined in several ways. When describing chemometric methods, RMSEP is the root mean square error of prediction, and RMSECV is the root mean square for cross-validation.
By applying the OPLS approach to the validation set of the two drug mixture, the RMSECV, RMSEP and R2 were determined. The results are shown in Table 3. Statistical parameters for simultaneous determination of VER and NAR by OPLS are shown in Tables 4 and 5.
The validity of calibration procedures can also be assessed by simultaneously estimating two drugs in the set. The mean percent recovery for two drug solutions combined using the OPLS method was found within acceptable ranges (Indrayanto et al. 2018). Recovery studies were conducted using standardized drugs added in certain amounts to a synthesized mixture of a specific concentration. After defining the sample volume or concentration in which it was to be added, known amounts of standard working solutions were added. The volume of the flask was then filled with methanol. Chemometric techniques were used to evaluate the recovery of the two drugs in the mixtures (Table 6).
In Table 4, mean % and root mean square deviation (RSD) values are shown. Recovery study results showed that the procedures had successfully been proved since the % root mean square deviation (RSD) was less than 2 and the recovery study was closer to 100%. Additionally, the results shows that the suggested techniques are correct and that excipients do not affect the assessment of two drugs. By looking at various drug concentrations, the linearity for VER and NAR was evaluated. The root mean square error of cross validation (RMSECV) was calculated which can be used to determine the error in the predicted concentrations (Van Wyngaard et al. 2021). Drug concentrations in various sets of mixtures used for method development were analyzed and predicted to validate the approaches. By comparing the predicted concentrations on the graph, the method’s ability for prediction was determined. Figures 3 and 4 explains variation of model that shows the peaks to be linear for both the drugs therefore the optimization of calibration set shows a good R2 value (0.997 & 1) and good prediction power Q2 value (0.210 & 0.128), minimum RMSEE (0.310 & 0.175), and RMSECV (1.226 & 0.665) for each drug. Figures 5 and 6 explains the predictive ability from which we can record the RMSEP value. It indicates OPLS- optimized technique, is good for both the drugs in the prediction set, indicating the method to be effective and has good predictive ability.
Model validation
The Hotelling T2 test and D.ModX were used to find weak and moderate outliers, and they encountered no deviation, indicating that no outliers were found (Shabbak et al. 2012). According to permutation analysis, all of the permuted Q2 were below 0, and R2 was below or about 1. Since Q2 and R2 were lower than the original values, it is likely that the model fitting was accurate and unlikely to have developed randomly. We can see all the samples that fall inside the Hotelling range by looking at the score plot. We get the conclusion that the model is effective since every optimisation strategy satisfy the requirements. The summary fit, score representation, Hotelling’s range, D.ModX, and permutation plot are displayed in Figs. 7, 8, 9, 10, 11.
Discussion
It was found that the lambda max value for both the drugs i.enaringin and verapamil was very near to each other. Since, iso-absorptive point was very near for the drugs, therefore, the Q-ratio method development was not processed.
Without a successful Q-ratio method, an innovative UV spectrophotometric technique based on chemometrics was developed (Darbandi et al. 2020). The chemometrics technique can solve many spectrum resolution problems. This technique can improve the signal-to-noise ratio, increase selectivity in determination, optimize experimental conditions, boost analytical efficiency, and give much information. Hence, it is used for the simultaneous determination of multi-component mixtures. Absorptivity and other parameters were checked for naringin and verapamil, and methanol was selected as a solvent for quantifying the drugs in the method. This method was linear over a range of naringin 0.05–50 ug/mL and verapamil 0.5–50 ug/mL with a correlation coefficient of 0.9978 and 0.9998, respectively.
The calibration model prepared for OPLS is optimal because it has a good correlation coefficient and low RMSEP and RMSECV for naringin. Naringin has good linearity in the range of concentrations of 0.5–50 mg/mL, with a value of 0.9987, and verapamil has a value of 1.
The calibration model was used to predict known synthetic mixtures where RMSEP 2. By identifying the minimum error, further assays, accuracy and precision tests were conducted. The RSD percentage for the intra-day and Inter-day precision was less than 2%, which indicates that this method is reproducible and repeatable (Kamal et al. 2015). This method shows good consistency in recovery ranging between 98.92 and 103.59% for VER and from 96.21 to 101.84% NAR. As saying the synthetic mixture revealed that it had a high percentage of purity. The mean assay for VER and NAR using the OPLS technique was 100.80%, respectively.
The chemometric studies mainly implicated for experimental design and optimization, data treatments, sample classification, calibration for determination of concentrations and model identification.Finally Based on the results of different assessments, we can formulate some hypotheses about the reproducibility of the methods. The suggested procedures were verified in compliance with ICH guidelines (I. H. T. Guideline 1994).
Conclusions
A simple, reproducible, and accurate chemometric technique was developed for the simultaneous estimation of NAR and VER. The technique is a great way to analyze commercial pharmaceuticals without interference or the need to physically separate them. The method solved the issue of interference and merging of mixture peaks. The results of this method were validated according to ICH Guidelines and met the criteria. This method is valid and may be used to perform routine quality controls on VERs and NARs.
Availability of data and materials
All necessary data generated or analyzed during this study are included in this manuscript. Any additional data could be available from the corresponding author upon request.
Abbreviations
- OPLS:
-
Orthogonal partial least square
- VER:
-
Verapamil
- NAR:
-
Naringin
- OPLS:
-
Orthogonal partial least square
- UV:
-
Ultraviolet–visible spectroscopy
- HPLC:
-
High-performance liquid chromatography
- UPLC:
-
Ultra performance liquid chromatography
- LC–MS/MS:
-
Liquid chromatography with tandem mass spectrometry
- FTIR:
-
Fourier transform infrared spectroscopy
- LDR:
-
Light dependent resistors
- RMSEP:
-
Root mean square error of prediction
- RMSECV:
-
Root mean square for cross-validation
- RSD:
-
Relative standard deviation
References
Abdallah FF, Darwish HW, Darwish IA, Naguib IA (2019) Orthogonal projection to latent structures and first derivative for manipulation of PLSR and SVR chemometric models’ prediction: a case study. PLoS ONE 14(9):e0222197
Akiyama T, Yamada T, Maitani T (2000) Analysis of enzymatically glucosylated flavonoids by capillary electrophoresis. J Chromatogr A 895:279–283
Alam P, Siddiqui N, Al-Rehaily A, Alajmi M, Basudan O, Khan T (2014) Stability-indicating densitometric high-performance thin-layer chromatographic method for the quantitative analysis of biomarker naringin in the leaves and stems of Rumex vesicarius L. J Planar Chromatogr Mod TLC 27:204–209
Auti P, Gabhe S, Mahadik K (2019) Bioanalytical method development and its application to pharmacokinetics studies on Simvastatin in the presence of piperine and two of its synthetic derivatives. Drug Dev Ind Pharm 45:664–668
Bhushan R, Gupta D (2005) Thin-layer chromatography separation of enantiomers of verapamil using macrocyclic antibiotic as a chiral selector. Biomed Chromatogr 19:474–478
Biancolillo A, Marini F (2018) Chemometric methods for spectroscopy-based pharmaceutical analysis. Front Chem 6:576
Cârcu-Dobrin M, Hancu G, Papp LA, Fülöp I, Kelemen H (2021) Development of a chiral capillary electrophoresis method for the enantioseparation of verapamil using cyclodextrins as chiral selectors and experimental design optimization. Symmetry 13(11):2186
Darbandi A, Sohrabi MR, Bahmaei M (2020) Development of a chemometric-assisted spectrophotometric method for quantitative simultaneous determination of Amlodipine and Valsartan in commercial tablet. Optik 218:165110
Dinc E, Baleanu D (2002) Spectrophotometric quantitative determination of cilazapril and hydrochlorothiazide in tablets by chemometric methods. J Pharm Biomed Anal 30:715–723
Hakim A, Loka IN, Prastiwi NW (2019) New method for isolation of Naringin compound from citrus maxima. Nat Resour 10:299
I. H. T. Guideline, Note for Guidance on Toxicokinetics (1994) The Assessment of Systemic Exposure in Toxicity Studies S3A, ICH harmonization for Better Health, Geneva, Switzerland
Indrayanto G (2018) Chapter five: validation of chromatographic methods of analysis: application for drugs that derived from herbs. In: Brittain HG (ed) Profiles of drug substances excipients and related methodology. Academic Press, pp 359–392
Ishii K, Furuta T, Kasuya Y (1997) Determination of naringin and naringenin in human urine by high-performance liquid chromatography utilized solid-phase extraction. J Chromatogr B Biomed Appl 704:299–305
Kamal A, Khan W, Ahmad S, Ahmad FJ, Saleem K (2015) Development and validation of high-performance liquid chromatography and high-performance thin-layer chromatography methods for the quantification of khellin in Ammi visnaga seed. J Pharm Bioallied Sci 7(4):308–313
Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, Li Q, Shoemaker BA, Thiessen PA, Yu B, Zaslavsky L, Zhang J, Bolton EE (2023) PubChem 2023 update. Nucleic Acids Res 51(D1):D1373–D1380
Mazumder R, Mahanti B, Mazumder S, Pal R, Chowdhury AK (2020) Improved comprehensive analytical method for assessment of satranidazole in drug and product. Fut J Pharmaceut Sci 6:202054
National Center for Biotechnology Information (2023). PubChem compound summary for CID 2520, Verapamil. Accessed 15 Jul 2023
Opinion S (2011) Scientific Opinion on the safety and efficacy of naringin when used as a sensory additive for all animal species. EFSA J 9:1–12
Prachi B, Asutosh Phatak SR (2013) Simultaneous determination of doxylamine succinate, pyridoxine hydeochloride and folic acid by chemometric spectrophotometry. Int J Pharma Bio Sci 4:738–749
Prasad S, Mandal I, Singh S, Paul A, Mandal B, Venkatramani R, Swaminathan R (2017) Near UV-visible electronic absorption originating from charged amino acids in a monomeric protein. Chem Sci 8(8):5416–5433
Ravi Y, Rajkamal B (2019) An improved Lc-Ms/Ms method development and validation for the determination of trandolapril and verapamil in human plasma. Int J Pharm Pharm Sci 11:91–95
Ríos-Reina R, Azcarate SM (2023) How chemometrics revives the UV-Vis spectroscopy applications as an analytical sensor for spectralprint (nontargeted) analysis. Chemosensors 11(1):8
Shabbak A, Habshah Midi H (2012) An improvement of the hotelling statistic in monitoring multivariate quality characteristics. Math Probl Eng 2012:15
Sharma AK, Bharti S, Ojha S, Bhatia J, Kumar N, Ray R, Kumari S, Arya DS (2011) Up-regulation of PPARγ, heat shock protein-27 and-72 by naringin attenuates insulin resistance, β-cell dysfunction, hepatic steatosis and kidney damage in a rat model of type 2 diabetes. Br J Nutr 106:1713–1723
Sonawane SS, Chhajed SS, Attar SS et al (2019) An approach to select linear regression model in bioanalytical method validation. J Anal Sci Technol 10:1
Trivedi L, Telrandhe R, Dhabarde D (2017) Differential spectrophotometric method for estimation and validation of Verapamil in Tablet dosage form. Int J Pharm Drug Anal 5:419–422
Tulasi I, Shivani G (2019) Bioavailability enhancers: an overview. Int J Adv Res 5:825–829
Van Wyngaard E, Blancquaert E, Nieuwoudt H, Aleixandre-Tudo JL (2021) Infrared spectroscopy and chemometric applications for the qualitative and quantitative investigation of grapevine organs. Front Plant Sci 12:723247
Vijayabaskar S, Mahalingam V (2017) Analytical method development and validation for the analysis of verapamil hydrochloride and its related substances by using ultra performance liquid chromatography. J Pharm Biomed Anal 137:189–195
Wen J, Qiao Y, Yang J, Liu X, Song Y, Liu Z, Li F (2012) UPLC-MS/MS determination of paeoniflorin, naringin, naringenin and glycyrrhetinic acid in rat plasma and its application to a pharmacokinetic study after oral administration of SiNiSan decoction. J Pharm Biomed Anal 66:271–277
Xiong X, Jiang J, Duan J, Xie Y, Wang J, Zhai S (2014) Development and validation of a sensitive liquid chromatography-tandem mass spectrometry method for the determination of naringin and its metabolite, naringenin, in Human Plasma. J Chromatogr Sci 52:654–660
Zou L, Li H, Ding X, Liu Z, He D, Kowah JAH, Wang L, Yuan M, Liu XA (2022) Review of The application of spectroscopy to flavonoids from medicine and food homology materials. Molecules 27:7766
Acknowledgements
The authors acknowledge Himalayan Pharmacy Institute, Majhitar, East Sikkim, for providing adequate facilities to perform the experiments and also thanks to Gitanjali College of Pharmacy, Lohapur-731237, Birbhum, West Bengal, India, Calcutta Institute of pharmaceutical Technology & A.H.S, Uluberia, Howrah and BCDA College of Pharmacy & Technology, Barasat, Kolkata for providing all necessary support.
Funding
The authors have no funding to report.
Author information
Authors and Affiliations
Contributions
SS and BS contributed to the technique, experimental work, original draft writing, and data analysis. SM and SC contributed to the conceptualization, instructions, reviewing and formal analysis of the study. RM drafted and edited the manuscript. BS supervised the work and NRB conceived and designed of the study. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sharma, S., Shrestha, B., Bhuyan, N.R. et al. Chemometric method development for the determination of naringin and verapamil. Bull Natl Res Cent 48, 13 (2024). https://doi.org/10.1186/s42269-024-01169-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s42269-024-01169-3