Abstract
Background
This work presents an experience done to evaluate the number of very small objects in the field of view of a stereo microscope, which are usually counted by direct observation, with or without the use of grids as visual aids. We intend to show that deep learning recent algorithms like YOLO v5 are adequate to use in the evaluation of the number of objects presented, which can easily reach the 1000 s. This kind of algorithm is open-source software, requiring a minimum of skills to install and run on a regular laptop. We further intend to show that the robustness of these kinds of approaches using convolutional neural networks allowed for the use of images of less quality, such as the images acquired with a cell phone.
Results
The results of training the algorithm and counting microalgae in cell phone images were assessed through human curation in a set of test images and showed a high correlation, showing good precision and accuracy in detections.
Conclusions
This is a low-cost alternative available worldwide to many more facilities than expensive cameras and high-maintenance rigid set-ups, along with software packages with a slow learning curve, therefore enlarging the scope of this technique to areas of knowledge where the conditions of laboratory and human work are a limiting factor.
Similar content being viewed by others
Background
When growing microalgae, being it at a laboratory or industrial scale, cell counting methods are essential for monitoring population growth. There are several alternative methods of estimating population size and growth, such as the optical density of the culture or in vivo fluorescence of chlorophyll a, but in many instances knowing the number of cells is necessary so that physiological processes and products may be referred in a per-cell basis (Guillard and Sieracki 2005).
The most common and simple counting method for microalgae cultures is by light microscopy using specific counting devices (counting chambers) selected based on the size of the organism and the cell concentration (Guillard 1973; Karlson et al 2010; Reguera et al 2016). This method has the advantage that the cells can be observed by the researcher, and the physiological status of the culture maybe assessed. However, it is also very time-consuming and tedious, particularly when a high number of cultures need to be examined, a common practice in laboratory experiments and industrial production units.
Already in the second half of the twentieth century, electronic devices aiming at automated counting systems started to be developed to help overcome these constraints (Parsons 1973). In the last decades, automated counting methods for cell cultures have been increasingly implemented and developed and there are now in the market several bench-top solutions available (e.g. CellDrop, DeNovix Inc., USA; Countess, Thermo Fisher Scientific, USA; EVE, NanoEntek, South Korea) characterized by having high efficiency and precision, and requiring low application times and low expertise. These methods are now starting to be used in microalgae research laboratories (e.g. Salbitani et al 2022). The main drawback of these automated methods is their initial high cost (above 15 000€) rendering them inaccessible to many laboratories, particularly in underdeveloped countries.
Notwithstanding, other high technological approaches and equipment are nowadays easily accessible. Cell phones are increasingly accessible and feature high-quality cameras, with images useful in many computer vision tasks such as object detection and recognition. In the last decades, the detection of objects using deep learning techniques has been intensively exploited, mainly due to its broad application (Cao et al. 2018; Viso.AI n. d.; Wang et al. 2018) and good results namely in almost all fields of biology (Webb 2018; Ching et al 2018), microscopy imagery (Sorzano et al 2009; Ito et al 2018; Xing et al 2018; Devan et al 2019) and microorganisms in general (Zhang et al. 2021). What motivated our interest in deep learning algorithms was the limited success of conventional image processing techniques applied to images where context is a decisive factor and the operator reasoning must be incorporated. In addition, inherent in deep learning is the possibility of data augmentation. Data augmentation is the introduction of small randomized changes to train data, in the form of radiometric and geometric alterations in dimensions, rotation, translation, and shearing. This is particularly interesting because whatever the number of train images we use, we cannot guarantee that all the possible positions, sizes, and aspects of the cells are contemplated in the training set.
The main objective of this work was to develop a low-cost cell counting method for microalgae cultures that were simultaneously easy, fast, and accurate. To achieve this objective, we used a stereo microscope and images acquired with a common cell phone, which were pre-processed in Matlab environment, with basic image processing operations available in any open-source image processing software (e.g. ImageJ), and a deep learning algorithm also open-source, running on a regular laptop.
Methods
Concerning microalgae strains and culture conditions in this study, two different strains of the marine dinoflagellate species Protoceratium reticulatum (Claparède & Lachmann) Bütschli 1885 (IO116-01 and IO116-02) were used both in the training and validation stage of the counting experiments. The cultures were obtained from the algae culture collection of the University of Lisbon (ALISU) and were grown under controlled laboratory conditions (Fitoclima 600PL, Aralab, Portugal), in L1 medium (Andersen et al 2005), at a salinity of 33, at 19 ± 1 °C under a 12:12 h light: dark cycle, 100–110 μmol photons m−2 s−1.
In what concerns sample preparation for cell counts, 3 ml culture samples were fixed with approximately 0.15 ml of Lugol’s solution (Karlson et al 2010). Immediately before filling the counting chamber samples were homogenized by gently rotating the flask 25 times. A sub-sample was then used to fill the chamber of a Palmer–Maloney counting slide (100 µl) (LeGresley and McDermott 2010). No dilution steps were used. The sample was allowed to settle for 5 min, and the chamber was placed under a stereo microscope at 10 × magnification (Zeiss Stemi 305, Germany). The images were then acquired through the eyepiece with a cell phone (Samsung M21, South Korea) equipped with a 48.0 MP camera (Samsung S5KGM1—f/2.0, 26 mm (wide), 1/2.0″, 0.8 µm). The image covered the whole area of the counting chamber, the equivalent of a100 µl culture sample.
For the image train process, a total of 6 images of 2250 × 4000 pixels at 24 bits were acquired from cultures in different phases of the growth curve to cover a variety of particle properties (e.g. range of cell sizes, cell debris, and thecal plates).
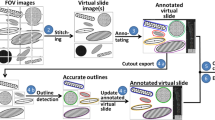
The pre-processing of the images was achieved in Matlab R2021a environment and consisted of four steps: first, a modified homomorphic filter was applied with a sigma of 11 to compensate for irregular illumination of the background; second, all images were histogram matched to one reference image, chosen for its ideal radiometric range; the third step consisted in producing a binary mask for each image with a global threshold, followed by morphological operations to consolidate the area of interest and eliminate surrounding structures included in the field of view (FOV); finally, this mask was applied to the processed image (Fig. 1).
The algorithm used is one of the latest developments in one-stage algorithms based on convolutional neural networks (CNNs), the 5th version since the introduction of the concept You Only Look Once (YOLO) (Redmon et al 2016). YOLO v5 was made publicly available in a GitHub repository in 2020 (GitHub Ultralytics n.d.). The algorithm has been retrained for the task using a transfer learning technique: since many basic features are common to all detection problems (edges, contrasts, forms, etc.), an already heavily trained network can be used to implement a new problem. The new discriminators will define the last layers of the CNN, tuning the detector according to the details of the specific problem. After the download and successful installation of YOLOv5, the algorithm was trained on our data set as described below. The set of images acquired was segmented into tiles of 800 × 800 pixels to accelerate training procedure, as it was our objective to use the most complete model of YOLOv5, the model x that uses a CNN with 476 layers.
The train demands a set of images with all the objects of interest identified with bounding boxes, and the respective list; the data set annotated in this way, called ground truth, is then split into train and validation subsets, between which the algorithm will converge to the best possible achievement in terms of precision (percentage of true positives correctly classified) and recall (percentage of true positives detected).
A metric usually considered in object detection applications is the mean average precision (mAP) that quantifies the stability and consistency of the model, within a confidence threshold related to the intersection-over-union (IoU) areas between the anchor boxes estimated from the train data and the bounding boxes predicted by the model in the annotated data. With a threshold of X, the box is assigned to an object of interest if the IoU quotient is above X and considered background in the opposite case. Non-maxima suppression ensures most multiple detections are avoided, by considering only the box with maximum probability in each set of overlapping boxes.
The annotation of a subset of images for train and validation purposes can be made with online tools, such as (Makesense.AI n.d.) used in the present work, with a user-friendly graphic interface. The images to be annotated (usually 30% of the images available, further split in 20% for train, 10% for validation) are uploaded to the site and the graphic tools available in the interface are used to draw boxes around all the objects of interest in each image, using zoom, correction, delete and pan functionalities. At the end, a text file for each image is exported in a user-defined format, with all the annotations (image coordinates for the boxes) made in that image. The images and corresponding text files are then distributed between validation and train, because YOLOv5 requires a fixed directory tree, with names that it will recognize to know where to find image and label files during the train stage. Once trained, the algorithm was applied to a test data set of 43 images of P. reticulatum cultures as described above. Results were assessed by manually verifying the false negatives and false positives in each image, and the performance of the model was evaluated based on precision and recall.
Results
The 6 images acquired were cropped in 72 initial tiles of 800 × 800 pixels, and the algorithm was trained with 21 annotated images, using 14 images for train and 7 for validation. We verify later that several images only contain the pieces surrounding the area of interest, so we tested on 43 remaining images containing 3659 microalgae cells. With a confidence threshold of 0.25 for inference, YOLOv5 detected 3681 objects, of which 100 were false positives, leaving out 78 false negatives, giving an overall precision of 97.4% and a recall of 97.9%. Other parameterizations for the confidence threshold (0.20, 0.30 and 0.35) were tried with less success, as the improvement in accuracy led to a decrease in recall performance. An IoU threshold of 0.25 had the best results, avoiding most multiple detections. The mAP@0.5 was 0.862.
With this trade-off in parameterization, among the 3681 particles identified 2.7% were wrongly detected (false positives) and 2.1% of the 3659 particles present were not detected (false negatives), which seems like reasonable numbers compared to human curation.
If we look at the results considering each image as a unity, 72% reach a precision of 95%, 74% attain 95% of recall and 67% attain both recall and precision better than 95% simultaneously. The result of the inference for the image that exemplifies the pre-processing is shown in Fig. 2.
Points of interest (red squares) detected by YOLOv5 in the image shown in Fig. 1
Inference over an 800 × 800 pixel image is computed in 1800 ms in the laptop described below, giving two or three outputs: a numerical output in the command window with the total count of detections and a file containing a copy of the image with all occurrences flagged (Fig. 2), as well as an optional text file featuring the coordinates of all the windows around the occurrences in image coordinates, allowing to extract every single particle for further processing if desired.
Discussion
The estimation of cell numbers when culturing microalgae is essential because the physiological processes under study or the target products to be produced usually need to be referred in a per-cell basis. Manual counting of microalgae cells for estimation of cell concentrations with acceptable accuracy levels consumes considerable human resources and is a tedious procedure sometimes with results affected by operator subjectivity and fatigue. This limits the number of samples that can be processed which may partly constraint the outputs of the work. Nevertheless, it is still used today in routine estimation of cell culture concentrations (e.g. Pereira et al 2016; Sheward et al 2017; Rocha et al. 2022) probably because of the lack of affordable, accurate and precise automatic alternatives in the market.
In the present work we provide the tools for the implementation of an automated cell counting method with no costs or only residual costs to the user. Compared to other automated cell counting methods routinely used in many laboratories, the methodology described is accessible in financial terms in facilities where a stereo microscope is available, as it only requires a regular laptop and a cell phone, and software available online without any associated costs. The learning curve is fast for anyone used to software with parameterizable interfaces.
The same methodology can be easily extended to other particles, requiring however a separate training stage for each kind of organism to be counted. The high number of epochs or iterations in the training stage is time-consuming, but it is done just once. As a reference, in our case, 700 iterations of model x, with images of 800 × 800 pixels took 21.2 h in a HP-Pavilion 17-cd1006np laptop with an Intel Core i7 processor, 16 GB SDRAM and a graphic unity NVIDIA GeForce RTX 2060 Max-Q 6 GB. In microalgae production units or when developing a particular laboratory experiment, the number of species that are cultured is relatively small, and this methodology could easily be implemented for all species of interest.
The annotation of the subset of images for train and validation purposes should be done by someone knowing well the characteristics of the organism to be counted. This will allow high accuracy output once the system is implemented. From the technical operator perspective, the system does not require an expensive training program since the only needed expertise is image acquisition using a standard cell phone and the basic informatic skills of any computer user to run a new image.
In addition, the method here proposed allows the counting of a higher number of cells in a significantly shorter time. This increases the confidence limits of the concentration estimates while allowing for a larger number of samples to be counted each day.
Conclusions
The present work describes a processing chain that allows the implementation of a particle counting procedure with application in the field of microalgae research and production. It allows monitoring population growth in microalgae cultures using a deep learning method with accurate results, and much less cost in both human labour, laboratory technical equipment and high-end image acquisition equipment. As with other automated cell counting methods, it is faster than using the light microscope and allows for the enumeration of high particle numbers, increasing statistical robustness (Guillard & Sieracki 2005). The human uncertainty involved in the tedious labour of counting a large number of cultures is eliminated, as well as the errors due to interruptions or fatigue.
Future work will include an evaluation of the performance of the same train in cultures of other microorganisms of similar gross morphology, which could lead to a portfolio of ready-to-run train files appropriated for the set of organisms most usually processed in each facility.
Availability of data and materials
Not applicable.
Abbreviations
- MARE:
-
Marine and Environmental Sciences Centre
- ARNET:
-
Aquatic Research Network
- FCUL:
-
Faculty of Sciences, University of Lisbon
- YOLO v5:
-
You Only Look Once version 5
- ALISU:
-
Algae culture collection of the University of Lisbon
- CNN:
-
Convolutional neural network
- mAP:
-
Mean average precision
- IoU:
-
Intersection-over-union
References
Andersen RA, Berges JA, Harrison PJ, Watanabe MM (2005) Appendix A-Recipes for freshwater and seawater media. In: Andersen RA (ed) Algal culturing techniques. Elsevier, Oxford, pp 429–538
Cao C, Liu F, Tan H, Song D, Shu W, Li W, Zhou Y, Bo X, Xie Z (2018) Deep learning and its applications in biomedicine. Genom Proteom Bioinf 16:17–32
Ching T, Himmelstein DS, Beaulieu-Jones BK, Kalinin AA, Do BT, Way GP et al (2018) Opportunities and obstacles for deep learning in biology and medicine. J R Soc Interface 15(141):20170387. https://doi.org/10.1098/rsif.2017.0387
Devan KS, Walther P, von Einem J, Ropinsky T, Kestler HA, Read C (2019) Detection of herpesvirus capsids in transmission electron microscopy images using transfer learning. Histochem Cell Biol 151:101–114
GitHub Ultralytics n.d. GitHub - ultralytics/yolov5: YOLOv5 in PyTorch > ONNX > CoreML > TFLite. Accessed 21 June 2021
Guillard RR (1973) Division rates. In: Stein JR (ed) Handbook of phycological methods: culture methods and growth measurements. Cambridge University Press, Cambridge, pp 289–311
Guillard RR, Sieracki MS (2005) Counting cells in cultures with the light microscope. In: Andersen RA (ed) Algal culturing techniques. Elsevier Academic Press, Oxford, pp 239–252
ImageJ n.d. ImageJ Accessed 6 June 2022
Ito E, Sato T, Sano D, Utagawa E, Kato T (2018) Virus particle detection by convolutional neural network in transmission electron microscopy images. Food Environ Virol 10:201–208
Karlson B, Godhe A, Cusack C, Bresnan E (2010) Introduction to methods for quantitative phytoplankton analysis. In: Karlson B, Cusack C, Bresnan E (eds) IOC Manuals and Guides, 55: Microscopic and molecular methods for quantitative phytoplankton analysis. UNESCO, Paris, pp 5–12
LeGresley M, McDermott G (2010) Counting chamber methods for quantitative phytoplankton analysis-haemocytometer, Palm-er-Maloney cell and Sedgewick-Rafter cell. In: Karlson B, Cusack C, Bresnan E (eds) IOC Manuals and Guides, 55: Microscopic and molecular methods for quantitative phytoplankton analysis. UNESCO, Paris, pp 25–30
Makesense.AI. n.d. Accessed 24 June 2021. https://www.makesense.ai/
Parsons TR (1973) Coulter Counter for phytoplankton. In: Stein JR (ed) Handbook of phycological methods: culture methods and growth measurements. Cambridge University Press, Cambridge, pp 345–358
Pereira H, Gangadhar KN, Schulze PSC, Santos T, de Sousa CB, Schueler LM, Custódio L, Malcata FX, Gouveia L, Varela JCS, Barreira L (2016) Isolation of a euryhaline microalgal strain, Tetraselmis sp. CTP4, as a robust feedstock for biodiesel production. Sci Rep 6:1–11. https://doi.org/10.1038/srep35663
Redmon J, Divvala S, Girshick R, Farhadi A (2016) You only look once: unified, real-time object detection. IEEE Conf on Computer Vision and Pattern Recognition (CVPR) 2016:779–788. https://doi.org/10.1109/CVPR.2016.91
Reguera B, Méndez S, Alonso R (2016) Quantitative analysis of microalgae: General considerations. In: Reguera B, Alonso R, Moreira A, Méndez S, Dechraoui-Bottein M-Y (eds) Guide for designing and implementing a plan to monitor toxin-producing microalgae, 2nd edn. Intergovernmental Oceanographic Commission (IOC) of UNESCO and International Atomic Energy Agency (IAEA), Paris and Vienna. IOC Manuals and Guides 59:35–43
Rocha GS, Parrish CC, Espíndola ELG (2022) Changes in photosynthetic parameters and lipid classes of N-limited Ankistrodesmus densus (Chlorophyceae) under cadmium exposure. J Appl Phycol. https://doi.org/10.1007/s10811-022-02859-z
Salbitani G, Carillo P, Di Martino C, Bolinesi F, Mangoni O, Loreto F, Carfagna S (2022) Microalgae cross-fertilization: short-term effects of Galdieria phlegrea extract on growth, photosynthesis and enzyme activity of Chlorella sorokiniana cells. J Appl Phycol 34:1957–1966. https://doi.org/10.1007/s10811-022-02769-0
Sheward RM, Poulton AJ, Gibbs SJ, Daniels CJ, Bown PR (2017) Physiology regulates the relationship between coccosphere geometry and growth phase in Coccolithophores. Biogeosciences 14(6):1493–1509. https://doi.org/10.5194/bg-14-1493-2017
Sorzano COS, Recarte E, Alcorlo M, Bilbao-Castro JR, SanMartín C, Marabini RJM (2009) Automatic particle selection from electron micrographs using machine learning techniques. J Struct Biol 167:252–260
Viso.AI. n.d. 83 Most Popular Computer Vision Applications in 2022 - viso.ai. Accessed Jan 2022
Wang J, Ma Y, Zhang L, Gao RX, Wu D (2018) Deep learning for smart manufacturing: methods and applications. J Manuf Syst 48(C):144–156
Webb S (2018) Deep learning for biology. Nature 554:555–557
Xing F, Xie Y, Su H, Liu F, Yang L (2018) Deep learning in microscopy image analysis: a survey. IEEE Trans Neural Netw Learn Syst 29:4550–4568
Zhang Y, Jiang H, Ye T, Juhas M (2021) Deep Learning for Imaging and Detection of Microorganisms. Trends Microbiol 29(7):569–572. https://doi.org/10.1016/j.tim.2021.01.006
Acknowledgements
Not applicable.
Funding
This study had the support of national funds through Fundação para a Ciência e Tecnologia (FCT), under the project LA/P/0069/2020 granted to the Associate Laboratory ARNET and UIDB/04292/2020+UIDB/04292/2020, granted to MARE (Marine and Environmental Sciences Centre).
Author information
Authors and Affiliations
Contributions
MCP contributed to conceptualization, methodology, and software; MB contributed to data curation and validation; AA contributed to formal analysis, review, and editing; all authors contributed to writing the final version; all authors have read and agreed to the published version of the manuscript and approved the submitted version.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Proença, M.C., Barbosa, M. & Amorim, A. Counting microalgae cultures with a stereo microscope and a cell phone using deep learning online resources. Bull Natl Res Cent 46, 278 (2022). https://doi.org/10.1186/s42269-022-00965-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s42269-022-00965-z