Abstract
Background
A common complication of any respiratory disease by a virus could be a secondary bacterial infection, which is known to cause an increase in severity. It is, however, not clear whether the presence of some opportunistic pathogens called pathobionts contributes to the severity of the disease. In COVID-19 patients, undetected bacterial co-infections may be associated with the severity of the disease. Therefore, we investigated the implications of bacterial co-infections in COVID-19 cases.
Results
This is a cross-sectional study that involved archived specimens collected from nasopharyngeal samples of 150 people for COVID-19 screening in Lagos. DNA extraction from the samples was carried out to determine the presence of five respiratory bacterial pathogens using nested real-time PCR, and data were analysed using the Chi-square test. Of the 150 samples collected, 121 (80.7%) were positive for SARs-CoV-2 infection and 29 were negative. The proportion of patients with bacteria co-infection in COVID-19-negative, asymptomatic, and mild cases were 93.1%, 70.7%, and 67.5%, respectively. There was no statistically significant difference between mild COVID-19 conditions and bacteria co-infection (p = 0.097). There was also no significant difference in the nasal carriage of Staphylococcus aureus, Mycoplasma pneumoniae, and Haemophilus spp. However, there was a statistically significant increase in the carriage of Moraxella catarrhalis and Chlamydophila pneumoniae among COVID-19-negative patients when compared with the positive patients (p value = 0.003 and 0.000 for Moraxella catarrhalis and Chlamydophila pneumoniae, respectively).
Conclusions
The current study shows that bacterial co-infection and superinfection with COVID-19 are not associated with mild and asymptomatic COVID-19 cases in our setting. However, given the high prevalence of Staphylococcus aureus and Mycoplasma pneumoniae among the mild COVID-19 cases seen in this study, early diagnosis and treatment of these bacterial co-infections are still encouraged to mitigate the effect on the severity of COVID-19.
Similar content being viewed by others
Background
The coronavirus disease 2019 (COVID-19) which is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is highly infectious. The disease emerged in Wuhan, China, and has become a pandemic (Shereen et al. 2020). Studies have shown that SARS-CoV-2and severe acute respiratory syndrome-like viruses (found in bats) are phylogenetically linked, and hence, bats could be the principal reservoir for SARS COV2 (Banerjee et al. 2019; Boni et al. 2020; Shereen et al. 2020). Epidemiologically, COVID-19 is currently a major pandemic with over 5.8 million deaths reported as of 15 February 2022 globally, which is of great concern (WHO Report 2022).
Secondary bacterial infections have been reported to be the cause of respiratory viral disease complications leading to increased morbidity in humans (Smith and McCullers 2014; Morris et al. 2017; Manohar et al. 2020). It was recently proposed that respiratory infections are linked to an imbalance of the nasopharyngeal microbiota (Man et al. 2017; Qin et al. 2020). Furthermore, increased bacterial colonization, immune dysfunction, and viral infection of the respiratory tract expose it to secondary bacterial infections (Morris et al. 2017). However, new data reveal that the microbial communities that live on our mucosal surfaces influence the rigor of our immune responses as well as the ecological connections that exist between hosts and infections (Chiu et al. 2017; Tay et al. 2020). In the last decade, there has been a lot of interest in figuring out how the microbial populations that live in our bodies regulate the balance between health and disease vulnerability, including infections. This suggests the notion that an acute viral infection disrupting normal microbial communities contributes to the development of post-viral bacterial pneumonia (Hanada et al. 2018).
Safaeyan et al. (2015) reported the presence of specific bacteria using qPCR technique in adult patients infected with influenza A, as well as healthy individuals. Staphylococcus aureus, Streptococcus pneumoniae, and Haemophilus influenzae were present in 12%, 24%, and 32% of infected patients, respectively, as compared to 5%, 11%, and 10% of uninfected patients.
It has been reported that pneumonia in older populations and young adults was associated with dysbiosis of the upper respiratory tract microbiome with bacterial overgrowth of a single species (de Steenhuijsen Piters et al. 2016).
It is, however, not clear whether the presence of some opportunistic pathogens called pathobionts contributes to the severity of the disease. Therefore, the study aimed at investigating and identifying bacterial pathogens or pathobionts in nasopharyngeal samples through direct amplification of their 16SrDNA genes from patients presenting symptoms of the COVID-19 could provide information regarding the probable relationship.
Methods
Ethics, study type, and study population
This study was submitted to the Institutional Review Board (IRB) of the Nigerian Institute of Medical Research (NIMR) and ethical approval was obtained from the (IRB/20/032) and written consent was obtained from the patients prior to sample collection.
This study was a cross-sectional study. A total of 150 nasopharyngeal samples obtained from participants who presented for COVID-19 testing during the period of May 2020 and August 2020 were retrieved from the NIMR biobank and included in the study.
Sample collection
This step was carried out once for each patient. Briefly, nasopharyngeal samples were obtained using a swab and then placed into Viral Transport Medium (VTM) and taken to a Biosafety level III laboratory within the Institute. Information on demographic characteristics and symptoms presented was recorded during the patient’s enrolment.
Reverse transcription real-time polymerase chain reaction (RT-PCR) for the detection of COVID-19
First, the extraction of the viral RNA was performed from the inactivated samples aliquots using QIAamp RNA extraction kit according to the manufacturer’s instructions (Qiagen, Maryland, USA). Next, one-step reverse transcriptase (RT) real-time (qPCR) was carried out to detect SARS-CoV-2 using qPCR assay designed by BGI, Shenzhen, China (Shaibu et al. 2021).
Molecular analysis for bacterial identification
The method of Curran et al. (2007) was adopted with slight modification for the molecular analysis of the bacteria. Bacterial DNA was extracted from the sample, and species-specific primers of 5 bacteria known to be associated with respiratory tract infections (S. aureus, M. catarrhalis, Haemophilus sp., C. pneumoniae, and M. pneumoniae) were used for the amplification of the bacterial DNA using PCR (Table 1). The first reaction of the nested PCR was performed using the conventional PCR method in a Peltier PTC-200, MJ Research Thermal Cycler at an initial denaturation of 95 °C for 3 min, 35 cycles of 95 °C for 30 s, 55 °C for 40 s, 72 °C for 45 s, final elongation at 72 °C for 10 min, and a final hold at 10 °C for 0 s. The final reaction volume for the first round of the nested PCR was 20 µl which contained 4 µl of extracted DNA. The primers used for the target genes nuc, uspA, p6, momp, and p1 are listed in Table 1. The target genes were amplified using conventional PCR and nested real-time PCR. Master Mix (EvaGreen Solis Biodyne) made of 1 mM of each dNTP, 1X PCR buffer, 7.5 mM of Mgcl 2, and 1U of thermostable Taq polymerase, and 1 µM of each primer set (forward and reverse). Molecular grade water was used to make up the reaction mixture.
The second reaction of the nested PCR was done using a BioRad CFX 96 Deep Well Real-Time system with HOT Firepol Evagreen ® qPCR supermix, 5× (cat no. 08-36-00008) according to the manufacturer’s instructions to identify the genetic expression in ct values of the genes specific to the bacteria using the primers. The supermix is a ready-to-use mix containing chemically modified Firepol® DNA Polymerase that enables hot start, optimized hot-start PCR buffer, Mgcl2, dUTPs, dNTPs, and Evagreen dye. The reaction was performed in a 20 µl reaction volume containing 0.1 µM Evagreen qPCR supermix (5×), 1 µM of each set of primers (forward and reverse), and 4 µl of DNA template from the first PCR reaction. The amplification was done at an initial denaturation of 95 °C for 5 min, followed by 30 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s. The final elongation was done at 72 °C for 5 min. The temperature was raised to 95 °C for 30 s, 65 °C for 30 s, and the melt curve was performed at 0.5 °C increments of 65–95 °C every 1 s before the cycle was terminated.
Quantitative nested real-time PCR
The method of Curran et al. (2007) was adopted with slight modification. Nested primers were designed targeting specific functional genes of a range of bacteria associated with respiratory infection (Table 1). The first round of each assay was carried out on a conventional thermal cycler (Peltier Thermal Cycler 200, MJ Research), while the second round of real-time PCR was carried out on an ABI 7000 system (Applied Biosystems, Warrington, UK). Each run included a negative control containing nuclease-free water as template. The 10-mL of PCR mastermix in the first-round contained two microlitres of either standard or sample DNA added to each reaction. The hot-start PCR cycling conditions were as follows: initial denaturation at 94 °C for 3 min; followed by 30 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s; with a final extension step at 72 °C for 5 min.
The second-round real-time PCR mastermix was as above with 1_ SYBR Green I (Sigma). The template for the second round was 0.2 mL of the first-round reaction. The hot-start PCR cycling conditions were as follows: initial denaturation at 95 °C for 5 min; followed by 30 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s; with a final extension step at 72 °C for 5 min. For melt-curve analysis, samples were denatured at 95 °C for 30 s, cooled at 65 °C for 30 s, and then slowly heated to 95 °C with continuous monitoring of fluorescence to determine the melting temperature of the DNA product. The results of the qPCR were analysed with the BioRad CFX Manager 3.1 software using the negative control to set the ct value.
Statistical analysis
Demographic data such as age and gender were retrieved from records, de-identified, and presented in frequencies and percentages. Chi-square test was used to determine the association between the participants’ gender and age group of the severity of COVID-19 cases. The distribution and prevalence of bacterial co-infections with COVID-19 cases were evaluated using Chi-square test. Multivariate analysis (binary logistic regression) was used to ascertain the effects of age and gender on the likelihood of bacterial infection associated with COVID-19 severity. Also, SPSS software (version 23.0) was applied for all statistical analyses. Significant statistical associations were considered at a p value less than or equal to 0.05.
Results
The prevalence of COVID-19 was higher in males than females. There is no statistically significant difference between the variables, gender, and severity of COVID-19 cases as p > 0.05. This implies that both males and females are equally likely to have severe cases of COVID-19. The association between participants’ age group and severity of COVID-19 cases is statistically significant as p ≤ 0.05, and this suggests that the variables, age group, and severity of COVID-19 cases are associated with each other. All the participants within the different age groups tend to exhibit dissimilar severe cases of COVID-19 (Table 2). The table depicts the severity level of cases such as mild, no symptoms as well as negative samples for the participant’s age group and gender.
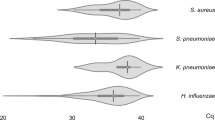
Generally, among study participants, the results showed that bacterial co-infection was present in 55.3% of the 150 samples with Chlamydophila pneumoniae being the most common bacterial co-infection among COVID-19 patients. Furthermore, there was a statistically significant increase in the carriage of Moraxella catarrhalis and Chlamydophila pneumoniae among COVID-19-negative patients when compared with the positive patients (p value was 0.003 and 0.000 for Moraxella catarrhalis and Chlamydophila pneumoniae, respectively) (Table 3). Of the 150 samples, 121 were positive for SARs-CoV-2 infection and 29 were negative. The COVID-19-negative patients had the highest prevalence of bacterial co-infections (93.1%) followed by asymptomatic COVID-19 patients (70.7%) and those presented with mild symptoms (67.5%). There was no statistically significant difference between COVID-19 severity and bacteria co-infection/superinfection (p = 0.097). The proportion of patient samples with at least one bacterial infection was found to be highest among those who presented with mild COVID-19 symptoms. There was a reduction in the proportion with a corresponding increase in the number of bacterial co-infections. The patients with mild symptoms were found to present the highest bacterial co-infections (1.2%) (Table 4). However, the patients with the highest prevalence of bacterial co-infections were those who tested negative at 93.1%. Overall, there was no association between the bacterial co-infections and COVID-19 cases.
Regression models:
Staphylococcus aureus Infection = 1.243 − 0.216age − 0.015gender − 0.217severity
Moraxella catarrhalis Infection = 0.578 + 0.088age + 0.923gender + 0.499severity
Chlamydophila pneumoniae Infection = − 0.334 + 0.035age + 0.507gender + 1.314severity
Mycoplasma pneumoniae Infection = 2.792 − 0.310age − 0.788gender − 0.101severity
Haemophilus spp. Infection = 3.135 + 0.176age + 0.321gender − 1.270severity
The logistic regression model is a good fit since the log-likelihood value of the models is high with the model for Staphylococcus aureus having the highest value. Similarly, the Chi-square test showed that the model fits well as p > 0.05. The model for Chlamydophila pneumoniae explained 13.3% (Nagelkerke R2) of the variance in bacterial infections and correctly classified 75.2% of bacterial infections (Table 5). For the Chlamydophila pneumoniae model, males were 1.7 times more likely to get infected with Chlamydophila pneumoniae than females (OR = 1.66, 95% C.I. 0.698–3.950, p > 0.05). In addition, patients with mild COVID-19 cases were 3.7 times more likely to get infected with Chlamydophila pneumoniae than asymptomatic patients (OR = 3.721, 95% C.I. 1.551–8.926, p < 0.05). Increasing age and gender was associated with an increased likelihood of COVID-19 patients getting infected with Chlamydophila pneumoniae. Similarly, increasing severity of COVID-19 cases was associated with an increased likelihood of COVID-19 patients getting infected with Chlamydophila pneumoniae (Table 6).
Discussion
In this study, we investigated the presence of bacterial co-infections in COVID-19 cases and analysed their epidemiological characteristics. There have been several reports of prevalence of COVID-19 to be higher in males than females (Jin et al. 2020; Abate et al. 2020; Huang et al. 2021). Data from this study revealed the prevalence of COVID-19 to be higher in males than females. This agrees with the findings of Sharifipour et al. (2020) where 58% of the COVID-19-positive patients were male and 42% were female. This could be attributed to the fact that females present a higher immune response than males (Takahashi et al. 2020). Also, gender-based lifestyles such as higher levels of smoking among men compared to women and women having a more responsible attitude towards the COVID-19 pandemic than men in their undertaking of preventive measures such as wearing face masks and frequent handwashing may be contributing factors (Bwire 2020).
Findings from this study revealed that age group 20–61 had the highest prevalence of COVID-19 particularly among those who presented with mild symptoms. This is not surprising as the working-class group falls within this age category and they could have been predisposed to the virus through commuting to work in comparison with the younger age group as well as the elderly who were mostly at home during the partial lockdown period at the time.
Bacterial co-infections have been linked to viral disease severity, both directly and indirectly as well as via immunological response (DaPalma et al. 2010; Alosaimi et al. 2020; Mirzaei et al. 2020). Several studies have reported the prevalence of influenza/bacterial co-infection as well as multiple viral co-infections (Alosaimi et al. 2020; D’Abramo et al. 2020; Ding et al. 2020; Konala et al. 2020). However, data on clinical significance of bacterial co-infections with COVID-19 are limited and the few available studies detected the bacterial pathogens through culture-based methods. Our study investigated the coexistence of 5 bacteria respiratory pathogens and COVID-19 simultaneously in order to understand the relationship between bacterial infection and a possible increase in severity of the disease. Previous studies have revealed that secondary bacterial co-infection is the main cause of death in patients with viral pneumonia (MacIntyre et al. 2018; Guo et al. 2020). In this study, bacterial co-infection was present in 55.3% of the study participants and the most common bacterial co-infection among COVID-19 patients was Chlamydophila pneumoniae.
Moraxella catarrhalis was observed in the study to be significantly more in SARS-CoV-2-negative cases. Other reports show prevalence of Moraxella catarrhalis to be higher in COVID-19-positive cases with less than 5% (Massey et al. 2020; Feldman and Anderson 2021). Our result is, however, in agreement with Calcagno et al. (2021) where Moraxella catarrhalis was higher in COVID-19 cases. The ubiquitous surface protein A2 (UspA2) on Moraxella catarrhalis is very important for its bactericidal activity as well as cross-reactive antibodies. It is usually a negative regulator of TLR3, and IL-1β consequently could reduce the antiviral defence function of the epithelium and cause an increase in viral infections susceptibility (Heinrich et al. 2016; Ysebaert et al. 2021).
Though Staphylococcus aureus, Mycoplasma pneumoniae and Haemophilus spp recorded the highest prevalence among the group which presented with mild COVID-19 symptoms, there was also no statistically significant difference in the nasal carriage of these bacteria among cases with both asymptomatic and mild symptoms. Our findings appeared to be in concordance with previous findings from Saudi Arabia but inconsistent with findings from China (Xing et al. 2020) where Mycoplasma pneumoniae and Legionella pneumophila were the most common bacteria detected among COVID-19 patients. This could be attributed to the diversity of geographical distribution of circulating respiratory bacteria. Nevertheless, in this study, Chlamydophila pneumoniae infection has been associated with mild COVID-19 symptoms. This, however, is a cause for concern and the bacterium has been reported as a common cause of acute respiratory infections with a seroprevalence of 34.1% in COVID-19 patients with fatalities (Du et al. 2020).
Our study has some limitations. First, only 5 bacteria were identified. Second, our study did not include severe and critical cases thereby limiting the highest severity level to patients with mild symptoms. Future studies to overcome these limitations need to be considered.
Conclusions
Our study highlights the importance of screening for co-infecting bacteria in COVID-19 patients, given the high prevalence of Chlamydophila pneumoniae infection. As there is a continuous increase in antimicrobial resistance, the detection of bacterial co-infections in COVID-19 cases shows the importance of early diagnosis and commencement of early treatment with antibiotics to reduce possible impact on the severity of the disease by hospitalization and associated mortality. Further studies of the respiratory tract microbiome in relation to respiratory illnesses (COVID-19) using a modern molecular approach such as next-generation sequencing (NGS) which would identify other pathobionts of the respiratory tract are recommended.
Availability of data and materials
The datasets used and/or analysed during the current study are included in the article.
References
Abate BB, Kassie AM, Kassaw MW, Aragie TG, Masresha SA (2020) Sex difference in coronavirus disease (COVID-19): a systematic review and meta-analysis. BMJ Open 10:e040129
Alosaimi B, Hamed ME, Naeem A, Alsharef AA, AlQahtani SY, AlDosari KM et al (2020) MERS-CoV infection is associated with downregulation of genes encoding Th1 and Th2 cytokines/chemokines and elevated inflammatory innate immune response in the lower respiratory tract. Cytokine 126:154895
Avadhanula V, Rodriguez CA, Devincenzo JP, Wang Y, Webby RJ, Ulett GC et al (2006) Respiratory viruses augment the adhesion of bacterial pathogens to respiratory epithelium in a viral species- and cell type-dependent manner. J Virol 80:1629–1636
Banerjee A, Kulcsar K, Misra V, Frieman M, Mossman K et al (2019) Bats and coronaviruses. Viruses 11(1):41
Boni MF, Lemey P, Jiang X, Lam TT, Perry BW, Castoe TA et al (2020) Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat Microbiol 5:1408–1417
Bwire GM (2020) Coronavirus: why men are more vulnerable to covid-19 than women? SN Compr Clin Med 1–3
Calcagno A, Ghisetti V, Burdino E, Trunfio M, Allice T, Boglione L et al (2021) Co-infection with other respiratory pathogens in COVID-19 patients. Clin Microbiol Infect 27(2):P297-298
Chiu L, Bazin T, Truchetet ME, Schaeverbeke T, Delhaes L, Pradeu T (2017) Protective microbiota: from localized to long-reaching co-immunity. Front Immunol 8:1678
Curran T, Coyle PV, McManus TE, Kidney J, Coulter WA (2007) Evaluation of real-time PCR for the detection and quantification of bacteria in chronic obstructive pulmonary disease. FEMS Immunol Med Microbiol 50(1):112–118
D’Abramo A, Lepore L, Palazzolo C, Barreca F, Liuzzi G, Lalle E (2020) Acute respiratory distress syndrome due to SARS-CoV-2 and Influenza A co-infection in an Italian patient: mini-review of the literature. Int J Infect Dis 97:236–239
DaPalma T, Doonan BP, Trager NM, Kasman LM (2010) A systematic approach to virus-virus interactions. Virus Res 149(1):1–9
de Steenhuijsen Piters WA, Huijskens EG, Wyllie AL, Biesbroek G, van den Bergh MR, Veenhoven RH et al (2016) Dysbiosis of upper respiratory tract microbiota in elderly pneumonia patients. ISME J 10:97–108
Ding Q, Lu P, Fan Y (2020) The clinical characteristics of pneumonia patients coinfected with 2019 novel coronavirus and influenza virus in Wuhan, China. J Med Virol 92(9):1549–1555
Du Y, Tu L, Zhu P, Mu M, Wang R, Yang P et al (2020) Clinical features of 85 fatal cases of COVID-19 from Wuhan: a retrospective observational study. Am J Respir Crit Care Med 201(11):1372–1379
Feldman C, Anderson R (2021) The role of co-infections and secondary infections in patients with COVID-19. Pneumonia 13(5)
Guo L, Wei D, Zhang X, Wu Y, Li Q, Zhou M et al (2020) Clinical features predicting mortality risk in patients with viral pneumonia: the MuLBSTA score. Front Microbiol 10:2752
Hanada S, Pirzadeh M, Carver KY, Deng JC (2018) Respiratory viral infection-induced microbiome alterations and secondary bacterial pneumonia. Front Immunol 16(9):2640
Heinrich A, Haarmann H, Zahradnik S, Frenzel K, Schreiber F, Klassert TE et al (2016) Moraxella catarrhalis decreases antiviral innate immune responses by down-regulation of TLR3viainhibition of p53 in human bronchial epithelial cells. FASEB J 30:2426–2434
Huang B, Cai Y, Li N, Li K, Wang Z, Li L et al (2021) Sex-based clinical and immunological differences in COVID-19. BMC Infect Dis 21:647
Ishizuka S, Yamaya M, Suzuki T, Takahashi H, Ida S, Sasaki T et al (2003) Effects of rhinovirus infection on the adherence of Streptococcus pneumoniae to cultured human airway epithelial cells. J Infect Dis 188:1928–1939
Jin JM, Bai P, He W, Wu F, Liu XF, Han DM et al (2020) Gender differences in patients with COVID-19: focus on severity and mortality. Front Public Health 8:152
Konala VM, Adapa S, Gayam V, Naramala S, Daggubati SR, Kammari CB et al (2020) Co-infection with infuenza A and COVID-19. Eur J Case Rep Intern Med 7(5):001656
MacIntyre CR, Chughtai AA, Barnes M, Ridda I, Seale H, Toms R, Heywood A (2018) The role of pneumonia and secondary bacterial infection in fatal and serious outcomes of pandemic influenza a(H1N1) pdm09. BMC Infect Dis 18(1):637
Man WH, de Steenhuijsen Piters WA, Bogaert D (2017) The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat Rev Microbiol 15:259–270
Manohar P, Loh B, Nachimuthu R, Hua X, Welburn SC, Leptihn S (2020) Secondary bacterial infections in patients with viral pneumonia. Front Med (lausanne) 5(7):420
Massey BW, Jayathilake K, Meltzer HY (2020) Respiratory microbial co-infection with SARS-CoV-2. Front Microbiol 11:2079
Mirzaei R, Goodarzi P, Asadi M, Soltani A, Aljanabi HAA, Jeda AS et al (2020) Bacterial co-infections with SARS-CoV-2. IUBMB Life 72(10):2097–2111
Morris DE, Cleary DW, Clarke SC (2017) Secondary bacterial infections associated with influenza pandemics. Front Microbiol 8:1041
Qin T, Geng T, Zhou H, Han Y, Ren H, Qiu Z et al (2020) Super-dominant pathobiontic bacteria in the nasopharyngeal microbiota as causative agents of secondary bacterial infection in influenza patients. Emerg Microbes Infect 9(1):605–615
Safaeyan F, Nahaei MR, Seifi SJ, Kafil HS, Sadeghi J (2015) Quantitative detection of Staphylococcus aureus, Streptococcus pneumoniae and Haemophilus influenzae in patients with new influenza A (H1N1)/2009 and influenza A/2010 virus infection. GMS Hyg Infect Control 10:06
Shaibu JO, Onwuamah CK, James AB, Okwuraiwe AP, Amoo OS, Salu OB et al (2021) Full length genomic sanger sequencing and phylogenetic analysis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in Nigeria. PLoS ONE 16(1):e0243271
Sharifipour E, Shams S, Esmkhani M, Khodadadi J, Fotouhi-Ardakani R, Koohpaei A et al (2020) Evaluation of bacterial co-infections of the respiratory tract in COVID-19 patients admitted to ICU. BMC Infect Dis 20:646
Shereen MA, Khan S, Kazmi A, Bashir N, Siddique R (2020) COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J Adv Res 24:91–98
Smith AM, McCullers JA (2014) Secondary bacterial infections in influenza virus infection pathogenesis. Curr Top Microbiol Immunol 385:327–356
Takahashi T, Ellingson MK, Wong P, Israelow B, Lucas C, Klein J et al (2020) Sex differences in immune responses that underlie COVID-19 disease outcomes. Nature 588:315–320
Tay MZ, Poh CM, Renia L, MacAry PA, Ng LFP (2020) The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol 20:363–374
Wang JH, Kwon HJ, Jang YJ (2009) Rhinovirus enhances various bacterial adhesions to nasal epithelial cells simultaneously. Laryngoscope 119:1406–1411
WHO Report (2022) https://covid19.who.int/table. Assessed on 15 Feb 2022
Xing Q, Li GJ, Xing YH, Chen T, Li WJ, Ni W et al (2020) Precautions are needed for COVID-19 patients with coinfection of common respiratory pathogens [Preprint] Lancet SSRN
Ysebaert C, Castado C, Mortier MC, Rioux S, Feron C, Di Paolo E et al (2021) UspA2 is a cross-protective Moraxella catarrhalis vaccine antigen. Vaccine 39(39):5641–5649
Acknowledgements
Not applicable.
Funding
This work was supported by the Nigerian Institute of Medical Research (NIMR) COVID-19 Research Support (Grant Number NMG-CIF-16-0034).
Author information
Authors and Affiliations
Contributions
OFD conceptualized the study. OFD, OSA, TIO, JCO, and RAA† performed DNA extraction and molecular analyses. MF, OO, and OO performed the molecular analyses. MF, EA, KAO, and JO performed statistical analyses. OAU, BGI, AAD, NO, and RAA reviewed the manuscript. OFD and OSA wrote the manuscript. RAA†† and BLS supervised the project, provided resources, and reviewed the draft. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was submitted to the Institutional Review Board (IRB) of the Nigerian Institute of Medical Research (NIMR), and ethical approval was obtained from the IRB in NIMR (IRB/20/032). Written consent was obtained from the patients prior to sample collection.
Consent for publication
Written informed consent for publication was obtained.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visithttp://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Davies-Bolorunduro, O.F., Fowora, M.A., Amoo, O.S. et al. Evaluation of respiratory tract bacterial co-infections in SARS-CoV-2 patients with mild or asymptomatic infection in Lagos, Nigeria. Bull Natl Res Cent 46, 115 (2022). https://doi.org/10.1186/s42269-022-00811-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s42269-022-00811-2