Abstract
Background
Antibiotic resistance has risen as a result of a variety of conditions, prompting researchers to look for new compounds that can combat multidrug-resistant organisms. Over the last two decades, chalcones have been proved to be attractive moieties in drug discovery. Various substituted acetophenones, propiophenones and 4-(Diphenylamino) benzaldehyde were combined, using the Aldol condensation reaction to obtain eight novel triphenylamine chalcones. The compound’s antimicrobial properties were investigated (in vitro). With the non-mutant X-ray Human cytochrome P450 21A2 Hydroxyprogesterone retrieved from Protein Data Bank (PDB: 5VBU), molecular docking experiments were also carried out to analyse the most favourable conformation and find the orientation that maximizes interaction and minimize energy.
Results
Eight novel triphenylamine chalcones were successfully synthesized and recrystallized using ethanol, the percentage yield of the compounds were between 30 and 92%. The activity against different pathogens revealed that, all synthesized compounds showed marked antimicrobial activity against the tested microorganisms. (E)-3-(4-(diphenylamino)phenyl)-1-(3′-nitrophenyl)prop-2-en-1-one (1b) showed the highest zone of inhibition against Aspergillus niger, measuring 30 mm. The minimum inhibitory concentration (MIC) results revealed that (E)-1-(4′-bromophenyl)-3-(4-(diphenylamino)phenyl)prop-2-en-1-one (1a), (E)-3-(4-(diphenylamino)phenyl)-1-(3′-nitrophenyl)prop-2-en-1-one (1b), (E)-1-(4′-chlorophenyl)-3-(4-diphenylamino)phenyl)prop-2-en-1-one (1c), (E)-3-(4-diphenylamino)phenyl)-1-(4′-fluorophenyl)prop-2-en-1-one (1d) and (E)-4-(3-(diphenylamino)phenyl)-1-(4-fluorophenyl)-2-methylbut-3-en-1-one (2d) had the lowest MIC and inhibit Aspergillus niger growth at 12.5 µg/ml. All the synthesized compounds showed minimum bactericidal concentration and minimum fungicidal concentration (MBC/MFC) effect against Escherichia coli, Pseudomonas aeruginosa, Bacillus subtilis, Candida albicans and Aspergillus niger at 50 µg/ml. The docking studies of the synthesized chalcones with the binding site of the Human cytochrome P450 21A2 Hydroxyprogesterone (PDB: 5VBU) reveal that the binding affinity of the synthesized chalcones was in the range of − 11.2 to − 9.4 kcal/mol and showed highest binding score compared to that of the standard drugs (Fluconazole and Ciproflaxacin), with docking scores of − 7.9 and − 7.3 kcal/mol, respectively.
Conclusions
The investigation reveals that compound 1b showed the highest ZOI of 30 mm, least MIC and MBC/MFC of 12.5 and 50 µg/ml against Aspergillus niger, respectively. Therefore, displayed better antifungal potential as compared to the rest of the compounds. The outcome of the docking analysis revealed that (E)-4-(3-(diphenylamino)phenyl)-1-(4′-hydroxyphenyl)-2-methylbut-3-en-1-one (2a) showed a better binding affinity of -11.2 kcal/mol, which is higher than the remaining compounds and the control drugs (fluconazole and ciproflaxacin).
Similar content being viewed by others
Background
The chemistry of chalcones has generated a bustle of research around the globe. The synthesis, chemical reactions, and biological applications of these compounds have captivated the attention of researchers (Singh et al. 2014; Kumara et al. 2017). Chalcones are characterized by a conjugated double bond on both benzene rings and a totally delocalized π-electron system. (Maidur and Patil 2018), which is dependent on the presence of other auxochromes. β-phenyl-α-benzoyl-ethylene is another name for chalcones (Ugwu et al. 2015). Chalcones are abundantly found in spices, tea, fruits, and vegetables as one of the principal classes of natural products (Kumar et al. 2021).
Claisen–Schmidt condensation of acetophenones with benzaldehydes is the most prevalent and efficient methods for producing chalcones. The yields are typically excellent, and reaction takes place at lower temperature. The duration for reaction varies from 3 to 20 h depending on the substitution pattern and the solvent/base system utilized, (Bukhari et al. 2012; Adnan et al. 2020).
Chalcones have been proved to be biologically active, some substituted derivatives, including heterocyclic analogues, have been found to exhibit strong biological characteristics that have been shown to inhibit microorganism development (Frazier 2020). Some chalcone derivatives have been proved to be poisonous to mammals (Gomes et al. 2009) and insects (Singh et al. 2019), as well as inhibiting enzymes (Agilandeshwari et al. 2016) and herbaceous plants (Mahapatra et al. 2019). Anti-inflammatory (Ugwu et al. 2015; Rashid et al. 2019), antifungal and antibacterial (Singh et al. 2019), antioxidant (Kumar et al. 2020; Singh et al. 2019; Vagish et al. 2021), anti-ulcer, antineoplastic, antispasmodic, antitumor (Ugwu et al. 2015), antimalarial (Reeta et al. 2019), antituberculosis (Matos et al. 2014), anti-helmintics (Matos et al. 2014) are just a few of the many biological activities linked to chalcones. Antimalarial action has been found for quinoline-based chalcones (Tomar et al. 2010; Karaman et al. 2010; Ugwu et al. 2015; Martelli et al. 2019).
Due to the continued resistance and resurgence of pathogens against available antibiotics, the constant search for potentially novel drug candidates has emerged. Simultaneously, researchers developed the concepts of protein purification and crystallography, allowing researchers to learn more about the protein and ligand interactions. Today, computational methodologies are pervading many facets of drug development (Walters et al. 1998; Bajorath, 2002; Jorgensen, 2004). Molecular docking investigations have become important tools in the search for new drugs candidates (Langer and Hoffmann, 2001). This technique represents atomic level interface with target protein, allowing us to define small molecule behaviour in target protein binding sites (McConkey et al. 2002). The process also determines the ligand structure, location, and orientation within the binding site, as well as the binding affinity (Kitchen et al. 2004; Prabhudeva et al. 2019).
Human cytochrome P450 21A2 Hydroxyprogesterone (PDB: 5VBU) is the major steroid 21-hydroxylase, converting progesterone to 11-deoxycorticosterone and 17α-hydroxyprogesterone to 11-deoxycortisol. Deficiency of this enzyme is involved in approximately 95% cases of human congenital adrenal hyperplasia, a disorder of adrenal steroidogenesis, Pradeep et al. (2015). To our knowledge, there is no report on synthesis, antibiotic evaluation and molecular docking studies of triphenylamine chalcones. Hence, present research intended to synthesize, screen antimicrobial potential and study the molecular docking of novel triphenylamine chalcones.
Methods
All reagents and solvents used in this research were obtained from Sigma-Aldrich (Germany). These were used as obtained without further purification. These include 4-Chloroacetophenone,
4-(Diphenylamino)benzaldehyde, 4-Bromoacetophenone, 3-Nitroacetophenone, 4-Flouroacetophenone, 4-Hydroxypropiophenone, 2-Bromopropiophenone, 4-Methoxypropiophenone, 4-Flouropropiophenone, 10% NaOH and Ethanol.
Synthesis of the novel triphenylamine chalcones analogues (1a–d, 2a–d)
To 25 ml ethanol taken in 100 ml round bottomed flask equipped with a magnetic stirrer, were transferred equimolar quantities of 4-(diphenylamino)benzaldehyde (0.4 g, 1.5 mmol) and substituted acetophenones/propiophenone (0.4 g, 2 mmol) and stirred for 30 min. A solution of sodium hydroxide (10 mL, 10%) was added dropwise while stirring. The temperature for the reaction was maintained between 20 and 24 ℃ using cold water bath for 4–5 h. The progress and completion of the reaction were monitored using thin-layer chromatographic technique (TLC). On completion, the reaction mixture was placed in a refrigerator for 10 h. Formation of precipitate was observed, which was filtered, washed severally with water (100 ml), dried in air and purify by recrystallization from ethanol (30 ml) to obtained a triphenylamine chalcone, (1a–d, 2a–d). Hongtian et al. (2019). Table 1 shows the structures of the synthesized triphenylamine chalcones and their percentage yields (Schemes 1, 2).
The synthesis of the target novel triphenylamine chalcones was achieved via the conventional Claisen–Schmidt condensation reaction, where different equimolar substituted acetophenones/propiophenones and 4-(Diphenylamino) benzaldehyde in the presence of NaOH (10%) were stirred. This reaction was originated by the abstraction of proton from the alpha-carbon of the acetophenones/propiophenones to generate the resonance stabilized enolate ion by the base. The second stage was the nucleophilic attack on the electron deficient carbonyl carbon of 4-(Diphenylamino) benzaldehyde, which results in the formation of a new C–C bond. This connects the alpha-carbon of the acetophenones/propiophenones to the aldehydic or carbonyl carbon of 4-(Diphenylamino) benzaldehyde to which form an intermediate. The reaction was completed by protonation and deprotonation by the hydroxyl ion of the base to form the targeted α,β-unsaturated triphenylamine chalcone.
Antimicrobial evaluation
The synthesized triphenylamine chalcones were screened for antimicrobial properties, against some selected clinical isolates, which include Pseudomonas aeruginosa, Staphylococus aureus, Bacillus substilis, Candida albicans, Salmonella typhi, Aspergillus Niger, Escherichia coli and Methicilin Resistant Staphylococus aureus (MRSA).
Zone of inhibition (ZOI)
The (ZOI) of the synthesized triphenylamine chalcones was carried out according to the procedure as described by Karou et al. (2006).
Minimum inhibitory concentration (MIC)
The (MIC) of the synthesized triphenylamine chalcones was determined by using the broth dilution method as reported by Bruton et al. (2007).
Minimum bactericidal/fungicidal concentration (MBC/MFC)
The MBC/MFC of the synthesized triphenylamine chalcones was determined according to the procedure, as described by CLSI, 2015 (CLSI = Clinical and Laboratory Standard Institute).
Molecular docking studies
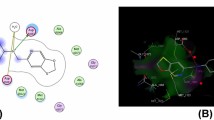
Molecular Docking Analysis for eight (8) ligands (novel triphenylamine chalcones analogues) and controls (Fluconazole and Ciproflaxacin) was carried out to examine the most favourable interaction and identify the orientation which maximizes interactions and minimizes energy with the target receptor (PDB: 5VBU) (www.rcsb.org) (Fig. 1).
Preparation of the receptor
The three-dimensional structure of target receptor (Fig. 1) retrieved from the PDB was prepared by removing the water molecules and other heteroatoms, before minimization for the docking study. Discovery Studio Visualizer software v.21.1.0.20298 was used for the preparation. The treated target receptor (Fig. 2) was saved in PDB file format and transferred to Pyrx software for docking.
Preparation of ligand
All the eight (8) ligands (triphenylamine chalcones analogues) and controls (Fluconazole and Ciproflaxacin) studied were designed and synthesized as mentioned earlier. ChemDraw ultra 8.0 software was used to generate the two-dimensional structures of the synthesized chalcones (Li et al. 2004). Spartan software (Spartan’20 v.1.1 /2020) was used to convert the 2D structures to 3D. Geometrical optimization using the AM1 semi-empirical method was performed on all the compounds using the Spartan software and saved as pdb files. Polar hydrogens were added before computing Gasteiger charges, using the BIOVIA DiscoveryStudio2021.
Molecular docking
PyRx virtual screening software was used to study the ligand-receptor interactions between the target receptor (PDB: 5VBU) and the eight (8) synthesized ligands (novel triphenylamine chalcones analogues) and controls (Fluconazole and Ciproflaxacin), while Discovery studio visualizer software was employed in visualizing and analysing the docking results.
Results
Results of antimicrobial studies
Results of molecular studies
See Figs. 3, 4, 5, 6, 7 and 8, Tables 5 and 6.
Discussion
Antimicrobial studies
All the synthesized compounds were shown to possess remarkable activities against the tested microbes, by showing a significant zone of inhibitions relative to that of the standard drugs used as shown in Table 2. Compound 1b showed prominent zone of inhibition of 30 mm against Aspergillus niger, while compounds 1a, 2d, 1c, 2b and 2c showed zone of inhibitions of 25 mm, 23 mm, 22 mm and 21 mm also, against Aspergillus niger. Compounds 1c, 1d, 2a, 2b and 2c each showed zone of inhibition of 20 mm against Bacillus subtilis, Pseudomonas aeruginosa, Candida albicans and Aspergillus niger. While, compounds 1a, 1b and 2a showed the lowest zone of inhibition of 13 mm each against Candida albicans, Staphylococus aureus and Escherichia coli. Thus, compounds 1a, 1b, 1c and 1d showed zone of inhibitions (25 mm, 30 mm and 23 mm) greater than the standard drugs (Fluconozole and ciprofloxacin) against Aspergillus nigger Singh et al. (2019).
Table 3 presents the results of the MIC and showed that compounds 1a, 1b, 1c, 1d and 2d possess the least MIC and inhibit the growth of Aspergillus niger at 12.5 µg/ml. While compounds 1b, 1c, 1d, 2a, 2b, 2c and 2d inhibit the growth of Candida albicans, Bacillus subtilis, MRSA, Escherichia coli, Pseudomonas aeruginosa and Aspergillus niger at 25 µg/ml. At 50 µg/ml, the growth of Pseudomonas aeruginosa, Bacillus subtilis, Candida albicans, MRSA, Salmonella typhi, Staphylococus aureus and Escherichia coli were inhibited by compounds 1a, 1b, 1c, 1d, 2a, 2b, 2c and 2d. Compounds 1a, 1b, 1c, 1d, 2a, 2b, 2c and 2d inhibit the growth of Escherichia coli, Pseudomonas aeruginosa, Salmonella typhi, Staphylococus aureus, Bacillus subtilis and MRSA at 100 µg/ml.
The results of MBC/MFC showed that compounds 1a, 1b, 1c, 1d, 2a, 2b, 2c and 2d completely killed Escherichia coli, Pseudomonas aeruginosa, Bacillus subtilis, Candida albicans and Aspergillus niger at 50 µg/ml, while a concentration of 100 µg/ml was required by compounds 1a, 1b, 1c, 1d, 2a, 2b, 2c and 2d to completely kill Escherichia coli, Pseudomonas aeruginosa, Salmonella typhi, MRSA, Staphylococus aureus, Bacillus subtilis and Candida albicans as shown in Table 4.

Molecular docking studies
Each ligand/chalcones was successively docked to the binding site of the receptor (PDB: 5VBU), in order understand the mode of interaction of the triphenylamine chalcones with the target receptor. The analysis of the docking investigations provided us with an insight into the interactional relation of the novel triphenylamine chalcone analogues and human cytochrome P450 21A2 Hydroxyprogestrerone complex.
Table 5 shows the results of the binding affinity of the synthesized triphenylamine chalcones/ligands, which ranges between − 11.2 and − 9.4 kcal/mol. Compound 2a showed the highest docking score of − 11.2 kcal/mol, followed by compounds/ligands 1b and 2b which showed the binding energy of − 10.7 kcal/mol. While compounds 1c, 2c, 2d and 1a showed the docking scores of − 10.4, − 10.3, − 10.3 and − 10.1 respectively. Compound/ligand 1d showed the least binding score of − 9.4. The eight (8) ligands (novel triphenylamine chalcones analogues) showed highest binding score compared to that of the standard ligand/compounds (fluconazole and ciproflaxacin), which showed the docking scores of − 7.9 and − 7.3 kcal/mol, respectively.
Ligand/triphenylamine chalcone (2a) showed the highest binding affinity (− 11.2 kcal/mol) compared to other compounds. The interaction of the ligand/ triphenylamine chalcone (2a) with the target receptor (PDB: 5BVU) is shown in Figs. 3, 4 and 5. The interaction was observed with one hydrogen, one electrostatic attraction and six hydrophobic interactions (2.5449, 4.0569, 3.73562, 4.34337. 5.2863, 5.48907, 5.44454 and 5.35682 Å) with (ARG427, LYS121, VAL470, TRP202, VAL360, LEU364, LEU108, LEU430) as shown in Figs. 6 and 7. The two-dimensional structural form of the ligand-receptor complex is shown in Fig. 8. One hydrogen bond interaction with ARG427 was formed, due to the presence of OH group in the triphenylamine chalcone/chalcone (2a). The electrostatic interaction of the ligand/triphenylamine chalcone was detected with LYS121. Also, the hydrophobic interactions were noticed with VAL470, TRP202, VAL360, LEU364, LEU108 and LEU430 of the target receptor. The areas which represent the both hydrogen bond and hydrophobic interactions between the target receptor and the ligand/triphenylamine chalcone are shown in Figs. 6 and 7. Therefore, ligand/triphenylamine chalcone (2a) fits perfectly into the binding site of the receptor (PDB: 5VBU). All type of interactions, amino acids and bond length are shown in Table 6.
Correlation of molecular docking studies and antimicrobial studies
The docking studies of the synthesized chalcones with the binding site of the Human cytochrome P450 21A2 Hydroxyprogesterone (PDB: 5VBU) reveal that the binding affinity of the synthesized chalcones was in the range of − 11.2 to − 9.4 kcal/mol and showed highest binding score compared to that of the standard drugs (fluconazole and ciproflaxacin), with docking scores of − 7.9 and − 7.3 kcal/mol, respectively. Compound 2a displayed best docking score of -11.2 kcal/mol. The binding affinity, hydrogen bond and hydrophobic interactions of the synthesized chalcones and the standard controls (fluconazole and ciproflaxacin) are summarized in Tables 5 and 6. Compared to fluconazole, compound (1b) showed similar residual interactions (hydrophobic) profiles with amino acid residues TRP 202, LEU 110 and VAL 470, (1a) TRP 202 and LEU 110, (2a) VAL 470 and (1c) TRP 202. Similarly, compounds 1b, 1d and 2d showed Hydrophobic interactions LEU 40, as shown by the standard drug ciproflaxacin. Therefore, the synthesized compounds showed similarities in the residual amino acid interactions with clinical drugs against the Human cytochrome P450 21A2 Hydroxyprogesterone enzyme. The docking investigations are in agreement with the in vitro antimicrobial assay results. Hence, compound 1b indicated higher antifungal potential by showing similarities in residual interactions with amino acids residue as shown by standard antifungal drug (fluconozole), while the antimicrobial investigation reveals that compound 1b showed the highest ZOI of 30 mm, least MIC and MBC/MFC of 12.5 and 50 µg/ml against Aspergillus niger, respectively.
Conclusions
Synthesis of eight novel triphenylamine chalcones was successively carried out via the conventional Claisen–Schmidt condensation reactions, and the compounds were successfully characterized using FT-IR and NMR spectroscopic analyses. The results of the antimicrobial screening of the synthesized chalcones as indicated by zone of inhibition (ZOI) showed that all the synthesized compounds possess remarkable activities against the tested microbes, by showing a significant zone of inhibitions relative to that of the standard drugs used. Compound 1b showed the highest ZOI of 30 mm, least MIC and MBC/MFC of 12.5 and 50 µg/ml against Aspergillus niger. The outcome of the docking studies revealed that compound 2a showed marked docking score with binding affinity of − 11.2 kcal/mol, which is higher relative to other compounds and the standard controls (fluconazole and ciproflaxacin). Therefore, compounds 1b and 2a which showed better antifungal and highest binding affinity could be potential candidates in drug design.
Availability of data and materials
Not applicable.
Abbreviations
- ZOI:
-
Zone of inhibition
- MIC:
-
Minimum inhibitory concentration
- MRSA:
-
Methicilin resistant Staphylococus aureus
- MBC:
-
Minimum bactericidal concentration
- MFC:
-
Minimum fungicidal concentration
- mm:
-
Millimetre
- µg/ml:
-
Microgram per mil
- PDB:
-
Protein data bank
- kcal:
-
Kilocalorie
- mol:
-
Mole
- β:
-
Beta
- α:
-
Alpha
- mmol:
-
Millimole
- g:
-
Gram
- TLC:
-
Thin layer chromatography
- mL:
-
Milli litre
- DMSO:
-
Dimethyl sulfoxide
- ℃:
-
Degree Celsius
- MHB:
-
Mueller hinton broth
- CFU:
-
Number of colonies times dilution factor
- 2D:
-
Two dimensional
- 3D:
-
Three dimensional
- rmsd:
-
Root mean square deviation
- TRP:
-
Tryptophan
- VAL:
-
Valine
- CYS:
-
Cysteine
- ARG:
-
Argenine
- LEU:
-
Leucine
- SER:
-
Serine
- ILE:
-
Isoleucine
- PRO:
-
Proline
References
Adnan D, Singh B, Mehta SK, Kumar V, Kataria R (2020) Simple and solvent free practical procedure for chalcones: an expeditious, mild and greener approach. Curr Res Green Sustain Chem 3:100041. https://doi.org/10.1016/j.crgsc.2020.100041
Agilandeshwari R, Meenatchi V, Meenakshisundaram SP (2016) Synthesis, growth, structure and characterisation of chalcone crystal: a novel organic NLO material. J Mol Struct 1118:356–366
Bajorath J (2002) Integration of virtual and high-throughput screening. Nat Rev Drug Discov 1(11):882–894
Bruton LL, Lazo JS, Parker KL (2007) The Pharmaceutical basis of therapeutics, 11th edn. Mc Graw-Hill Medical Publishing Division, New York
Bukhari SN, Jasamai M, Jantan I (2012) Synthesis and biological evaluation of chalcone derivatives. Mini Rev Med Chem 1(2):1394–1403
Frazier Z (2020) Screening a library of chalcone derivatives for antibacterial properties via kirby bauer disk diffusion. Undergraduate Theses, p 44
Gomes A, Neuwirth O, Freitas M, Couto D, Ribeiro D, Figueiredo AGPR, Silva AMS, Seixas RSGR, Pinto DCGA, Tomé AC (2009) Synthesis and antioxidant properties of new chromone derivatives. Bioorg Med Chem 17:7218–7226
Hongtian Z, Lei T, Chenghong Z, Baochu W, Pingrong Y, Dian H, Lifang Z, Yang Z (2019) Synthesis of chalcone derivatives: inducing apoptosis of HepG2 cells via regulating reactive oxygen species and mitochondrial pathway. Front Pharmacol. https://doi.org/10.3389/fphar.2019.01341
Jorgensen WL (2004) The many roles of computation in drug discovery. Science 303(5665):1813–1818
Karaman H, Gezegen MB, Gürdere A, Ceylan M (2010) Screening of biological activities of a series of chalcone derivatives against human pathogenic microorganisms. Chem Biodivers 7:400–408
Karou D, Savadogo A, Canini A, Yameogo S, Montesano C, Simpore J, Colizzi V, Traore AS (2006) Antibacterial activity of alkaloids from Sida acuta. Afr J Biotechnol 5(2):195–200
Kitchen DB, Decornez H, Furr JR, Bajorath J (2004) Docking and scoring in virtual screening for drug discovery: methods and applications. Nat Rev Drug Discov 3(11):935–949
Kumar AD, Vagish C, Renuka N, Lokeshwari DM, Kumar KA (2020) Green synthesis of novel pyrazoline carbothioamides: a potent antimicrobial and antioxidant agents. Chem Data Collect 28:100445. https://doi.org/10.1016/j.cdc.2020.100445
Kumar A, Rout L, Achary LSK, Mohanty SK, Nayak PS, Barik B, Dash P (2021) Solvent free synthesis of chalcones over graphene oxide-supported MnO2 catalysts synthesized via combustion route. Mater Chem Phys 259:124019. https://doi.org/10.1016/j.matchemphys.2020.124019
Kumara K, Naveen S, Dileep Kumar A, Ajay Kumar K, Lokanath NK, Warad I (2017) (E)-1-(1,3-Benzodioxol-5-yl)-3-[4-(dimethylamino)-phenyl]prop-2-en-1-one. IUCrData 2:162029. https://doi.org/10.1107/S2414314616020290
Langer T, Hoffmann RD (2001) Virtual screening: an effective tool for lead structure discovery? Curr Pharm Des 7(7):509–527
Li X, Li Y, Cheng T, Liu Z, Wang R (2010) Evaluation of the performance of four molecular docking programs on a diverse set of protein-ligand complexes. J Comput Chem 31(11):2109–2125
Mahapatra DK, Bharti SK, Asati V, Singh SK (2019) Perspectives of medicinally privileged chalcone based metal coordination compounds for biomedical applications. Eur J Med Chem 174:142–158
Maidur SR, Patil PS (2018) (2018): “Z-scan studies of third-order nonlinear optical and optical limiting properties of chalcones doped Poly(methyl methacrylate) thin films for visible laser protection. Opt Mater 84:28–37
Martelli LSR, Viera LCC, Paixao MW, Zukerman-Schpector J, de Souza JO, Aguiar ACC, Oliva G, Guido RVC, Correa G (2019) Organocatalytic asymmetric vinylogous 1,4-addition of α, α-Dicyanoolefins to chalcones under a bio-based reaction media: discovery of new Michael adducts with antiplasmodial activity. Tetrahedron 75:3530–3542. https://doi.org/10.1016/j.tet.2019.05.022
Matos MJ, Vasquez-Rodriguez S, Uriarte E, Santana L (2014) Potential pharmacological uses of chalcones: a patent review (from June 2011–2014). Expert Opin Ther Patents 25:3–16
McConkey BJ, Sobolev V, Edelman M (2002) The performance of current methods in ligand-protein docking. Curr Sci 83:845–855
Prabhudeva MG, Vivek HK, Ajay Kumar K (2019) Synthesis of novel pyrazole carboxamides using reusable catalyst as antimicrobial agents and molecular docking studies. Chem Data Collect 20:100193. https://doi.org/10.1016/j.cdc.2019.100193
Pradeep SP, Chunxne W, Michae RW, Peter FG, Martin E (2015) Human cytochrome P4450 21A2, the major steroid 21-hydroxylase. J Biol Chem 290(21):13128–131143
Rashid H, Xu Y, Ahmad N, Muhammad Y, Wang L (2019) Promising anti-inflammatory effects of chalcones via inhibition of cyclooxygenase, prostaglandin E2, inducible NO synthase and nuclear factor κb activities. Bioorg Chem 87:335–365. https://doi.org/10.1016/j.bioorg.2019.03.033
Rajendran V, Rangarajan TM, Singh RP, Singh M (2019) Synthesis of novel chalcones through palladium-catalyzed C-O cross-coupling reaction of bromo-chalcones with ethyl acetohydroxamate and their antiplasmodial evaluation against Plasmodium falcipuram in vitro. Bioorgan Chem 86:631–640. https://doi.org/10.1016/j.bioorg.2019.02.016
Singh P, Anand A, Kumar V (2014) Recent developments in biological activities of chalcones: a mini review. Eur J Med Chem 85:758–777
Singh G, Arora A, Kalra P, Maurya IK, Ruizc CE, Estebanc MA, Sinha S, Goyal K, Sehgal R (2019) A strategic approach to the synthesis of ferrocene appended chalcone linked triazole allied organosilatranes: antibacterial, antifungal, antiparasitic and antioxidant studies. Bioorg Med Chem 27:188–195
Tomar V, Bhattacharjee G, Kamaluddin K, Rajakumar S, Srivastava K, Puri SK (2010) Synthesis of new chalcone derivatives containing acridinyl moiety with potential antimalarial activity. Eur J Med Chem 45:745–751
Ugwu DI, Ezema BE, Okoro UC, Eze FU, Ekoh OC, Egbujor MC, Ugwuja DI (2015) Synthesis and pharmacological applications of chalcones: a review. Int J Chem Sci 13:459–500
Vagish CB, Kumar AD, Kumara K, Vivek HK, Renuka N, Lokanath NK, Kumar KA (2021) Environmentally benign synthesis of substituted pyrazoles as potent antioxidant agents, characterization and docking studies. J Iran Chem Soc 18:479–493. https://doi.org/10.1007/s13738-020-02042-6
Walters WP, Stahl MT, Murcko MA (1998) Virtual screening - an overview. Drug Discov Today 3:160–178
Acknowledgements
The authors would like to express their gratitude to the staff and laboratory technologists Department of Chemistry, Ahmadu Bello University Zaria, Kaduna State. Nigeria, for giving the resources that allowed this study to be completed.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
AT carried out the synthesis, the antimicrobial studies and the molecular docking studies and produces the manuscript. RG guides the synthesis and review the manuscript. ORAI guides the synthesis of the novel chalcones. JDH guides the synthesis, antimicrobial studies and the review of the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tukur, A., Habila, J.D., Ayo, R.GO. et al. Design, synthesis, docking studies and antibiotic evaluation (in vitro) of some novel (E)-4-(3-(diphenylamino)phenyl)-1-(4-methoxyphenyl)-2-methylbut-3-en-1-one and their analogues. Bull Natl Res Cent 46, 60 (2022). https://doi.org/10.1186/s42269-022-00745-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s42269-022-00745-9