Abstract
Background
Data on the prevalence, genotypes and antibiotic resistance patterns of colonizing and infection-associated Staphylococcus aureus (S. aureus) strains both in humans and animals in Tanzania are scarce. Given the wide range of infections caused by S. aureus and the rise of methicillin-resistant S. aureus (MRSA) globally, this review aims at collecting published data on S. aureus bacterium to improve our understanding of its epidemiology in Tanzania.
Main body
We carried out a systematic review of scientific studies reporting on prevalence, antibiotic resistance and genotyping data pertaining to S. aureus in human and animal infection and colonization. The literature extracted from electronic databases such as PubMed and Google Scholar was screened for eligibility and relevant articles were included. The review is limited to manuscripts published in English language between the years 2010 and 2020. A total of 45 studies conducted in 7 of the 9 administrative zones in Tanzania were reviewed to gather data on S. aureus prevalence in humans and animals. Prevalence in humans ranged from 1 to 60%. Antibiotic resistance patterns of S. aureus isolated from colonized humans showed high resistance rates against co-trimoxazole (46%) and erythromycin (41%) as compared to reports from studies conducted outside Africa. The review suggests an increased MRSA prevalence of up to 26% as compared to 6–16% reported in previous years. Genotypic data reviewed suggested that MRSA predominantly belonged to ST88. The prevalence of S. aureus in animal studies ranged from 33 to 49%, with 4 to 35% of MRSA isolates. Most studies reported low antibiotic resistance levels, with the exception of penicillin (85%) and ampicillin (73%).
Conclusion
The prevalence of S. aureus and MRSA in Tanzania is rising, although clear variations between different geographic areas could be observed. Non-susceptibility to commonly prescribed antibiotics in community-associated S. aureus is of concern. Research strategies to ameliorate our knowledge on S. aureus epidemiology should employ regular antibiotic resistance surveillance, antimicrobial stewardship as well as genotypic characterization.
Similar content being viewed by others
Background
Antimicrobial resistance (AMR) is a global concern estimated to account for approximately 700,000 deaths each year (O’Neill 2016). If no appropriate measures are taken to slowdown the progression of this epidemic it is estimated that by 2050 it will cost the world around 10 million lives per annum (O’Neill 2016). The lack of development of new antimicrobial agents in the pipeline further emphasizes the necessity to reduce dependency on antibiotics by implementing infection control strategies (O’Neill 2016; WHO 2014).
According to the 2014 World Health Organization (WHO) report, most regions stated over 50% bacterial resistance against third-generation antibiotics, particularly methicillin-resistant S. aureus (MRSA) clones in hospitals (WHO 2014). Furthermore in 2017, the WHO listed antibiotic-resistant bacteria including Staphylococcus aureus (S. aureus) as a priority bacterium to guide research, discovery and development of new antibiotics (WHO 2017). The report further recommended special attention to be directed towards the identified resistant bacteria due to their ability to rapidly develop resistance against multiple antibiotic classes hence limiting therapeutic options (WHO 2017).
Staphylococcus aureus is an old bacterium discovered in the eighteenth century, antibiotic resistance in the bacterium against penicillin was described in the 1950’s, the penicillin-resistant staphylococci inactivates penicillin function by an enzyme called penicillinase or beta-lactamase which degrades the β-lactam ring in penicillin, thus altering the shape of penicillin and preventing its binding to the penicillin binding proteins (Haddadin et al. 2002). Following penicillin failure in treating S. aureus infections, methicillin was introduced. Methicillin was particularly effective upon its introduction into clinical therapy due to its ability to resist the action of β-lactamase. However, methicillin resistance in S. aureus was also reported only a year after its introduction. Strains of S. aureus resistant to methicillin or oxacillin or other β-lactam compounds are still termed MRSA, only to honour the historic role of methicillin that used to effectively treat staphylococcal infections, and it is to methicillin that the resistance was first described (Jevons 1961). Several terms are used to describe different MRSA strains associated with outbreaks in different settings based on strains’ genomic background and level of virulence (Zetola et al. 2005). MRSA strains associated with hospital-acquired infections are commonly abbreviated to HA-MRSA, while strains causing community-associated infection are abbreviated to CA-MRSA and those associated with livestock are abbreviated as LA-MRSA.
Staphylococcus aureus is both a commensal and potentially harmful pathogen in humans and animals. The bacterium can give rise to a variety of infections ranging from mild skin and soft tissue infections to more serious and complex diseases such as pneumonia, septicaemia, infective endocarditis and other deep-seated infections (e.g. osteomyelitis) in humans as well as mastitis and necrotic infections in a variety of animal hosts. Staphylococcus aureus can also colonize the skin and approximately 30% of the human population is found to be transiently colonized if nasal swabs are examined microbiologically.
High levels of MRSA have been reported across Africa, ranging between 43 and 72%, as reported in Cameroon, South Africa and Ethiopia (Founou et al. 2017). The vast majority of clinical S. aureus/MRSA data in Africa is associated with hospital acquired infection (HAI) affecting individuals with healthcare-related risk such as hospitalization, surgery and underlying chronic diseases (David 2010). Information on community-acquired S. aureus/MRSA infections (CAI) causing diseases in people with no healthcare-associated risk is also available on the continent even though to a lesser extent. Moreover, very limited data on livestock acquired S. aureus/MRSA infections (LAI) whereby S. aureus clones of animal origins colonize or cause infections in humans have been published in Africa (Founou et al. 2017). Further information on the antibiotic-resistant clones occurring in both human and animal are very limited, hindering comprehensive understanding of the epidemiology of the bacterium (Lozano et al. 2016).
Tanzania, a developing country located along the East African coast, housing a population of about 57 million people, of which 36% is engaged in livestock keeping as the major source of livelihood. The potential for human–animal contact is very high, especially in tribes that cohabit with their animals such as Maasai people. The aim of this review is to summarize literature reporting on the prevalence, antibiotic resistance of S. aureus in Tanzania. Furthermore, reported genotypic characterization of S. aureus will be reviewed to provide a more nuanced profile of S. aureus genetic diversity in colonization and infections both in animal and human hosts as well as their possible relationship.
Main text
Methods
Eligibility criteria
A systematic review of Tanzanian scientific studies reporting on prevalence, antimicrobial resistance and genotypic characterization of S. aureus from human and animal sources published between 2010 and 2020 was performed. Information sought for in the publications included commensal and clinical S. aureus recovered from different infections including invasive and non-invasive. Animal studies reporting on S. aureus recovered from animals and their products were also considered. All studies that recovered less than five S. aureus isolates as well as publications that were written in a language other than English were excluded from this review.
Information sources and search strategies
PubMed database was researched in November 2018 and February 2021 by a librarian using Boolean operators “AND” and “OR” to identify studies fitting our inclusion criteria. The following search terms were used “Staphylococcus aureus OR S. aureus AND antimicrobial susceptibility OR antibiotic resistance AND prevalence AND Tanzania.” Additionally search words such as “Molecular typing AND S. aureus AND antibiotic resistance AND Tanzania” and “Staphylococcus aureus OR S. aureus AND antimicrobial susceptibility OR antibiotic resistance AND prevalence AND Tanzania” were used in Google scholar to identify eligible articles. The bibliographies of all eligible documents were hand-searched for additional publications eligible for review.
Data extraction and appraisal process
The Joana Briggs Institute checklist was applied to appraise and review the quality of each study accessed by two independent reviewers. A data extraction form was designed to capture required information such as author, year of publication, study period, methodology, MRSA identification strategies, S. aureus and MRSA prevalence, antibiotic resistance patterns as well as genotypic information. Two reviewers were involved in the process whereby the first reviewer (TM) extracted the information and the second reviewer (TK) double checked the information to eliminate possible bias. All disagreement raised during the critical appraisal process were resolved through reviewers discussions. All extracted literature was analysed using reference manager ENDNOTEX7. This study followed the standardized scientific writing format of the Preferred Reporting Items for systematic reviews and meta-analyses (PRISMA) guidelines. The study has not taken on any meta-analysis due to the heterogeneity of the studies under review; nevertheless, mean resistance rates and prevalence were calculated to help present the results better.
Scope of the study
This review is limited to prevalence, antibiotic resistance patterns and genetic typing information [i.e. specified resistance and virulence genes, Staphylococcus protein A (spa) typing and multi-locus sequence typing] of S. aureus retrieved from human and animal hosts.
Results
PubMed search resulted in 18 eligible articles followed by an additional 13 articles included from the Google scholar search. Rigorous reference list review supplemented another 14 eligible publications making a total of 45 (Fig. 1).
Data concerning S. aureus prevalence, antimicrobial resistance and genotyping were extracted from studies performed in 7 zones of the 9 administrative zones in Tanzania. Table 1 describes the distribution of the published articles review in their consecutive zones. The majority of the publications reviewed were from regions in the eastern and lake zones (i.e. 40% and 33%, respectively), showing lack of S. aureus researched data in the other parts of the country.
The majority of the publications were reported in the Eastern and Lake Zones. S. aureus data of animal origin have been poorly represented throughout the country.
Prevalence, antibiotic resistance and genotyping of S. aureus isolated from humans
A total of 45 studies reported on prevalence, antimicrobial resistance and genetic characteristics of S. aureus in humans published between 2010 and 2020 in Tanzania as indicated in Table 1.
Prevalence of S. aureus in humans
Staphylococcus aureus prevalence was reported in 39 human-related studies, reporting a prevalence ranging from 6 to 69%. Higher infection rates were typically observed in SSTI’s, nevertheless the bacterium was also implicated in other infections. Furthermore, it was notably observed that S. aureus colonization ranged around 10–60% in the three different zones that reported on colonization. The summary of S. aureus prevalence in colonization and different infections in human, as well as the geographic distributions reported in Tanzania between 2010 and 2020, is summarized in Table 2.
Antibiotic resistance in human S. aureus isolates
The primary method for establishing S. aureus antibiogram in the reviewed work was done phenotypically using the Kirby-Bauer disc diffusion test along with Clinical Laboratory Standard Institute (CLSI) guidelines. Kirby-Bauer is an antimicrobial susceptibility test based on the size of inhibition zones of microbial growth in a lawn culture around discs impregnated with the antimicrobial drug (Hudzicki 2009).
Twenty-four studies contributed information on antibiotic susceptibility of S. aureus bacteria. MRSA in most cases was identified by resistance against cefoxitin; however, one study used methicillin as their identification disc. As summarized in Table 3, extremely high resistance rates against β-lactams, i.e. penicillin (87–99%) and ampicillin (67–92%), were observed.
Apart from high resistance to the reported β-lactams, trimethoprim/sulphamethoxazole (co-trimoxazole) and erythromycin showed average resistance of 54% and 47%, respectively. Notably individual analysis of antibiotics susceptibility in isolates collected from colonization showed lower rates of resistance compared to S. aureus isolated from clinical infections, as summarized in Table 3.
Resistance rates of 50% (co-trimoxazole), 45% (clindamycin), 37% (erythromycin) and 32% (gentamicin) were observed in SSTIs. Unlike in commensal and SSTI-related isolates, blood born S. aureus showed higher resistance variation against most commonly used antibiotics (i.e. erythromycin, gentamicin, co-trimoxazole and clindamycin) ranging from 42 to 66%.
MRSA detection in all reported reviewed studies had an average prevalence of 21%, nevertheless 7 studies reported on specific resistance patterns of MRSA isolates sighting higher rates against other antibiotics (clindamycin, erythromycin and co-trimoxazole) ranging from 50 to 100% (Joachim et al. 2018, 2017; Geofrey et al. 2015; Moyo et al. 2014; Moremi et al. 2014) apart from the expected β-lactams antibiotic resistance.
Even though most articles reported 100% susceptibility to vancomycin, Geofrey et al. 2015; Kayange et al. 2010; Seni et al. 2019a observed vancomycin resistance of above 10% in their respective studies (not demonstrated in Table 3).
Genotypic characterization of human S. aureus isolates
Five human-related studies were reviewed (summarized in Table 4). MSSA genotypic characterization by Staphylococcal protein A (spa) typing in one of the studies revealed 13 different spa types including one new spa type t10779. The most common spa types were t714 and t148 (associated with ST72) followed by t084 (ST15 and ST18) and t223 (ST 22). Furthermore, spa types t314 (ST121), t084 (ST15 and ST18) and t223 were reported to be shared by human and animal isolates in this study (Katakweba et al. 2016). In a more recent study whereby patients were swabbed for S. aureus carriage on admission, after their hospital stay as well as wound swabs for those who had SSTIs, the study further swabbed HCW attending the patients in question. Taking these groups into consideration 60 S. aureus were characterized by MLST, as in the other studies ST distribution was diverse. Eight STs were detected in the 17 isolates from admission of which ST8 (4/17) and ST5 (4/17) were dominant. Nine STs were detected among 13 acquired isolates typed of which ST152 (3/13) and ST5 (2/13) were predominant. Moreover in the 12 S. aureus SSTI isolates, eight STs were detected predominated by ST152 (3). Subsequently in a study which used the whole genome sequencing and multi-locus sequence typing (MLST) in their analysis reported 13 different sequence types predominated by ST8 (23%) followed by ST1 (13.3%) and ST152 (10%) (Kumburu et al. 2017). Unfortunately, the limited number of publications on genotypic MSSA data could not reveal dominance in any particular identified strain.
MRSA characterization was reported in three of the five publications (Moremi et al. 2019, 2012; Kumburu et al. 2017; Moremi et al. 2019) screened for the mecA resistance conferring gene as well as the Panton–Valentine leukocidin (pvl) virulence gene by conventional PCR technique. All analysed 24 MRSA isolates harboured the mecA gene hence concordantly agreeing 100% with the phenotypic results. Of note 16.7% of the isolates also harboured the pvl gene. Further characterization of the isolates by MLST and spa typing was done. These typing methods categorized the isolates in four sequence types. ST88 predominated by 52.2% (n = 13), followed by ST1719 at 29.2% (n = 7), ST8 and ST 1820 were assigned to 3 and 1 isolates, respectively. Of the 4 pvl positive isolates three belonged to ST88 and one to ST1820 (a single locus variant of ST 88). All isolates were ultimately divided into two clonal complexes, i.e. CC8 and CC88 (Moremi et al. 2012). In a subsequent study by Moremi et al. (2019) whereby conventional PCR methods were used so screen for mecA and pvl an account on the characterization of some of the MRSA analysed was given. The study reported that one MRSA from a healthcare worker nasal carriage belongs to ST88 and was pvl positive; furthermore, they reported that 2 SSTI-related MRSA were characterized to belong to ST612 (Moremi et al. 2019). Kumburu et al. (2018) on the other hand characterized MRSA using whole genome sequencing method. Of the 30 isolates analysed, 33.3% (n = 10) were confirmed to harbour the mecA resistance conferring gene. Among the identified 10 MRSA, 6 belonged to ST8 and 2 belonged to ST239, the remaining two had unknown sequence type. Furthermore, none of the MRSA strains harboured the pvl or toxin shock syndrome (tst) virulence genes (Moremi et al. 2016).
Nurjadi et al. 2014 while researching on trimethoprim resistance genes in S. aureus isolated from Sub-Saharan Africa determined that 100% of the Tanzanian S. aureus in the study which were phenotypically trimethoprim resistant in fact harboured trimethoprim conferring resistance genes (Nurjadi et al. 2014).
Prevalence, antibiotic resistance and genotyping of S. aureus isolated from animals
As indicated in Table 5, few studies (n = 5) have reported on prevalence, antimicrobial resistance and genetic characteristics of S. aureus in animals published between 2010 and 2018 in Tanzania.
Prevalence of S. aureus in animals
Five publication reported on prevalence of S. aureus in animals whereby the most examined sample in animals was milk (n = 4). Prevalence of S. aureus varied in different geographic areas. Kashoma et al. (2018) and Mohammed et al. (2018) reported the highest prevalence at 49% and 41% in Morogoro, respectively. Suleiman and Mdegela (2018) did not fall far back when observing a prevalence of 37% describing subclinical mastitis in cows on Unguja Island. The lowest prevalence of 33% was recorded in a study aiming at assessing raw milk quality in Arusha and Meru District (Ngasala et al. 2015). Generally prevalence of S. aureus seemed to be much lower in swabs taken from nasal cavities. Kashoma et al. (2018) who took nasal swabs from cows observed a S. aureus prevalence of 28% whereas Katakweba et al. (2016) observed a prevalence of 4% and 11% in pig and dog nasal swabs, respectively.
MRSA detection in milk-associated studies varied tremendously ranging from 4 to 35% (Mohammed et al. 2018; Kashoma et al. 2018), even though both studies were conducted in Morogoro township. Furthermore, MRSA prevalence in isolates recovered from cow nasal cavities was repowered at 16% (Suleiman and Mdegela 2018), whereas no MRSA was detected in pigs and dogs nasal swabs (Kashoma et al. 2018).
Antibiotic resistance in animal originated S. aureus isolates
Four studies described antibiotic resistance in S. aureus using different antibiotic pallets which included cephalexin, gentamicin, kanamycin, neomycin, tetracycline, streptomycin, amoxicillin, cephalexin, clindamycin, vancomycin, ampicillin, co-trimoxazole, oxacillin and cefoxitin. This review, however, focused on the most reported antibiotics in this area as indicated in Table 6.
Most of the reviewed studies reported low antibiotic resistance levels with exception of penicillin and ampicillin which showed mean resistances rates of 85% and 73%, respectively. Suleiman and Mdegela (2018) also further observed resistance against amoxicillin and cephalexin at a rate of 47% and 30%, which was unfortunately not reported by any of the other reviewed studies. Moreover, Tetracycline, co-trimoxazole and cephalexin were reported to have a resistance proportion of 30%, 32% and 30%, respectively.
MRSA detection in all studies was confirmed by resistance against cefoxitin and/or oxacillin, whereby mean resistance rates of 10% and 17% were observed, respectively. An average of 8% resistance against vancomycin was observed, reported in 3/5 studies as described in (Table 6).
Genotypic characterization of animal originated S. aureus isolates
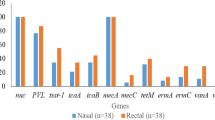
Genotypic data retrieved from 2 of the 5 studies were reviewed. In both, real-time- PCR (qPCR) was used to confirm S. aureus as well as screening for mecA resistance conferring gene. Katakweba et al. (2016) additionally screened for mecC, a recently discovered mecA homologue said to have the potential to be mis-categorized as methicillin-sensitive S. aureus (MSSA). None of the two resistance conferring genes were detected in this study; however, S. aureus was confirmed in 4 pigs and 11 dog samples, respectively. Further spa typing of the pig originating MSSA characterized them as spa type t131 associated with ST80. Additionally isolates of dog origin were characterized as spa types t314 (ST121), t084 (ST15 and ST18) and t223 (ST22) all of which were also identified in human nasal isolates reported in the same study.
In the study by Mohammed et al. (2018), both coagulase positive (n = 46) and coagulase negative (n = 2) S. aureus were detected in their milk samples. mecA gene was detected in two coagulase positive and one coagulase negative S. aureus isolate making this study the first in Tanzania to report on coagulase negative S. aureus harbouring the mecA gene. Of the 3 MRSA isolates phenotypically detected in the study, one was genotypically coagulase negative S. aureus characterized as spa type t2603, whereas the other two MRSA’s were spa un-typable.
Discussion
The publications under review showed that S. aureus was isolated in a number of infections including SSTI’s, bloodstream, otitis media, respiratory tract and urinary tract infections staying true to characteristically causing a wide range of infections.
This review observed a S. aureus average prevalence ranging from 1 to 45% in human (Fig. 2). However, particularly higher infection rates were typically observed in SSTI’s.
Distribution of S. aureus prevalence in colonization and clinical infections in humans. Based on reviewed publications in Tanzania from 2010 to 2020, slight zonal variations in prevalence of different infections could be observed. Blood born S. aureus was more prevalent in the Lake zone. Collectively the review revealed that S. aureus was most frequently implicated in association with SSTIs. Prevalence of S. aureus in human colonization in all reported zones was generally less than 30% with exception to the northern zone where the prevalence was uncharacteristically high
Furthermore, it was notably observed that S. aureus colonization ranged between 28 and 45% (Fig. 2) indicative of considerable circulation of both hospital- and community-associated S. aureus in the country. Considering the wide range of infections S. aureus can cause especially in various vulnerable populations such as children, elderly and the immune compromised individuals which was also observed in this review, there is need to emphasize the necessity to establish a reliable and sustainable surveillance system to monitor the S. aureus bacterium countrywide.
This review discovered that the antibiotic resistance patterns in colonization strains recorded uncharacteristically high proportions. Mean resistance rates of 41%, 27% and 46% against erythromycin, co-trimoxazole and tetracycline could be observed, respectively, whereas resistance rates of the same antibiotics reported in colonization strains in Europe and some parts of Asia did not exceed 20% (Heijer et al. 2013; Lestari et al. 2008). Contrary to resistance pattern in clinical S. aureus observed in this review were in accordance with other reports around Africa (Shittu and Lin 2006; Onwubiko and Sadiq 2011). As in many parts recorded resistance against β lactams was high. Resistance rates ranging from 30 to 65% against erythromycin, gentamicin, co-trimoxazole and clindamycin were also described in this review. The affected antibiotics are readily available in the community and also commonly used for empirical treatment of different cases of bacterial suspected infections. A notable presence of resistance against these antibiotics is an indication of their failure in treatment and a need for broader class antibiotics which are expensive and not accessible for most Tanzanians.
The two zones that reported on MRSA prevalence showed immense variation in their studies; however, on average prevalence was reported to be around 26% (Fig. 2). More than a third of the included studies reported prevalence above 25%. This shows that MRSA prevalence has risen in the last 10 years. Abdulgader's review on MRSA prevalence on the African continent categorized Tanzania as belonging to countries with low MRSA prevalence ranging from 6 to 16% between the years 2001 and 2009 (Abdulgader et al. 2015). This observed abrupt increase should be taken seriously by employing active antibiotic stewardship as well as directing more research efforts towards understanding and preventing the spread of these strains.
A wide spectrum of MSSA spa types and sequence types (ST) were identified in the reviewed studies. With the limited information gathered no dominance could be reported. Nevertheless, most pvl positive MSSA clones associate with skin infections identified in this review belonged to ST152 which concurs with the findings from other African-based studies (Ruffing et al. 2017). Genetic characterization of MRSA strains has managed to categorize them into five predominant sequence types, i.e. ST8, ST1719, ST 1820, ST239, ST612 and ST88. Most characterized MRSA were pvl negative consistent with the findings by Abdulgader et al. (2015) who suggested that Africa is predominated by pvl negative MRSA belonging to ST88. Even though some important information was apparent in this review, data concerning MSSA and MRSA genotyping is still limited, Kumburu et al. (2018) was the only study that reported on other virulence factors apart from pvl, hence the scarcity of knowledge on S. aureus virulence factors circulating in our communities. Furthermore, genotypic characterization studies predominantly focused on screening for the mecA gene; the β-lactam-associated resistance gene, neglecting other resistant markers for other commonly used antibiotic in the country. Katakweba et al. (2016) was the only study that screened for mecC gene; a mecA homologue which has been identified in other studies to be associated with causing infection in humans with animal contact (Petersen et al. 2013). This homologue can easily be misdiagnosed as MSSA hence causing consequences on patient management as well as on antimicrobial resistance surveillance strategies. In order to make informed decisions about disease prevention and management, there is a need for extended surveillance as well as genotypic based research in Tanzania.
Studies reporting on prevalence, antimicrobial resistance and genetic characteristics of S. aureus in animals published between 2010 and 2020 in Tanzania were few; only 5 studies were included in this review, scantly representing a small area in the country. As in human, animal S. aureus isolates seem to harbour highly resistance rates against to penicillin and ampicillin antibiotics. Mean resistance rates against oxacillin (17%) and tetracycline (30%) were further able to describe the effects of overusing oxytetracycline previously stated as the most used antibiotic in the livestock keeping business in Tanzania (Caudell et al. 2017).
Staphylococcus aureus is one of the leading causes of bovine mastitis which explains the fact that the majority of publications included in this review concerning S. aureus in animals addressed the pathogen in clinical and sub-clinical mastitis or in regards to milk production quality. This has been instrumental in the reviews failure to make a link in describing either animals or humans acting as potential S. aureus reservoir for each other, as well as the effect of such in both public and animal health. Very few studies on genomic S. aureus characterization were available for review and did not suffice in showing such linkage. Even so one publication reported on the genetic characteristics managed to confirm S. aureus strains belonging to the same spa type found to colonize both dogs and humans. Unfortunately, the humans and animals involved in this study were epidemiologically unrelated hence making it impossible to link the two. These findings nevertheless present a clear possibility for the S. aureus strain to infect across host species.
The primary method for establishing S. aureus antibiogram in the reviewed work was done phenotypically by using the Kirby-Bauer disc diffusion test along with CLSI guidelines. This method has been reported to have inherent shortcomings such as being highly dependent on experimental conditions that may affect end results (Reller et al. 2009). With this fact in mind the Kirby Bauer antibiogram results would have to be confirmed by another test of a different principle to prevent over reporting of resistance, particularly in the confirmation of MRSA and Vancomycin resistant S. aureus strains. In this review, four studies had MRSA confirmed genotypically which showed 100% agreement with the phenotypic Kirby-Bauer disc diffusion test. This agreement between the methods should encourage the use of the Kirby-Bauer disc diffusion test as an antibiotic sensitivity monitoring strategy in the country, since it is the most available and affordable method in the Tanzanian setting.
Information about prevalence, antibiotic susceptibility and genotypic characteristics of S. aureus originating from different hosts and sources in Tanzania still remains scarce. It is evident that the majority of the publications included in this review are from research institutions or tertiary hospitals affiliated to universities with health-related focuses hence most information could be derived from three focal points MNH in Dar es Salaam, BMC Mwanza and, KCMC Moshi, respectively (i.e. Northern, Lake and Eastern zones). According to the Tanzania National Bureau of Statistics the focal regions represented in this review earn higher income per capital compared to the zones whereby no information could be gathered. This implies that most of the data obtained for review are based in relatively well to do regions (Tanzania National Bureau of Statistics 2018). No published data between 2010 and 2020 on the epidemiology of S. aureus were available for the southern and western parts of the country, which are home to some of the most impoverished regions in Tanzania (refer to Fig. 3). It is well known that poverty struck areas also face other challenges as poor healthcare facilities and access, lack of basic needs such as food, proper housing and sanitation, which in turn leaves the population ridden by different infectious diseases (Alvarez-Uria et al. 2016). Furthermore, since the income gained by the poor is mainly for subsistence, the tendency to resort to self-treatment and or consulting traditional healer is very high (Green et al. 2015) all of which are known indicators associated with driving antibiotic resistance (Byarugaba 2004).
Tanzania administrative zones. Modified map adopted from Suleiman 2018 (Suleiman 2018). Tanzania regions are classified into 9 zones: 1. Eastern Zone ( Morogoro, Pwani and Dar es Salaam) 2. Northern Zone (Arusha, Kilimanjaro and Tanga); 3. Lake Zone (Kagera, Mwanza, and Mara); 4. Western Zone (Kigoma,Tabora and Shinyanga); 5. Central Zone (Dodoma, Manyara and Singida); 6. South West Highlands (Katavi, Mbeya and Rukwa); 7. Southern Highlands Zone (Iringa, Njombe and Ruvuma); 8. Southern Zone (Lindi and Mtwara) and 9. Zanzibar Zone
Limitations
The data from this review cannot be generalized as true prevalence, antibiotic resistance patterns or genotypic characterization of S. aureus bacterium in Tanzania, there is lack of S. aureus studies representation from other zones. High inter-study variations such as type of specimen analysed, objective of studies, time frame as well as methods employed including criteria set for resistance/MRSA confirmation may have influenced the outcome.
Methods used to genetically characterize S. aureus in this review were very divergent from simple PCR to whole genome sequencing. The data collected in these methods had different focuses which made comparing the results between studies difficult. Since the main objective in characterizing S. aureus or any other bacteria for that matter is understanding the genetic similarities and differences in conferring antibiotic resistance, strain type as well as virulence factors, it would therefore be useful to have a common guideline that allowing inter-laboratory comparability to gather more reliable and holistic data meant to guide treatment and infection prevention strategies in the country.
Conclusions
With the insight this review has given it is evident that prevalence of S. aureus and MRSA in Tanzania is rising, although clear variations between different geographic areas could be observed. Furthermore, non-susceptibility to commonly prescribed antibiotics in community-associated S. aureus is also of concern hence the need to emphasize further collection of inf epidemiology (susceptibility patterns and genotypic characterization) data of all bacteria of public health and animal health importance in Tanzania to gain a comprehensive description of the burden of the bacteria as well as enable proper strategies guiding empirical infections treatment and management in the country.
Availability of data and materials
Not applicable.
Abbreviations
- AMR:
-
Antimicrobial resistance
- AST:
-
Antibiotic susceptibility testing
- BMC:
-
Bugando medical centre
- CAI:
-
Community-acquired infection
- CLSI:
-
Clinical laboratory standard institute
- CNS:
-
Coagulase negative Staphylococcus
- HAI:
-
Hospital acquired infection
- HCW:
-
Healthcare workers
- ICU:
-
Intensive care unit
- KCMC:
-
Kilimanjaro Christian Medical Centre
- LAI:
-
Livestock acquired infections
- MLST:
-
Multi-locus sequence typing
- MNH:
-
Muhimbili National Hospital
- MRSA:
-
Methicillin-resistant Staphylococcus aureus
- MSSA:
-
Methicillin-sensitive Staphylococcus aureus
- ND:
-
Not done
- NR:
-
Not reported
- NS:
-
Not Screened
- OM:
-
Otitis media
- RCH:
-
Reproductive and child health
- Spa:
-
Staphylococcus protein A
- SSI:
-
Surgical site infection
- SSTI’s:
-
Skin and soft tissue infections
- ST:
-
Sequence type
- UTI:
-
Urinary tract infection
- WHO:
-
World Health Organization
References
Abdulgader SM, Shittu AO, Nicol MP, Kaba M (2015) Molecular epidemiology of methicillin-resistant Staphylococcus aureus in Africa: a systematic review. Front Microbiol 6:348
Ahmed M, Mirambo MM, Mushi MF, Hokororo A, Mshana SE (2017) Bacteremia caused by multidrug-resistant bacteria among hospitalized malnourished children in Mwanza, Tanzania: a cross sectional study. BMC Res Notes 10(1):62
Alvarez-Uria G, Gandra S, Laxminarayan R (2016) Poverty and prevalence of antimicrobial resistance in invasive isolates. Int J Infect Dis 52:59–61
Bloch EM, West SK, Mabula K, Weaver J, Mrango Z, Munoz B et al (2017) Antibiotic resistance in young children in Kilosa District, Tanzania 4 years after mass distribution of azithromycin for trachoma control. Am J Trop Med Hyg 97(3):815–818
Byarugaba DK (2004) A view on antimicrobial resistance in developing countries and responsible risk factors. Int J Antimicrob Agents 24(2):105–110
Caggiano S, Ullmann N, De Vitis E, Trivelli M, Mariani C, Podagrosi M et al (2017) Factors that negatively affect the prognosis of pediatric community-acquired pneumonia in district hospital in Tanzania. Int J Mol Sci 18(3):623
Caudell MAQM, Subbiah M, Call DRRC, Roulette JW et al (2017) Antimicrobial use and veterinary care among agro-pastoralists in Northern Tanzania. PLoS ONE 12(1):e0170328
Chalya PL, Mabula JB, Dass RM, Ngayomela IH, Chandika AB, Mbelenge N et al (2012) Major limb amputations: a tertiary hospital experience in northwestern Tanzania. J Orthop Surg Res 7:18
Chochua S, D’Acremont V, Hanke C, Alfa D, Shak J, Kilowoko M et al (2016) Increased nasopharyngeal density and concurrent carriage of Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis are associated with pneumonia in febrile children. PLoS ONE 11(12):725
David MZ (2010) Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev 23:616–687
den Heijer CD, van Bijnen EM, Paget WJ, Pringle M, Goossens H, Bruggeman CA et al (2013) Prevalence and resistance of commensal Staphylococcus aureus, including meticillin-resistant S. aureus, in nine European countries: a cross-sectional study. Lancet Infect Dis 13(5):409–415
Founou RC, Founou LL, Essack SY (2017) Clinical and economic impact of antibiotic resistance in developing countries: a systematic review and meta-analysis. PLoS ONE 12(12):e0189621
Geofrey A, Abade A, Aboud S (2015) Methicillin-resistant Staphylococcus aureus (MRSA) colonization among intensive care unit (ICU) patients and health care workers at Muhimbili national hospital Dar Es Salaam, Tanzania. Pan Afr Med J. https://doi.org/10.11604/pamj.2015.21.211.420
Gilyoma JM, Mabula JB, Chalya PL (2013) Animal-related injuries in a resource-limited setting: experiences from a Tertiary health institution in northwestern Tanzania. World J Emerg Surg 8(1):7
Green JA, Norris P, Bukhari NI (2015) Self-medication, home remedies, and spiritual healing: common responses to everyday symptoms in Pakistan AU—Anwar, Mudassir. Health Psychol Behav Med 3(1):281–295
Haddadin AS, Fappiano SA, Lipsett PA (2002) Methicillin resistant Staphylococcus aureus (MRSA) in the intensive care unit. Postgrad Med J 78(921):385–392
Hudzicki J (2009) Disk diffusion susceptibility test protocol. Kirby-Bauer. Amecan Society for Microbiology, Washington
Jevons MP (1961) “Celbenin”—resistant Staphylococci. Br Med J 1(5219):124–125
Joachim A, Moyo SJ, Nkinda L, Majigo M, Mmbaga E, Mbembati N et al (2017) Prevalence of methicillin-resistant Staphylococcus aureus carriage on admission among patients attending regional hospitals in Dar es Salaam, Tanzania. BMC Res Notes 10(1):417
Joachim A, Moyo SJ, Nkinda L, Majigo M, Rugarabamu S, Mkashabani EG et al (2018) Nasal carriage of methicillin-resistant Staphylococcus aureus among health care workers in tertiary and regional Hospitals in Dar es Salam, Tanzania. Int J Microbiol 2018:7
Kashoma IP, Lalata EP, Maiga CJ, Mtemela BO, Medardus JJ (2018) Prevalence and antimicrobial susceptibility profiles of Staphylococcus aureus from cow’s milk, nasal and environmental swabs in selected dairy farms in Morogoro Tanzania. Tanzan Vet J 30(2):61–75
Kassam NA, Damian DJ, Kajeguka D, Nyombi B, Kibiki GS (2017) Spectrum and antibiogram of bacteria isolated from patients presenting with infected wounds in a Tertiary Hospital, northern Tanzania. BMC Res Notes 10(1):757
Katakweba AS, Muhairwa AP, Espinosa-Gongora C, Guardabassi L, Mtambo MM, Olsen JE (2016) spa typing and antimicrobial resistance of Staphylococcus aureus from healthy humans, pigs and dogs in Tanzania. J Infect Dev Ctries 10(2):143–148
Kayange N, Kamugisha E, Mwizamholya DL, Jeremiah S, Mshana SE (2010) Predictors of positive blood culture and deaths among neonates with suspected neonatal sepsis in a tertiary hospital, Mwanza-Tanzania. BMC Pediatr 10(1):1
Kazimoto T, Abdulla S, Bategereza L, Juma O, Mhimbira F, Weisser M et al (2018) Causative agents and antimicrobial resistance patterns of human skin and soft tissue infections in Bagamoyo, Tanzania. Acta Trop 186:102–106
Kinabo GD, van der Ven A, Msuya LJ, Shayo AM, Schimana W, Ndaro A et al (2013) Dynamics of nasopharyngeal bacterial colonisation in HIV-exposed young infants in Tanzania. Trop Med Int Health TM IH 18(3):286–295
Kiponza R, Balandya B, Majigo MV, Matee M (2019) Laboratory confirmed puerperal sepsis in a national referral hospital in Tanzania: etiological agents and their susceptibility to commonly prescribed antibiotics. BMC Infect Dis 19(1):690
Kumburu HH, Sonda T, Mmbaga BT, Alifrangis M, Lund O, Kibiki G et al (2017) Patterns of infections, aetiological agents and antimicrobial resistance at a tertiary care hospital in northern Tanzania. Trop Med Int Health TM IH 22(4):454–464
Kumburu HH, Sonda T, Leekitcharoenphon P, van Zwetselaar M, Lukjancenko O, Alifrangis M, et al (2018) Hospital epidemiology of methicillin-resistant staphylococcus aureus in a tertiary care hospital in Moshi, Tanzania, as determined by whole genome sequencing. BioMed Res Int 2018:2087693
Lestari ES, Severin JA, Filius PMG, Kuntaman K, Duerink DO, Hadi U, Wahjono H, Verbrugh HA (2008) Antimicrobial resistance among commensal isolates of Escherichia coli and Staphylococcus aureus in the Indonesian population inside and outside hospitals. Eur J Clin Microbiol Infect Dis 27:45–51
Lozano C, Gharsa H, Ben Slama K, Zarazaga M, Torres C (2016) Staphylococcus aureus in animals and food: methicillin resistance, prevalence and population structure. A review in the African continent. Microorganisms 4(1):12
Mahende C, Ngasala B, Lusingu J, Butichi A, Lushino P, Lemnge M et al (2014) Aetiology of acute febrile episodes in children attending Korogwe District Hospital in north-eastern Tanzania. PLoS ONE 9(8):e104197
Makani J, Mgaya J, Balandya E, Msami K, Soka D, Cox SE et al (2015) Bacteraemia in sickle cell anaemia is associated with low haemoglobin: a report of 890 admissions to a tertiary hospital in Tanzania. Br J Haematol 171(2):273–276
Manyahi J, Matee MI, Majigo M, Moyo S, Mshana SE, Lyamuya EF (2014) Predominance of multi-drug resistant bacterial pathogens causing surgical site infections in Muhimbili National Hospital, Tanzania. BMC Res Notes 7:500
Marwa KJ, Mushi MF, Konje E, Alele PE, Kidola J, Mirambo MM (2015) Resistance to cotrimoxazole and other antimicrobials among isolates from HIV/AIDS and Non-HIV/AIDS patients at Bugando Medical Centre, Mwanza, Tanzania. AIDS Res Treat 2015:8
Mawalla B, Mshana SE, Chalya PL, Imirzalioglu C, Mahalu W (2011) Predictors of surgical site infections among patients undergoing major surgery at Bugando Medical Centre in Northwestern Tanzania. BMC Surg 11:1–7
Mbunda F, McHembe MD, Chalya PL, Rambau P, Mshana SE, Kidenya BR et al (2012) Experiences with surgical treatment of chronic lower limb ulcers at a tertiary hospital in northwestern Tanzania: a prospective review of 300 cases. BMC Dermatol 12:17
Mhada TV, Fredrick F, Matee MI, Massawe A (2012) Neonatal sepsis at Muhimbili national hospital, Dar es Salaam, Tanzania; aetiology, antimicrobial sensitivity pattern and clinical outcome. BMC Public Health 12:904
Mikomangwa WP, Bwire GM, Kilonzi M, Mlyuka H, Mutagonda RF, Kibanga W et al (2020) The existence of high bacterial resistance to some reserved antibiotics in tertiary hospitals in Tanzania: a call to revisit their use. Infect Drug Resist 13:1831–1838
Mohammed J, Ziwa MH, Hounmanou YMG, Kisanga A, Tuntufye HN (2018) Molecular typing and antimicrobial susceptibility of methicillin-Resistant Staphylococcus aureus isolated from bovine milk in Tanzania. Hindawi Int J Microbiol 2018:1–6
Moremi N, Mshana SE, Kamugisha E, Kataraihya J, Tappe D, Vogel U et al (2012) Predominance of methicillin resistant Staphylococcus aureus—ST88 and new ST1797 causing wound infection and abscesses. J Infect Dev Ctries 6(8):620–625
Moremi N, Mushi MF, Fidelis M, Chalya P, Mirambo M, Mshana SE (2014) Predominance of multi-resistant gram-negative bacteria colonizing chronic lower limb ulcers (CLLUs) at Bugando Medical Center. BMC Res Notes 7(1):211
Moremi N, Claus H, Mshana SE (2016) Antimicrobial resistance pattern: a report of microbiological cultures at a tertiary hospital in Tanzania. BMC Infect Dis 16(1):756
Moremi N, Claus H, Vogel U, Mshana SE (2019) The role of patients and healthcare workers Staphylococcus aureus nasal colonization in occurrence of surgical site infection among patients admitted in two centers in Tanzania. Antimicrob Resist Infect Control 8:102
Moyo S, Aboud S, Kasubi M, Maselle SY (2010) Bacteria isolated from bloodstream infections at a tertiary hospital in Dar es Salaam, Tanzania–antimicrobial resistance of isolates. S Afr Med J S Afr Tydskr Geneeskd 100(12):835–838
Moyo SJ, Aboud S, Blomberg B, Mkopi N, Kasubi M, Manji K et al (2014) High nasal carriage of methicillin-resistant Staphylococcus aureus among healthy Tanzanian under-5 children. Microb Drug Resist (larchmont NY) 20:82–88
Mpogoro FJ, Mshana SE, Mirambo MM, Kidenya BR, Gumodoka B, Imirzalioglu C (2014) Incidence and predictors of surgical site infections following caesarean sections at Bugando Medical Centre, Mwanza, Tanzania. Antimicrob Resist Infect Control 3:25
Mushi MF, Mwalutende AE, Gilyoma JM, Chalya PL, Seni J, Mirambo MM et al (2016) Predictors of disease complications and treatment outcome among patients with chronic suppurative otitis media attending a tertiary hospital, Mwanza Tanzania. BMC Ear Nose Throat Disord 16:1
Mwambete KD, Eulambius M (2018) High prevalence of antibiotic-resistant otitis media-associated bacterial flora of asymptomatic people living with HIV at Morogoro Hospital, Tanzania. J Int Assoc Provid AIDS Care 17:2325958218759761
Ngasala JHNH, Madundo M, Mtambo A (2015) Assessment of raw milk quality and stakeholders’ awareness on milk-borne health risks in Arusha City and Meru District, Tanzania. Trop Anim Health Prod 47:927–932
Ngocho JSAC, Sariko M, Mmbaga BT, Kibiki GS (2015) Bacterial etiology of respiratory tract infections among ambulatory school children in Moshi Municipality, Tanzania. Sci J Public Health 3(5):625–632
Nurjadi D, Olalekan AO, Layer F, Shittu AO, Alabi A, Ghebremedhin B et al (2014) Emergence of trimethoprim resistance gene dfrG in Staphylococcus aureus causing human infection and colonization in sub-Saharan Africa and its import to Europe. J Antimicrob Chemother 69(9):2361–2368
Okamo B, Moremi N, Seni J, Mirambo MM, Kidenya BR, Mshana SE (2016) Prevalence and antimicrobial susceptibility profiles of Staphylococcus aureus nasal carriage among pre-clinical and clinical medical students in a Tanzanian University. BMC Res Notes 9(1):1–6
O’Neill J (2016) Tackling drug-resistant infections globally: final report and recommendations
Onken A, Said AK, Jorstad M, Jenum PA, Blomberg B (2015) Prevalence and antimicrobial resistance of microbes causing bloodstream infections in Unguja, Zanzibar. PLoS ONE 10(12):e0145632
Onwubiko NE, Sadiq NM (2011) Antibiotic sensitivity pattern of Staphylococcus aureus from clinical isolates in a tertiary health institution in Kano, Northwestern Nigeria. Pan Afr Med J 8(4):1–7
Petersen A, Stegger M, Heltberg O, Christensen J, Zeuthen A, Knudsen LK et al (2013) Epidemiology of methicillin-resistant Staphylococcus aureus carrying the novel mecC gene in Denmark corroborates a zoonotic reservoir with transmission to humans. Clin Microbiol Infect 19(1):E16–E22
Reller LB, Weinstein M, Jorgensen JH, Ferraro MJ (2009) Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clin Infect Dis 49(11):1749–1755
Ruffing U, Alabi A, Kazimoto T, Vubil DC, Akulenko R, Abdulla S et al (2017) Community-associated Staphylococcus aureus from sub-Saharan Africa and Germany: a cross-sectional geographic correlation study. Sci Rep 7(1):154
Seni J, Mwakyoma AA, Mashuda F, Marando R, Ahmed M, DeVinney R et al (2019a) Deciphering risk factors for blood stream infections, bacteria species and antimicrobial resistance profiles among children under five years of age in North-Western Tanzania: a multicentre study in a cascade of referral health care system. BMC Pediatr 19(1):32
Seni J, Tito JN, Makoye SJ, Mbena H, Alfred HS, van der Meer F et al (2019b) Multicentre evaluation of significant bacteriuria among pregnant women in the cascade of referral healthcare system in North-western Tanzania: bacterial pathogens, antimicrobial resistance profiles and predictors. J Glob Antimicrob Resist 17:173–179
Shittu AO, Lin J (2006) Antimicrobial susceptibility patterns and characterization of clinical isolates of Staphylococcus aureus in KwaZulu-Natal province, South Africa. BMC Infect Dis 6:125
Silago V, Mushi MF, Remi BA, Mwayi A, Swetala S, Mtemisika CI et al (2020) Methicillin resistant Staphylococcus aureus causing osteomyelitis in a tertiary hospital, Mwanza, Tanzania. J Orthop Surg Res 15(1):95
Suleiman R (2018) Local and regional variations in conditions for agriculture and food security in Tanzania: a review
Suleiman TSKE, Mdegela RH (2018) Prevalence of bovine subclinical mastitis and antibiotic susceptibility patterns of major mastitis pathogens isolated in Unguja island of Zanzibar, Tanzania. Trop Anim Health Prod 50(2):259–266
Tanzania National Bureau of Statistics (2018) Highlights for the third quarter (July–September) gross domestic product. United republic of Tanzania, National Bureau of Statistics Ministry of Finance and Planning
Thriemer K, Ley B, Ame S, von Seidlein L, Pak GD, Chang NY et al (2012) The burden of invasive bacterial infections in Pemba, Zanzibar. PLoS ONE 7(2):e30350
WHO (2014) Antimicrobial resistance: Global report on surveillance Geneva. WHO, Geneva
WHO (2017) Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. WHO, Geneva
Zetola N, Francis JS, Nuermberger EL, Bishai WR (2005) Community-acquired meticillin-resistant Staphylococcus aureus: an emerging threat. Lancet Infect Dis 5(5):275–286
Acknowledgements
Not applicable.
Funding
This research was conducted under the PhD fellowship given by the Consortium for Advanced Research Training in Africa (CARTA). CARTA is jointly led by the African Population and Health Research Center and the University of the Witwatersrand and funded by the Carnegie Corporation of New York (Grant number B 8606.R02), Sida (Grant number 54100113), the DELTAS Africa Initiative (Grant number 107768/Z/15/Z) and Deutscher Akademischer Austauschdienst (DAAD). The DELTAS Africa Initiative is an independent funding scheme of the African Academy of Sciences (AAS)’s Alliance for Accelerating Excellence in Science in Africa (AESA) and supported by the New Partnership for Africa’s Development Planning and Coordinating Agency (NEPAD Agency) with funding from the Wellcome Trust (UK) and the UK government. The statements made and views expressed are solely the responsibility of the Fellow.
Author information
Authors and Affiliations
Contributions
TM and TK were responsible for the concept, design, reviewing studies for the manuscript: JM contributed in literature search strategies and data extraction method: MB, RM and MM took part in revising important contents of the manuscript: TM drafted the manuscript: TK and SLB critically reviewed the manuscript for publication. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mzee, T., Kazimoto, T., Madata, J. et al. Prevalence, antimicrobial susceptibility and genotypic characteristics of Staphylococcus aureus in Tanzania: a systematic review. Bull Natl Res Cent 45, 162 (2021). https://doi.org/10.1186/s42269-021-00612-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s42269-021-00612-z