Abstract
Background
Fruit rot disease is one of the most important factors limiting the production potential of Bhut Jolokia (Capsicum chinense Jacq.), which is known as one of the hottest chillies in the world. The management strategies are highly dependent on synthetic chemicals which are causing a detrimental impact on the environment. Considering the factors, this study focuses on exploring potential endophytic microflora from native chilli cultivars which can suppress the fruit rot pathogen.
Results
Endophytic microflora occurs ubiquitously in plants that possess various plant-benefiting abilities. A total of 34 endophytic isolates were obtained from different chilli cultivars. These endophytic isolates were subjected to screening in vitro for their potential to suppress the incitant pathogen Colletotrichum gloeosporioides (the causal agent of chilli fruit rot), which was confirmed based on cultural, morphological, pathogenicity and molecular studies. The preliminary screening yielded four bacterial endophytic isolates capable of suppressing the pathogen which was found non-pathogenic to Bhut Jolokia plant. On the basis of morphological, biochemical and molecular identification, the four most promising isolates were identified as Bacillus velezensis, B. mycoides, B. altitudinis and B. cereus, respectively, and used for further in vitro tests. B. velezensis showed the highest inhibition (68.67%) on mycelial growth of C. gloeosporioides, followed by B. mycoides (65.33%), B. altitudinis (52.89%) and B. cereus (45.33%). Among the compatible combination, the highest efficacy (56.00%) was found in the combination of B. velezensis and B. altitudinis.
Conclusion
From the present study, it can be concluded that B. velezensis and B. mycoides alone and in combination can be used as potential biocontrol agent in managing the fruit rot of Bhut Jolokia considering their performance in field conditions.
Similar content being viewed by others
Background
India is known for its production of a variety of spices which add a plethora of flavours to Indian foods. Out of the spices, Chilli is one of the major components. India is known to be the world’s largest producer, exporter and consumer of Chilli and contributes a total of 36% to global production (Sahitya et al. 2014). Northeast India is blessed with a variety of chillies cultivars which are grown indigenously. Bhut Jolokia (Capsicum chinense Jacq.) is one of the native cultivars considered one of the hottest chillies in the world. It has been registered in Guinness World Records 2006 as the world’s hottest Chilli after being reported to have a Scoville rating of 1,001,304 SHUs (Bosland and Baral 2007). In the world now it stands in seventh position.
In Northeast India, Bhut Jolokia is used to make many dishes. Starting from pickles to different cuisines to beverages, the people of the Northeast always find ways to spice up their food. Different start-ups are trying to exploit the potential of Bhut Jolokia. Not only dishes Bhut Jolokia is use for medicinal purposes since old ages. Deorani and Sharma (2007) reported the beneficial effect on ailments like boils, headaches and night blindness. The capsaicin present in Bhut Jolokia helps relieve asthma and gastrointestinal abnormalities and can dilate bronchial blood vessels and chronic congestion (Sarwa et al. 2012).
Despite having so much potential it is always challenged by various biotic factors. Diseases are among the factors due to which the crop’s full potential is never achieved. Chilli plant suffers from nearly 83 different kinds of diseases, out of which 40 are caused by fungi (Rangaswami et al. 1998). Fruit rot or anthracnose, a major fungal disease, alone contributes to a loss of about 10–50% in both yield and quality of the fruit (Pakdeevaraporn et al. 2005). The disease is also responsible for reducing the productivity and market value of the Bhut Jolokia crop (Manandhar et al. 1995).
The management perspective of the disease is limited to synthetic chemicals and indiscriminate use of chemicals leads to irreversible detrimental impact on the environment. The use of naturally occurring microbiomes as an alternative management strategy and increasing productivity can be a solution. The endophytic microflora can play an important role in developing host plant resistance against targeted diseases.
Endophytes are present ubiquitously in all living plants for at least a part of the life cycle without causing any visible symptoms of the disease (White et al. 2000). Endophytes are known to have several effects or can change the functions of plants during their life cycle (Hardoim et al. 2008). They are considered potent plant inoculants that can enhance plant abilities to deal with biotic and abiotic stresses (Singh et al. 2011). Their ability to manage diseases is a research topic that has become popular recently due to the need to substitute chemical alternatives. The ability of endophytes can be classified mainly into one or more functional groups, i.e. microbes that can (1) reduce abiotic stress in plants, (2) shield the host plant from biotic stress, and (3) improve the plant's nutritional status by boosting nitrogen and phosphorus levels (Singh et al. 2019). Endophytes are found to be involved in the production of biochemicals same as plants which increase their survivability inside the plant ecosystem (Caruso et al. 2020). These mutualistic association of endophytes leads to the better survivability of plants in the ecosystem, providing beneficial impact to plant life without causing any apparent harm.
There have been no to very less reports of fruit rot or anthracnose disease occurring in native chilli cultivars of Northeast India. Also, there is not much information available as to why the infection of fruit rot is less in native chillies of Northeast India. The presence of endophytic microflora can be a reason for the less occurrence of fruit rot disease in those chillies. Considering all these, this research focused on the biocidal activities of endophytes isolated from local cultivars of chilli against fruit rot pathogen.
Methods
Isolation of the pathogen
Fruit samples of Bhut Jolokia showing prominent symptoms of fruit rot disease were collected from Horticultural Experimental Farm, Assam Agricultural University (AAU), Jorhat, Assam. The infected Bhut Jolokia fruits were washed thoroughly with sterile water. Small bits were taken considering that the bits contained both infected and healthy portions. The bits were surface sterilized using 1% sodium hypochlorite (NaOCl) solution for 30 s. To remove traces of sodium hypochlorite, the bits were then rinsed with sterile water for three times. Excess water was soaked using sterilized bloating paper. The bits were aseptically placed in a Petri plate containing potato dextrose agar media (PDA) supplemented with streptomycin @ 100 ppm. Incubation was occurred at 27 ± 1 °C for three days to observe mycelial growth.
Purification and preservation of pathogen
To obtain pure culture, hyphal tip from the mycelial growth of the pathogen was aseptically transferred to a sterilized Petri plate containing PDA media. Incubation was at 27 ± 1 °C until full growth. The pure culture was then preserved in slants at 4 °C refrigerated condition for further study.
Cultural, morphological and molecular studies of pathogen
Cultural studies
For the cultural studies actively growing culture having complete growth in a 9-cm Petri plate was taken. The observations on colony colour, elevation, type of margin, etc., were recorded. The colour of the colony was recorded taking into account reference to the Royal Horticultural Society (RHS) colour chart.
Morphological studies
Morphological studies of the pathogen were done through modified slide culture technique described by Riddell in (1950) and through microscopic studies. Hyphal and conidial characteristics of the pathogen were studied.
Molecular studies
Fungal genomic DNA was extracted using the protocol described by Aamir et al. (2015) with slight modification. For the extraction, 250 μl of pre-warmed extraction buffer (containing 2% CTAB, 100 mM Tris–Cl, 20 mM EDTA and 5 M NaCl) was taken in 1.5-ml Eppendorf tube. A 100–300 mg fungal mat was taken from 7-day-old culture and transferred it to an Eppendorf tube containing extraction buffer. The fungal mat was properly ground in the Eppendorf tube with the help of a sterilized micropestle till a homogenized mixture was obtained. The mixture was vortex for 10 s and incubated at 65 °C for 1 h. The mixture was added with an equal volume of chloroform: Isoamyl alcohol (24:1) and centrifuged at 13,000 rpm for 15 min at 4 °C. The aqueous layer was removed without disturbing the protein layer and transferred to a new Eppendorf tube. The aqueous layer was mixed with 0.1 volume of sodium acetate and 0.6 volume of ice-cold isopropanol. The reaction mixture was then incubated at − 20 °C overnight. The reaction mixture was centrifuged at 13,000 rpm for 10 min at 4 °C. The obtained supernatant was discarded, and the pellet was washed with 70% ethanol. The reaction was centrifuged at 10,000 rpm for 4 min at room temperature. The obtained supernatant was discarded, and the pellet was left for air drying at room temperature. The pellet obtained was resuspended in 100 μl T.E (Tris EDTA) and preserved at – 20 ℃ for further study.
PCR amplification
The amplification of genomic DNA was used ITS-1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS-4 (5′-TCCTCCGCTTATTGATATGC-3′) primers. The PCR (Polymerase chain reaction) cycle followed was 94℃ for 1 min, 94 ℃ for 35 s, 57 ℃ for 1 min, 72 ℃ for 1 min and 72 ℃ for 10 min for 30 cycles.
Sequencing and phylogenetic analysis
The extracted DNA was sent for partial sequencing at Eurofins Analytical Services India Private Limited, Bengaluru, Karnataka. The result obtained was searched in public domain database. Basic Local Alignment Search Tool (BLAST) was used to find homolog sequences in the National Centre for Biotechnology Information (NCBI) database. The phylogenetic analysis of the sequence and the phylogenetic tree was prepared using CLUSTALW tool in MEGA X software.
Pathogenicity test of the pathogen
The pathogenicity test was performed on detach healthy fruit of Bhut Jolokia following the protocol described by Tovar-Pedraza et al. (2020). Healthy Bhut Jolokia fruit samples were brought to laboratory and washed in running tap water. The sample were washed in running water and then surface sterilized using 1% NaOCl for 40 s and 70% ethanol for 1 min, followed by three times rinsing with sterile water. A 5-cm mycelium disc from actively growing pathogen culture was placed on Bhut Jolokia fruit wounded with sterilized needle. For control non-colonized PDA disc was transferred. The inoculated samples were incubated at room temperature (27 ± 1 °C) till development of symptoms.
Collection of chilli samples for endophyte isolation
For the isolation of the endophytic microflora, fruit, leaf, stem and root samples of different landraces of chillies were collected from different districts of Assam. Various locally available landraces are considered for the present investigation, viz. Krishna Jolokia (Capsicum annum), Mem Jolokia (Capsicum frutescens), Dhan Jolokia (Capsicum frutescens), and Black chilli (Capsicum annum). The samples were thoroughly washed under tap water and used for isolation within 48 h of sample collection.
Isolation of endophytes
For the isolation of the endophytes, the protocol of McInroy and Kloepper (1995) was followed with slight modification. Samples of fruit, leaf, stem and roots from different chilli cultivars were brought to the laboratory of the Department of Plant Pathology, AAU, Jorhat. The samples were washed properly in running tap water to remove dust particles adhering to the surface. The samples were rubbed with 99.9% ethanol and 1 g of the sample was taken for isolation. The isolation was occurred in laminar air flow system equipped with HEPA filter where the samples were washed with 70% ethanol and rinsed three times. The samples were dipped in 1% NaOCl + 0.1% Tween 80 for one min and rinsed with double sterile water. From the final wash, 1 ml was pipetted out and poured into a sterile Petri plate where sterile media was poured. This is served as the sterility check. If any microbial growth was observed within four days of incubation, the isolates obtained were discarded. After final wash, the samples were triturated in sterilized pestle and mortar and diluted with 10 ml of 0.1 M potassium buffer (pH 7.0). The triturate was serially diluted in potassium buffer solution up to 107 times. 1 ml of diluted triturate was pipetted into a sterile Petri plate and suitable media was poured. The plates were then incubated at 27 ± 1 °C (two days for bacterial endophytes and four days for fungal endophytes).
Based on the cultural characteristics, representative colonies from the diluted plates were transferred to new media plates (PDA for fungal endophytes and Nutrient agar for bacterial endophytes). Pure culture of the colonies was preserved at 4 °C refrigerated condition for further study.
Preliminary screening of endophytes
Initially, mycelial disc of pathogenic fungus was taken and transferred to the centre of fresh PDA plate and incubated at 27 ± 1 °C for 48 h. For screening of endophytic bacteria for antagonistic properties, four different isolates of endophytic bacteria were taken and made streak leaving 1 cm away from the periphery on the same 48-h-old pathogenic fungus culture plate. Similarly, for screening fungal isolates, four different mycelial discs of endophytic fungal isolates were inoculated at four sides leaving 1 cm away from the periphery on the same 48-h-old pathogenic fungus culture plate. For control, only the mycelial disc of pathogenic fungi was incubated alone.
In vitro screening of endophytes
The most prominent bacterial endophytes were evaluated for their ability to suppress the pathogen following the protocol of Utkhede and Rahe, (1983). Mycelial disc of seven days old pathogenic fungus culture was inoculated and incubated at 27 ± 1 °C for 48 h. The endophytic bacterial isolate was inoculated through line of streak on both sides leaving 1 cm away from periphery of plate on the same 48-h-old pathogenic fungus culture plate. For control only mycelial disc of pathogenic fungus was incubated alone. The plates were then incubated at 27 ± 1 °C till the control plate attained full growth.
The per cent inhibition over control was calculated for each treatment using the following formula (Nwachukwu and Umechuruba 2001).
where I = Per cent inhibition, C = Radial growth diameter of pathogen in control, and T = Radial growth diameter of pathogen in treatment.
Mutual compatibility of selected endophytic isolates
For the mutual compatibility test, perpendicular streaks of two bacterial isolates were made in nutrient agar (NA) plates and incubated at 27 ± 1 °C for 48 h. Compatible isolates grow together, and incompatible combinations inhibit each other. The experiment followed completely randomized block (CRD) design with five replications. Incompatible combinations obtained in the compatibility test were excluded from further studies.
In vitro efficacy of mutually compatible isolates
To study the presence of synergistic effect in mutually compatible combinations of bacterial endophytes, in vitro test was done over the mycelial growth of pathogenic fungus following the protocol of Zegeye et al. (2011) with slight modification. The design of the experiment followed completely randomized design (CRD) with five replications.
Pathogenicity test of promising endophytes
The pathogenicity test of the most promising endophytic isolates was done on the Bhut Jolokia plant to ensure the endophytic isolates are non-pathogenic to the Bhut Jolokia plant. The test was conducted through spraying method, following the method suggested by Larran et al. (2016) and the root inoculation method suggested by Munif et al. (2011). The roots of one-month-old Bhut Jolokia plants were trimmed and washed with sterile water. The plants were then dipped in bacterial suspension 1 × 106 CFU/ml for 60 min and replanted in pots and kept in greenhouse conditions (22 ± 2 °C). The leaves of the plant were spread with bacterial suspension after wounding. For control instead of bacterial suspension, sterile water was taken. Observations were made from the 3rd to the 14th days of inoculation.
Characterization of selected bacterial endophytic isolates
For the morphological and cultural characterization, colony characteristics of selected bacterial endophytic isolates were studied.
For the molecular characterization, the genomic DNA of bacterial isolates was extracted and partial sequencing of 16 S rDNA was done at Eurofins Analytical Services India Private Limited, Bengaluru, Karnataka.
Extraction of selected bacterial genomic DNA
For the extraction of bacterial genomic DNA, the protocol of Cardinal et al. (1997) was followed with modification. 1.5 ml of bacterial culture was taken in a 2-ml Eppendorf tube from the broth culture prepared 24 h before the start of the process. The Eppendorf tube containing the bacterial culture was subjected to centrifuge at 12,000 rpm for 2–5 min. The supernatant was discarded and 500 μl of Lysis buffer (containing 10 mM Tris–Cl, 1 mM EDTA and 0.5% SDS) was added and vortex thoroughly. The homogenized mixture was then incubated at 65 °C for 20 min followed by shaking at an interval of 5 min. 250 μl of NaCl solution was added to the reaction mixture and incubated at − 20 °C for 20 min. After incubation, centrifugation was done at 12,000 rpm at 10 min at room temperature. The supernatant obtained was transferred to a new centrifuge tube. An equal volume of chilled chloroform: isoamyl alcohol (24:1) was added to the reaction mixture and was mixed till the mixture turned milky. The mixture was subjected to centrifuge at 12,000 rpm for 10 min at 4 °C. The aqueous layer was transferred to a new centrifuge tube without disturbing the protein layer. To that, 2.5 volume of chilled 99.9% ethanol was added and incubated at -20 °C overnight. The reaction mixture was centrifuged again at 12,000 rpm for 10 min at 4 °C. The supernatant obtained was discarded, and the pellet obtained was washed with 70% ethanol. The reaction mixture was centrifuged at 12,000 rpm for 3 min at room temperature. The supernatant was discarded and the obtained pellet was dissolved in 100 μl TE (Tris- EDTA) buffer and kept at − 20 °C for further studies.
PCR amplification
PCR amplification of the extracted bacterial genomic DNA was studied using universal primers U16SF (5′-AGAGTTTGATCMTGGCTCAG-3′) and U16SR (5′-TACGGYTACCTTGTTACGACTT-3′). For amplification, 94 ℃ for 3 min, 94 ℃ for 30 s, 55 ℃ for 1 min, 72 °C for 1 min 30 s and 72 °C for 7 min were followed for 35 cycles.
Sequencing and phylogenetic analysis
The extracted bacterial DNA was sent for partial sequencing of 16S rDNA at Eurofins Analytical Services India Private Limited, Bengaluru, Karnataka. The result obtained was searched in the public domain database NCBI using the Basic Local Alignment Search Tool (BLAST). The phylogenetic analysis of the sequence and the phylogenetic tree was prepared using the CLUSTALW tool in MEGA X software.
Statistical analysis
The experiment in Table No. 02 was analysed by using Fisher’s method of analysis of variance (ANOVA), and data interpretation was carried out following Gomez and Gomez (1984). Duncan’s multiple range test (DMRT) at p ≤ 0.05 was used to separate treatment means. Critical differences were calculated to compare the different treatments among themselves.
Results
Cultural, morphological and molecular studies of the pathogen
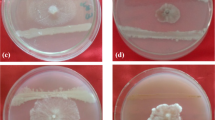
For cultural studies, the pure culture of the pathogen (PFC Jorhat) was studied on potato dextrose agar (PDA) media. The culture was observed to have flat to cottony colonies with dull white to pale grey colour from the front side and yellowish colour from the reverse side (Fig. 1(A) and (B)). There was the presence of salmon-coloured conidial masses near the centre (Fig. 1(C)). Morphological studies were done using microscopy. The hyphae were found to be hyaline, branched and septate. The conidia were found to be hyaline, cylindrical and obtuse at the apex with one or two oil globules at the centre. The size of the conidia was found to be 12.5 μm (length) and 5 μm (width). The presence of appressoria and characteristics of conidia preliminarily confirmed the pathogen as Colletotrichum gloeosporioides (Fig. 2(A) and (B)). For further confirmation, molecular studies were done.
Molecular studies
The concentration of the extracted genomic DNA of the pathogen was found to be 154.79 ng/μl, and the OD value (260/280) was 1.92. The extracted genomic DNA was subjected to PCR amplification, and the amplified product showed a band length of 600 bp in gel electrophoresis (Fig. 3). The sequence recovery of ITS region of the pathogen designated as PFC Jorhat isolate through partial sequencing was also 600 bp. The recovered sequence was compared with NCBI database using nucleotide BLAST tool and showed maximum homology (100%) with Colletotrichum gloeosporioides Tezpur isolate accession number KF053200.1 (Fig. 4).
Phylogenetic studies
The sequence PFC Jorhat isolate was aligned using CLUSTALW using Mega-X for the phylogenetic relationship. The phylogenetic tree was prepared using neighbourhood joining method with 1000 bootstrap replication which showed maximum homology with C. gloeosporioides Tezpur isolate, accession number KF053200.1 (Fig. 4).
Pathogenicity test
The symptoms of the lesion were observed from the 5th day of inoculation of the pathogen disc on detached fruit (Fig. 5). There was no symptom development in the control inoculated with sterile PDA disc. Re-isolation of the pathogen was done which showed the development of fungal colony with characteristic dull white to pale grey colony with conidial mass formation at the centre. The microscopic studies confirm the presence of the pathogen.
Isolation of chilli endophytic microflora
Healthy fruit, leaf, stem and root samples were collected from different cultivars of chilli from four different districts of Assam, viz. Jorhat, Golaghat, Nagaon and Diphu. A total of 34 endophytic microbes were isolated from fruit, leaf, stem and root samples of chilli and characterized on the basis of colony characteristics, viz. texture, growth pattern, color, shape of margin and viscosity of microbe (Table 1).
Observing the table it can be said that the numbers of endophytic isolates are relatively more in roots than the stem, leaf and fruits. All the endophytic isolates were preserved as a pure culture for further study.
Preliminary screening
All the endophytic isolates were subjected to preliminary screening against C. gloeosporioides to observe the presence of antagonistic properties. Out of 34 isolates, four bacterial endophytic isolates, viz. PBEDL1, PJE5B, PGE4B and PM5A, were found to be the most promising which were then subjected to further study.
In vitro efficacy of selected endophytic isolates against mycelial growth of C. gloeosporioides
The four most promising bacterial endophytic isolates were subjected to in vitro screening to observe their potential to suppress the mycelial growth of C. gloeosporioides. The bacterial endophytes significantly differ in their potential to inhibit the radial growth of pathogenic fungi. The isolate PBEDL1 showed maximum inhibition (68.67%) on mycelial growth of C. gloeosporioides over control followed by PJE5B (65.33%), PGE4B (52.89%) and PM5A (45.33%) (Fig. 6).
Mutual compatibility
From the various combinations of selected bacterial endophytic isolates, three combinations, viz. PBEDL1+PGE4B, PBEDL1+PM5A and PGE4B+PM5A, showed, viz. PBEDL1+PJE5B, PGE4B compatible reaction, while other three combinations +PJE5B and PM5A+PJE5B showed incompatible reaction.
In vitro efficacy of mutually compatible isolates
Out of the three compatible reactions, the highest inhibition (56.00%) of mycelial growth of C. gloeosporioides over control was recorded from the combination PBEDL1+PGE4B, followed by PBEDL1+PM5A (48.22%) and PGE4B+PM5A (44.67%) (Fig. 6). There was non-synergistic effect recorded from the combinations (Table 2).
Pathogenicity test of promising endophytes
As the bacterial endophytes were isolated from different chilli cultivars apart from Bhut Jolokia, the pathogenicity test of the endophytic isolates was tested to check whether the isolates possess any pathogenic ability towards Bhut Jolokia. From the observations recorded, there was no development of lesions, necrotic spots, hallowing or any kind of hypersensitive reaction in the inoculated plants along with the control. The lack of symptoms in the inoculated plants showed the non-pathogenic nature of the isolated bacterial endophytes.
Morphological and cultural characterization of selected bacterial endophytes
The morphological and cultural characteristics of the bacterial endophytes were recorded by observing various colony characteristics which are detailed in the table below (Table 3).
Molecular characterization of selected bacterial endophytic isolates
For the identification of bacterial endophytes, molecular characterization was studied. The concentration of the extracted genomic DNA was found to be 230.65 (PBEDL1), 186.19 (PJE5B), 331.02 (PGE4B) and 230.12 ng/μl (PM5A) and OD value (260/280) was 1.79 (PBEDL1), 1.86 (PJE5B), 1.92 (PGE4B) and 1.84 (PM5A). The PCR and gel electrophoresis using U16SF and U16SR primers results showed the band length of all the isolates were nearly 1450 bp approximately (Fig. 7). The sequence recovery after partial sequence of selected bacterial isolates was PBEDL1 (1416 bp), PJE5B (1,304 bp), PGE4B (1,437 bp) and PM5A (1,174 bp). On comparing the sequence in the public domain of NCBI using nBLAST showed maximum homology of PBEDL1 isolate with Bacillus velezensis, China isolate, accession number MT626060.1 (99.65%) followed by isolate PJE5B with Bacillus mycoides (96.98%), Himachal isolate, accession number KF475859.1, isolate PGE4B with Bacillus altitudinis (96.11%), China isolate, accession number MN710447.1 and isolate PM5A with Bacillus cereus (95.52%), Karnataka isolate, accession number MW227464.1 (Figs. 8, 9, 10 and 11).
Phylogenetic analysis
The sequence obtained was aligned in CLUSTALW using MEGA X software for phylogenetic analysis. The phylogenetic tree was prepared using neighbourhood joining using 1000 bootstrap replications. The phylogenetic tree of isolate PBEDL1 showed maximum similarity with B. velezensis Portugal isolate, accession number MZ234619.1, isolate PJE5B with B. mycoides Himachal isolate, accession number KF475859.1, isolate PGE4B with the B. altitudinis China isolate, accession number MN710447.1 and isolate PM5A with the B. cereus Karnataka isolate, accession number MW227464.1 (Figs. 8, 9, 10 and 11).
On the basis of morphological, cultural, biochemical and molecular characteristics, the four bacterial endophytic, viz. PBEDL1, PJE5B, PGE4B and PM5A, were identified as Bacillus velezensis, B. mycoides, B. altitudinis and B. cereus, respectively, which were found effective in suppressing the mycelial growth against fruit rot pathogen of Bhut Jolokia C. gloeosporioides. The present investigation showed the in vitro efficacy of endophytes where B. velezensis showed the highest inhibition (68.67%) followed by B. mycoides (65.33%), B. altitudinis (52.89%) and B. cereus (45.33%). From the compatible reaction, the combination of (B. velezensis+B. altitudinis) showed the highest efficacy (56.00%), followed by B. velezensis+B. cereus (48.22%) and then (B. cereus+B. altitudinis) (44.67%).
All the sequences of isolates were deposited in GenBank for accession number. The obtained accession numbers are presented in Table 4.
Discussion
In the present investigation, the causal agent of the fruit rot disease of Bhut Jolokia was identified as Colletotrichum gloeosporioides on the basis of morphological, cultural and molecular studies. This was in conformity with the findings of many researchers (Dutta 2020) who confirmed C. gloeosporioides as the most common and dominant one causing fruit rot disease of Bhut Jolokia in Northeast India. The cultural characteristics are in conformity with the reports of Dev et al. (2017) who described the colony characteristics as flat mycelium with regular margin and zonation. Kimaru et al. (2018) reported that the colony colour of the fungus varied from white to dark grey and the growth of the colony varied from flat, fluffy, raised and sparse on cultured media. Ganagadevi and Muthumary (2008) reported that the conidia are hyaline one-celled, straight, cylindrical, obtuse at ends and measuring 9 to 24 × 3 to 4.5 μm. Kimaru et al. (2018) also reported similar results where they found that the conidia of the fungus were cylindrical and hyaline measuring about 10.3 to 18.2 × 3.0 to 5.8 μm. Both the results were in tune with the present investigation.
The present investigation showed the non-pathogenic nature of the isolated bacterial endophytes. The lack of hypersensitive reaction in plants inoculated with endophytes was also reported by Larran et al. (2016) on 30-day-old wheat plants inoculated with 10 endophytic microbes. Similarly, Bacilio-Jiménez et al. (2001) also reported that two endophytic isolates Corynebacterium flavescens and Bacillus pumilus from rice rhizoplane showed a lack of hypersensitivity in the tobacco plant.
From the present study, the four most promising bacterial endophytes that can suppress the mycelial growth of C. gloeosporioides all belong to the genus Bacillus and are identified as B. velezensis, B. mycoides, B. altitudinis and B. cereus, respectively, on the basis of morphological, cultural, biochemical and molecular studies. The antimicrobial properties of the genus Bacillus were reported by many researchers. Chitarra et al. (2003) reported the presence of a number of antifungal compounds produced by Bacillus spp. which disrupt the fungal membrane-like release of chitin-binding proteins and Chitinase affecting the cell polarity.
Bacillus velezensis reported producing a wide range of antimicrobial compounds as secondary metabolites in the form of Surfactin, Plantazolicin, Bacilysin, and Iturin which are effective against many microorganisms (Kim et al. 2021).
The efficacy of B. altitudinis as a biocontrol agent has been established against various diseases including root rot disease of Vigna radiate (Sunar et al. 2015), Phytophthora rot of soybean (Lu et al. 2017) and potato common scab (Li et al. 2019).
Chauhan et al. (2016) studied the antifungal activities of Bacillus cereus and found two lipopeptides, surfactin and fengycin which are effective against many plant pathogens. Huang et al. (2005) also reported the presence of two chitinases (ChiCW and ChiCH) showing inhibitory activity of conidial germination of major fungal pathogen causing lily leaf blight, i.e. Botrytis elliptica.
Conclusion
The present investigation concluded that the potential of four bacterial endophytes, viz. Bacillus velezensis, B. mycoides, B. altitudinis and B. cereus, respectively, suppressed the fruit rot pathogen of Bhut Jolokia. The result of the present investigation explores the possibility of using endophytes as an eco-friendly and alternative solution against synthetic chemicals. Considering the performance of endophytes in field conditions, the bacterial endophytes B. velezensis and B. mycoides can be used as potential biocontrol agent in managing the fruit rot disease of Bhut Jolokia.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author upon reasonable request. All data generated or analysed during this study are included in this published article. The sequencing data generated and analysed in this study are available in the NCBI Sequence Read Archive database (https://www.ncbi.nlm.nih.gov/), Accession Number: OM202512, OM202620, OM203143, OM203114 and OM203119.
Abbreviations
- % :
-
Per cent
- °C :
-
Degree Celsius
- Approx.:
-
Approximately
- BLAST:
-
Basic Local Alignment Search Tool
- bp:
-
Base pair
- CD :
-
Critical difference
- CFU:
-
Colony forming unit
- CRD:
-
Completely randomized design
- CTAB:
-
Cetyl trimethyl ammonium bromide
- DMRT:
-
Duncan's multiple range test
- DNA:
-
Deoxyribonucleic acid
- E:
-
East
- EDTA:
-
Ethylene diamine tetraacetic acid
- et al. :
-
Et alia
- Fig.:
-
Figure
- g:
-
Gram
- h:
-
Hours
- HEPA:
-
High efficiency particulate air
- i.e.:
-
Id est (That is)
- IAA:
-
Indole acetic acid
- ITS:
-
Internal transcribed spacer
- kb:
-
Kilo base pair
- l:
-
Litre
- LC-MS:
-
Liquid chromatography–mass spectrometry
- M:
-
Molar
- min:
-
Minute
- ml:
-
Millilitre
- N:
-
North
- NaCl:
-
Sodium chloride
- NaOCl:
-
Sodium hypochlorite
- NCBI:
-
National Centre for Biotechnology Information
- ng/μl:
-
Nanogram per microliter
- nm:
-
Nanometre
- Nos.:
-
Numbers
- OD:
-
Optical density
- PBEDL1:
-
Bacillus velezensis
- PCR:
-
Polymerase chain reaction
- PDA:
-
Potato dextrose agar
- PFC Jorhat:
-
Colletotrichum gloeosporioides
- PGE4B:
-
Bacillus altitudinis
- pH:
-
Hydrogen ion concentration
- PJE5B:
-
Bacillus mycoides
- PM5A:
-
Bacillus cereus
- RH:
-
Relative humidity
- rpm:
-
Revolutions per minute
- S.Ed:
-
Standard error of deviation
- SDS:
-
Sodium dodecyl sulphate
- sp.:
-
Species
- T.E buffer:
-
Tris EDTA buffer
- TBE:
-
Tris Borate EDTA
- Tris-Cl:
-
Tris Hydrochloride
- UV:
-
Ultraviolet
- var.:
-
Variety
- viz.:
-
Videlicet (Namely)
- w/v:
-
Weight by volume
References
Aamir S, Sutar S, Singh SK, Baghela A (2015) A rapid and efficient method of fungal genomic DNA extraction, suitable for PCR based molecular methods. Plant Pathol Quar 5(2):74–81
Bacilio-Jiménez M, Aguilar-Flore S, del Valle MV, Pérez A, Zepeda A, Zenteno E (2001) Endophytic bacteria in rice seeds inhibit early colonization of roots by Azospirillum brasilense. Soil Biol Biochem 33(2):167–172
Bosland WP, Baral BJ (2007) Bhut Jolokia: the world’s hottest known chile pepper is a putative naturally occurring interspecific hybrid. HortScience 42(2):222–224
Cardinal MJ, Meghrous J, Lacroix C, Simard RE (1997) Isolation of Lactococcus lactis strains producing inhibitory activity against Listeria. Food Biotechnol 11(2):129–146
Caruso G, Abdelhamid MT, Kalisz A, Sekara A (2020) Linking endophytic fungi to medicinal plants therapeutic activity. Case Study Asteraceae Agric 10(7):286
Chauhan AK, Maheshwari DK, Kim K, Bajpai VK (2016) Termitarium-inhabiting Bacillus endophyticus TSH42 and Bacillus cereus TSH77 colonizing Curcuma longa L.: isolation, characterization, and evaluation of their biocontrol and plant-growth-promoting activities. Can J Microbiol 62(10):880–892
Chitarra GS, Breeuwer P, Nout MJR, Van Aelst AC, Rombouts FM, Abee T (2003) An antifungal compound produced by Bacillus subtilis YM 10–20 inhibits germination of Penicillium roqueforti conidiospores. J Appl Microbiol 94(2):159–166
Deorani SC, Sharma GD (2007) Medicinal plants of Nagaland. Published by M/S Bishen Singh Mahendra Pal Singh, Dehra Dun
Dev D, Somasekhara YM, Prasannakumar MK (2017) Cultural characterization of Colletotrichum gloeosporioides causing anthracnose of pomegranate. Indian Phytopathol 70(4):493–495
Dutta S (2020). Management of fruit rot of Capsicum chinense Jacq. With fungal bio-formulations. M.Sc. (Agri) Thesis, Department of Plant Pathology, AAU, Jorhat, Assam
Gangadevi V, Muthumary J (2008) Isolation of Colletotrichum gloeosporioides, a novel endophytictaxol- producing fungus from the leaves of a medicinal plant. Justicia Gendarussa Mycologia Balcanica 5(1):1–4
Gomez KA, Gomez AA (1984) Statistical procedures for agricultural research. Willy, Singapore
Hardoim PR, van Overbeek LS, van Elsas JD (2008) Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol 16(10):463–471
Huang CJ, Wang TK, Chung SC, Chen CY (2005) Identification of an antifungal chitinase from a potential biocontrol agent, Bacillus cereus. BMB Rep 38(1):82–88
Kim YS, Lee Y, Cheon W, Park J, Kwon HT, Balaraju K, Kim J, Jeon Y (2019) Characterization of Bacillus velezensis AK-0 as a biocontrol agent against apple bitter rot caused by Colletotrichum gloeosporioides. Sci Rep 11(1):1–14
Kimaru SK, Monda E, Cheruiyot RC, Mbaka J, Alakonya A (2018) Morphological and molecular identification of the causal agent of anthracnose disease of avocado in Kenya. Int J Microbiol 4:1–10
Lakshmi S, Deepthi R, Kasim D, Suneetha P, Krishna MSR (2014) Anthracnose a prevalent disease in capsicum. Res J Pharm Biol Chem Sci 5(3):1583–1604
Larran S, Simon MR, Moreno MV, Siurana MS, Perelló A (2016) Endophytes from wheat as biocontrol agents against tan spot disease. Biol Control 92:17–23
Li B, Wang B, Pan P, Li P, Qi Z, Zhang Q, Shi C, Hao W, Zhou B, Lin R (2019) Bacillus altitudinis strain AMCC 101304: a novel potential biocontrol agent for potato common scab. Biocontrol Sci Tech 29(10):1009–1022
Lu X, Zhou D, Chen X, Zhang J, Huang H, Wei L (2017) Isolation and characterization of Bacillus altitudinis JSCX-1 as a new potential biocontrol agent against Phytophthora sojae in soybean [Glycine max (L.) Merr.]. Plant Soil 416(1):53–66
Manandhar JB, Hartman GL, Wang TC (1995) Anthracnose development on pepper fruits inoculated with Colletotrichum gloeosporioides. Plant Dis 79(4):380–383
McInroy JA, Kloepper JW (1995) Population dynamics of endophytic bacteria in field-grown sweet corn and cotton. Can J Microbiol 41(10):895–901
Munif A, Hallmann J, Sikora RA (2013) The influence of endophytic bacteria on Meloidogyne incognita infection and tomato plant growth. J Int Soc Southeast Asian Agric Sci 19(2):68–74
Nwachukwu EO, Umechuruba CI (2001) Antifungal activities of some leaf extracts on seed-borne fungi of African yam bean seeds, seed germination and seedling emergence. J Appl Sci Environ Manag 5(1):29–32
Pakdeevaraporn P, Wasee S, Taylor PWJ, Mongkolporn O (2005) Inheritance of resistance to anthracnose caused by Colletotrichum capsici in Capsicum. Plant Breeding 124(2):206–208
Rangaswami G, Mahadevan A (1998) Diseases of crop plants in India. PHI Learning Pvt Ltd, Delhi
Riddell RW (1950) Permanent stained mycological preparations obtained by slide culture. Mycologia 42(2):265–270
Sarwa KK, Kira J, Sahu J, Rudrapal M, Debnath M (2012) A short review on Capsicum chinense Jacq. J Herb Med Toxicol 6(2):7–10
Singh LP, Gill SS, Tuteja N (2011) Unraveling the role of fungal symbionts in plant abiotic stress tolerance. Plant Signal Behav 6(2):175–191
Singh SK, Singh PP, Gupta A, Singh AK, Keshri J (2019) Tolerance of heavy metal toxicity using PGPR strains of Pseudomonas species. In: PGPR amelioration in sustainable agriculture, pp. 239–252
Sunar K, Dey P, Chakraborty U, Chakraborty B (2015) Biocontrol efficacy and plant growth promoting activity of Bacillus altitudinis isolated from Darjeeling hills. India J Basic Microbiol 55(1):91–104
Tovar-Pedraza JM, Mora-Aguilera JA, Nava-Diaz C, Lima NB, Michereff SJ, Sandoval-Islas JS, Camara MPS, Teliz- Ortiz D, Leyva-Mir SG (2020) Distribution and pathogenicity of Colletotrichum species associated with mango anthracnose in Mexico. Plant Dis 104(1):137–146
Utkhede RS, Rahe JE (1983) Interactions of antagonist and pathogen in biological control of onion white rot. Phytopathology 73(6):890–893
White JF Jr, Reddy PV, Bacon CW (2000) Biotrophic endophytes of grasses: a systematic appraisal. Marcel Dekker, Microbial endophytes. New York, pp 49–62
Zegeye ED, Santhanam A, Gorfu D, Tessera M, Kassa B (2011) Biocontrol activity of Trichoderma viride and Pseudomonas fluorescens against Phytophthora infestans under greenhouse conditions. J Agric Technol 7(6):1589–602
Acknowledgements
Not applicable.
Funding
This study was done as a Post-graduate degree research work under the Department of Plant Pathology at Assam Agricultural University supported by the Directorate of Post Graduate Studies, Assam Agricultural University.
Author information
Authors and Affiliations
Contributions
PPS conceived and designed the study, drafted the manuscript, and analyzed and interpreted the data under the guidance of HKD; PPS and HKD acquired the data; and HKD and TN revised the manuscript critically for important intellectual content. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All the authors have given their consent to publish the submitted manuscript as an ‘Original paper’ in EJBPC.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sarmah, P.P., Nath, H.K.D. & Nath, T. Endophytic Bacillus spp. from native chilli cultivars and their effect against fruit rot pathogen of Bhut Jolokia (Capsicum chinense Jacq.). Egypt J Biol Pest Control 33, 119 (2023). https://doi.org/10.1186/s41938-023-00763-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41938-023-00763-3