Abstract
Background
The gram pod borer (Helicoverpa armigera Hubner), Bihar hairy caterpillar (Spilosoma obliqua Walker), tobacco caterpillar (Spodoptera litura Fabricius) and root knot nematode (Meloidogyne incognita (Kofoid and White) Chitwood) are the major insect pest and nematodes infesting mungbean, urdbean and other legumes. Bacillus thuringiensis (Bt) is a promising biocontrol agent that helps in managing the above pest. Deploying Bt biopesticides instead of chemical pesticides in legume cultivation enhances the soil health substantially. The present study envisages morphological, molecular cum biochemical characterization, entomotoxicity and nematotoxicity potential of four indigenous Bt strains (Ak2.IIPR, F8.IIPR, F5.IIPR and F6.IIPR) along with HD1 (standard check from BGSC).
Results
The SEM micrograph analysis of above four Bt strains along with F1.IIPR revealed the presence of different combinations of insecticidal crystal proteins, viz. cuboidal crystal (CC), bipyramidal crystal (BC), spherical crystal (SC) and irregularly pointed crystal (IC). This was further confirmed by PCR amplification of Cry1, Cry2Aa, Cry2Ac, VIP3A and chi36 gene-specific primers and their phylogenetic analysis. F8.IIPR and Ak2.IIPR recorded the highest toxicity index against second-instar S. obliqua larvae (292.56 and 174.55), H. armigera (150.00 and 113.65) and S. litura (210.71 and 114.19) based on probit analysis. F5.IIPR (91.67%) and F8.IIPR (89.00%) recorded the highest juvenile mortality against M. incognita, followed by Ak2.IIPR (70.33%). The protein profiling of supernatant of terrific broth inoculated with F8.IIPR indicated the presence of VIP3A and Cry1I.
Conclusion
The present study concluded that F8.IIPR and Ak2.IIPR were potential Bt strains isolated from Indo-Gangetic plains which is having both entomocidal and nematicidal properties. Further, these strains were deposited at national repository, ICAR-NBAIM with the following accession no.: NAIMCC-SB-065 and NAIMCC-SB-066, respectively. These strains can be formulated as microbial biopesticide.
Similar content being viewed by others

Background
Mungbean (Vigna radiata (L.) R. Wilczekvar. radiata) is an important legume crop cultivated in South East Asia predominantly. The global area is 7.3 million hectares (Mha) with an average yield of 721 kg/ha (Nair and Schreinemachers 2020). In India, it is cultivated in 4.52Mha with a production of 2.51 million tons during 2019–2020 and an average yield was 544.25 kg/ha (Anonymous 2022). There is an ample scope to increase mungbean productivity by reducing the losses inflicted by key insect pest and nematodes. It is affected by an array of insect pest and nematodes. Polyphagous lepidopteran insects such as Helicoverpa armigera Hubner (Lepidoptera: Noctuidae), Spodoptera litura Fabricius (Lepidoptera: Noctuidae) and Spilosoma obliqua Walker (Lepidoptera: Erebidae) were infesting severely in legume crops in India, Philippines, Cambodia, Lao PDR and Indonesia (Sequeros et al. 2021).
Root knot nematodes (RKN), Meloidogyne spp. are capable of damaging a broad range of crops and causing serious yield losses. It causes heavy yield loss amounting to 18–90% in mungbean under congenial environmental conditions (Singh et al. 2020). This genus has around 98 species among which Meloidogyne incognita (Tylenchida: Heteroderidae) is very common and it causes root galls in almost all vascular plants (Jones et al. 2013). This is very difficult to control due to their quick generation time, high reproduction rate, sedimentary and endophytic nature (Engelbrecht et al. 2018). So, the main issue in the present scenario is to control these biotic factors so that crop yield losses can be prevented. Chemical insecticides and nematicides are generally used for managing this pest and RKN, but due to the negative impact of these pesticides on environmental and non-targets, the need for efficient biocontrol agents is increasing (Masson et al. 1998).
Bacillus thuringiensis Berliner (Bt) is an eco-friendly, target-specific microbial biological control agent. It helps in sustainable management of insects and nematodes. In India, only four Bt kurstaki strains (HD1 serotype 3a3b, Z-52 3a3b, A-97 serotype H-3a and DOR Bt-1 serotype-3a3b3c) were registered for commercial use with CIBRC (Central Insecticides Board and Registration committee). Out of four Bt strains, only DOR Bt-1 is indigenous Bt strain and most of the available Bt products (Delfin, Dipel, Thuricide, Halt, Biobit, Bactospeine, Agree, etc.) were produced from imported Bt strains (Ramanujam et al. 2014). Native strains were known for their higher toxicity and greater efficiency in managing the native insect pests because of coexistence and evolution (Hassan et al. 2021). The present experiments were carried out to study the entomotoxic and nematotoxic potential of indigenous Bt strains isolated from Indo-Gangetic plains against three polyphagous lepidopteran pests and root knot nematode, M. incognita infesting mungbean. Mungbean and other legumes were known for its role in enhancing the soil microbial diversity and thereby improving soil health by biological N fixation, P solubilization, organic carbon enhancement, etc.; when Bt biopesticides were deployed in place of synthetic chemical insecticides it will help conserving the microbial diversity in legume-based cropping systems (Ananda et al. 2022). Endospores and CRY proteins are the major active ingredients in any Bt biopesticides, and the cry protein alone accounts for more than 20–30% of total cell mass of Bt (Agaisse and Lereclus 1995). Endospore formation and cry protein accumulation occur during the lag phase of Bt growth. Classification of CRY proteins can be done on the basis of amino acid sequence and host specificity (Asokan et al. 2012). The only exception in cry protein is Cry1I because it is an excretory protein and it is active against both lepidopteran and coleopteran (Tailor et al. 1992). Hence, in the present study, five native Bt strains insecticidal genes were subjected to polymerase chain reaction (PCR) screening, scanning electron micrograph (SEM) analysis and bioassay against major pests for documenting its insecticidal genes diversity and its efficacy.
Methods
Morphological and biochemical characterization
The physio-morphological characteristics, viz. colony color, appearance, growth characters and spore morphology, were recorded 7 days after incubation at 30 °C, following standard identification protocols (Logan et al. 2009) for 12 native Bt strains maintained in Bio-ecology Lab, ICAR-Indian Institute of Pulses Research (ICAR-IIPR), Kanpur, India. Similarly, biochemical characterizations for 12 Bt isolates along with four standard Bt strains, viz. Bt subsp. kurstaki HD1, Bt subsp. aizawi HD133, Bt subsp. israelensis ONR60A and Bt subsp. morrisoni biovar tenebrionis (kindly supplied by Dr D. R. Zeigler from BGSC), was done with Hi-Bacillus kit (KB013) from M/s HiMedia Laboratories Pvt. Ltd., Mumbai, India, as per the instructions provided.
Hemolytic assay
Himedia sheep blood agar base with fresh sheep erythrocytes (RBC) was used to carry out this assay as per the instructions provided. After incubation at 30 °C for 24 h, the Petri plates were examined for the presence or absence of hemolytic zone surrounding the bacterial colonies (Ichikawa et al. 2008).
Spore crystal mixture (SCM) extraction and Bt endospore enumeration
A loop full of pure culture of four potential Bt strains (Ak2.IIPR, F8.IIPR, F5.IIPR and F6.IIPR and HD1) based on preliminary study (Sujayanand et al. 2021) were inoculated in a 250-ml conical flask containing 100 ml of nutrient broth and incubated in a shaker for 4 days at 30 °C. The spore crystal mixture (SCM) was isolated as described by Zothansanga et al. (2016). The SCM was serially diluted up to 10−9 cfu/ml, and its colony-forming units (CFU) were estimated by spread plate technique by replicating each dilution thrice and the plates were incubated at 30 °C overnight.
Scanning electron micrograph (SEM) analysis of SCM
The spore crystal mixture of Bt strains, viz. Ak2.IIPR, F8.IIPR, F5.IIPR, F6.IIPR and F1.IIPR, were diluted from 10−4 to 10−6 cfu/ml according to the endospore and crystal concentration in the respective strain. Then, the respective strains were subjected to SEM analysis as described by El-kersh et al. (2012) with slight modifications given in the following. For SEM analysis, a small aliquot of diluted SCM was taken and smeared on a cover glass, followed by vacuum evaporation for 45 min. The cover glass was then placed on carbon stub and subjected to sputter coating with gold (Hitachi, MC 1000) for 5 min. The gold-coated sample was placed in chamber of SEM (Hitachi, S-3400N), and secondary electron/backscattered electron micrographs were recorded by passing 1500kv current. The SEM micrographs were taken at ICAR-National Bureau of Agriculturally Important Microorganism (ICAR-NBAIM), Mau, India.
Insect bioassay
The toxicity potential of four native Bt strains (identified through range finding test), viz. Ak2.IIPR, F8.IIPR, F5.IIPR and F6.IIPR, was compared along with the standard Bt strain, i.e., Bt subsp. kurstaki HD1, by artificial diet contamination method (Li and Bouwer 2012). All the bioassay experiments were laid out in completely randomized design (CRD). Three polyphagous lepidopteran pests of mungbean, viz. Bihar hairy caterpillar (BHC), S. obliqua, pod borer (PB), H. armigera, and tobacco caterpillar (TC), S. litura, were maintained in Bio-ecology Lab, ICAR-Indian Institute of Pulses Research, Kanpur. The second-instar larvae of the above-mentioned three insects were subjected to insect bioassay by artificial diet contamination method with different SCM concentrations (4 for BHC; 5 for PB; and 3 for TC) of Bt strains. The insect mortality was recorded up to 7 days after treatment, and percent mortality for each strain was estimated. The lethal concentration of each Bt strain SCM (in cfu/ml) against these insects was estimated using probit analysis in SAS 6.2 by using PROC GLM procedure. The potency of the strains was compared as per the following formula (Sun and Jhonson 1960):
Toxicity index (T.I) of Bt strain = [LC50 of standard strain/LC50 of test strain] × 100.
Chitinolytic activity and bioassay against root knot nematodes (RKN)
Colloidal chitin preparation
One hundred grams of chitin flakes (shrimp shell, Sigma) was added slowly to 1.1.8 l concentrated HCl in a glass jar, and it was mixed gently for 3 h on a magnetic stirrer. This solution was then filtered by using eight layers of cheese cloths to 5 l of pre-chilled, distilled water with constant mixing to allow colloidal chitin to precipitate. This was incubated in a refrigerator at 4 °C for overnight. A dense white precipitate was formed and centrifuged at 10,000 rpm for 10 min at 4 °C. The precipitate was then washed in cold, distilled water repeatedly until the pH of the wash reached 5.5. The supernatant was discarded, and the colloidal chitin was then kept in a refrigerator for future use (Hsu and Lockwood 1975).
Chitinolytic activity
Colloidal chitin-enriched broth was prepared by adding 6 g Na2HPO4, 3 g KH2PO4, 1 g NH4Cl, 0.5 g NaCl, 0.5 g yeast extract and 5 g colloidal chitin in 1 l of distilled water. Bt strains were inoculated in 50 ml of autoclaved chitin-enriched broth in a 100-ml Erlenmeyer flask and incubated at 26 °C for two days. The suspensions were centrifuged, and the supernatant was filtered through MILLEX GV bacterial filter 0.22 µm (Merck). Colloidal chitin suspensions containing 1% (weight per volume) purified colloidal chitin in 100 mM of acetate buffer at pH 5.0 were used as chitin substrate (Vessey and Pegg 1973). The chitinolytic activity of five Bt strains, viz. Ak2, F8, F5, F4 and F1, was evaluated by using bacterial cultural filtrates containing crude chitinase solution and colloidal chitin suspension as a substrate which was incubated at 37 °C for 12 h, and spectrophotometric OD value of the solution was recorded at 420 nm.
RKN bioassay
One milliliter of cultural filtrates of all the strains with three replications was taken in 12-well tissue culture plates and distilled water as the control. Hundred freshly hatched second-stage juveniles (J2) were inoculated in each treatment. After 48 h, the dead nematodes were counted and kept separately in water to confirm whether juveniles were dead or nematostatic. The percent mortality was calculated based on the following formula:
Percentage J2 mortality = (No. of dead J2 /Total No. of juveniles) × 100.
The single egg mass culture of root knot nematode maintained at Nematode Biocontrol Laboratory, ICAR-IIPR, Kanpur, India, was deployed for challenging the chitinase supernatant obtained from the above-mentioned strains against J2 stage of root knot nematodes (RKN), i.e., Meloidogyne sp. The mortality was recorded for up to 7 days after treatment, at a time interval of every 24 h.
DNA isolation, PCR amplification of gyrB, Cry, Vip3Aa and chi36 genes and its phylogeny
The DNA of 12 Bt strains was extracted using Nucleopore®g DNA fungal bacterial mini kit (NP-7006D). The DNA gyrase gene (gyr B) was amplified for the 12 Bt strains as reported by Subbanna et al. (2018), except a slight modification in the annealing temperature which was set up at 50 °C for the current experiment.
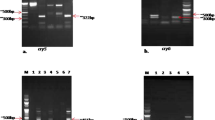
The cry gene content of six Bt strains (Ak2.IIPR, F8.IIPR, F5.IIPR, F6.IIPR, F4.IIPR and F1.IIPR) was identified by using oligonucleotide primers, viz. Gralcry1, Gralcry2, spe-cry2Aa, spe-cry2Ab and spe-cry2Ac, through PCR analysis as reported by Mendoza et al. (2012). The PCR conditions, followed for amplifying, were slightly modified from Vilas-Bos and Lemos (2004) for specific cry gene amplification with respect to annealing temperature between 50 and 53.7 °C according to the Bt strain requirement. The vip3 gene was amplified with primers and the PCR conditions as reported by Franco-Rivera et al. (2004) for five Bt strains (Ak2.IIPR, F8.IIPR, F5.IIPR, F6.IIPR and F1.IIPR).
The exochitinase (chi36) gene was amplified by using the primers reported by Arora et al. (2003). The PCR program for amplifying the exochitinase gene is given as follows: 94 °C for 5 min, 94 °C for 1 min, 50.2 °C for 1 min, 72 °C for 1 min and 30 s for 35 cycles and 72 °C for 10 min. The program will yield an amplicon with a size of 1083 bp. The PCR products were subjected to Sanger dideoxy sequencing by M/s Eurofins (P) ltd and M/s Xceleris (P) ltd. The above gene sequences were trimmed using BLAST analysis with the help of Bioedit and Finch TV 1.4.0 and then submitted to the NCBI database. The phylogenetic analysis of the four genes, viz., DNA gyrase, cry 1, cry2 and vip3A, was performed using MEGA7 by a neighbor-joining method.
VIP3 protein profiling
A 12-h-old nutrient broth culture of F8.IIPR was used as mother inoculum for vegetative insecticidal protein (VIP3) production in 100 ml of Terrific broth incubated for 19 h. The VIP3 protein present in the supernatant was precipitated, purified and dialyzed as described by Sattar et al. (2008). The concentrated protein was stored at − 20 °C until it was subjected to SDS–PAGE analysis.
Results
Morphological attributes and biochemical characteristics of 12 Bt strains
The growth of 12 Bt strains in nutrient agar (NA) was recorded 7 days after incubation at 25 °C in a BOD incubator. The highest width recorded was 1.06 cm (RoT61.IIPR), followed by 0.96 cm (Pa2.IIPR) and 0.9 cm (J3.IIPR, Ak2.IIPR) (Table 1). The lowest width (0.56 cm) was recorded from SgH5.IIPR, F4.IIPR and F6.IIPR. Nine strains showed irregular, continuous growth, whereas two strains recorded discontinuous growth (F4.IIPR and RoT61.IIPR) and one strain recorded regenerative growth. All the strains had exhibited off-white color. Almost majority of Bt strains were opaque with moist appearance, except Ak2.IIPR and F4.IIPR, wherein they have dry opaque appearance. Four Bt strains had shown lobate margin, while four Bt strains had undulated margins. All 12 Bt strains were gram positive, were sporulating and formed crystal protein (Table 1). The biochemical reactions performed with Hi-Bacillus kit showed that all the 16 (12 native Bt + 4 standard Bt) were catalase positive (Table 2), while for malonate test, all the Bt strains were negative, except J3.IIPR and F4.IIPR, and for Voges–Proskauer’s test, all strains were negative. Regarding utilization of citrate as the sole carbon source, all the isolated strains were negative, except J3.IIPR, F4.IIPR and F8.IIPR. For ONPG test, only F8 was positive and the remaining strains showed a negative reaction. Nitrate reduction test was found to be positive for all the strains, except F8.IIPR and J3.IIPR. Three Bt strains, viz. Ak2.IIPR, SgH5.IIPR and F6.IIPR, were negative for arginine production. For sugar reduction test, all the strains were negative for mannitol and arabinose, except F8.IIPR, which showed a positive reaction, whereas for glucose all the strains were positive, except F1.IIPR, F4.IIPR and HD1.
Hemolytic assay
No hemolytic zone was recorded from 12 Bt strains in sheep blood agar. Hence, these are concluded as non-hemolytic Bt strains.
SCM extraction and endospore enumeration
The spore crystal mixtures of four Bt strains (Ak2, F8, F5 and F6) and standard strain (HD1) were extracted and stored at − 20 °C for assessing their insecticidal potential and characterizing their crystal protein shape and endospore morphometrics. The endospore enumeration of each strain in different dilutions showed variation among the strains. All the tested four Bt strains recorded a higher number of endospores (at a dilution of 10−7 and 10−9 cfu/ml) than the standard strain. The highest number of endospores (1650 cfu/ml) at 10−9 was recorded from F8.IIPR, followed by Ak2.IIPR, F6.IIPR and F5.IIPR, whereas the lowest endospore count (495 cfu/ml) was recorded from standard strain, HD1 (Fig. 1).
SEM analysis of endospore and crystal protein
The electron micrographs of the five Bt strains revealed the presence of different crystal morphologies in single strain such as presence of cuboidal crystal (CC), bipyramidal crystal (BC), spherical crystal (SC) and irregularly pointed crystal (IC) in the case of F8.IIPR (Fig. 2b). Whereas Ak2.IIPR (Fig. 2a), F6.IIPR (Fig. 2d) and F1.IIPR (Fig. 2e) had three crystal morphologies, viz. BC, CC and SC, F5.IIPR (Fig. 2c) had only two crystal morphologies, viz. CC and BC. The endospore length varied from 1.63 to 2.36 μm; similarly, breadth varied from 0.79 to 1.18 μm. There existed a significant variation among the Bt strains with respect to endospore length and breadth (Fig. 2f). Ak2.IIPR had the largest endospore, while F6.IIPR had the smallest endospore with respect to breadth of endospore.
Insect bioassay
The second-instar Bihar hairy caterpillar, S. obliqua was challenged with SCM of four native Bt strains and Btk HD1. The lowest LC50 was recorded from F8.IIPR (1.99 × 103 cfu/ml), followed by Ak2.IIPR (3.34 × 103 cfu/ml) and F5.IIPR (5.18 × 103 cfu/ml) (Table 3). The HD1 recorded a LC50 of 5.18 × 103 cfu/ml against S. obliqua. The toxicity index was the highest (292.96) for F8.IIPR, followed by Ak2.IIPR (174.55) and F5.IIPR (112.55). Among the four Bt strains screened against the second-instar H. armigera larvae, the lowest LC50 was recorded from F8.IIPR (2.22 × 103 cfu/ml), followed by Ak2.IIPR (2.93 × 103 cfu/ml), while two strains, viz. F6.IIPR (5.06 × 103 cfu/ml) and F5.IIPR (6.82 × 103 cfu/ml), had recorded LC50 value higher than the standard check, Btk HD1 (3.33 × 103 cfu/ml). This showed that two Bt strains were superior to the Btk HD1 in killing H. armigera. Further, the toxicity index also indicated that F8.IIPR is highly toxic (150.00), followed by Ak2.IIPR (113.65), F6.IIPR (65.81) and F5.IIPR (48.83).
Based on the preliminary screening of Bt SCM against S. litura, only two Bt strains, viz. F8.IIPR and Ak2.IIPR, had promising results (data not shown). Among the two Bt strains screened against the second-instar S. litura, F8.IIPR had recorded the lowest LC50 (2.52X103cfu/ml), followed by Ak2.IIPR (4.65X103cfu/ml). The standard check (HD1) had recorded a LC50 of 5.31 × 103 cfu/ml. F8.IIPR recorded the highest toxicity index (210.71) against S. litura, followed by Ak2.IIPR (114.19).
Chitinolytic activity and bioassay against root knot nematodes
The five Bt strains ability to hydrolyze the colloidal chitin by producing a clear zone was recorded on colloidal chitin agar media (Sujayanand et al. 2019). Chitinase activity of Bt strains was measured in the presence of 0.2% colloidal chitin and measured by the Schales method. Chitinase activity was measured in U/mg. The maximum chitinase production was observed for F8.IIPR, followed by Ak2.IIPR. In chitinolytic assay, and the highest activity was recorded from F8.IIPR (0.525), followed by F4.IIPR (0.522), F5.IIPR (0.488), F1.IIPR and Ak2.IIPR. The percent mortality of root knot nematode juveniles revealed that F5.IIPR recorded the highest mortality, followed by F8.IIPR and Ak2.IIPR (Table 4).
DNA gyrase, Cry, Vip3Aa and chi36 genes phylogeny
The genomic DNA of 12 Bt strains was extracted and stored at − 20 °C. The DNA gyrase gene amplification yielded an amplicon of 342 bp for the 12 Bt strains. Further phylogenetic analysis indicated that F8.IIPR, Ak2.IIPR, F1.IIPR, F5.IIPR and F4.IIPR were more close to gyrase gene of Bt kurstaki BGSC 4D20 strain (Fig. 3). These strains produced bipyramidal protein. Whereas three Bt strains (B1.IIPR, RoT61.IIPR and SgH5.IIPR) showed similarity to Bt canadensis, two showed similarity to Bt pondicheriensis, one Bt strain (F6.IIPR) was more similar to B. tyoso strain, while J3.IIPR showed similarity to Bt entomocidus.
Phylogenetic analysis for DNA gyrase gene of 12 Bacillus thuringiensis strains. Evolutionary relationships of taxa. The evolutionary history was inferred using the neighbor-joining method [1]. The optimal tree with the sum of branch length = 0.68590967 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches [2]. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Kimura two-parameter method [3] and are in the units of the number of base substitutions per site. The analysis involved 41 nucleotide sequences. Codon positions included were 1st + 2nd + 3rd + noncoding. All positions containing gaps and missing data were eliminated. There were a total of 119 positions in the final dataset. Evolutionary analyses were conducted in MEGA7 [4]
The cry1 subtype for the six Bt strains [F8.IIPR (MK855068), Ak2.IIPR (MK843970), F6.IIPR (MK843971), F1.IIPR (MK855069), F5.IIPR (OM141596) and F4.IIPR (OM141595)] was screened by sequencing an amplicon size of 990 bp further phylogenetic analysis and also concluded as cry1A (Fig. 4a). Of the six different Bt strains screened for cry2 subtype having 1249 bp from gral cry2 primers, the sequence results yielded mixed reaction. Hence, we had gone for the gene-specific primers, viz., cry2Aa (460 bp), cry2Ab (771 bp) and Cry2Ac (841 bp). Five Bt strains, viz. F8.IIPR (MN548386 and MN548387), Ak2.IIPR (MT210572 and MT210570), F6.IIPR (MT210573 and MT210571), F1.IIPR (MT210575 and MT210576) and F5.IIPR (MT210574 and OM141597), had both cry2Aa (460 bp) and cry2Ac (841 bp), while F4.IIPR had only cry2Ac (MT210577) (Fig. 4b).
Phylogenetic analysis for cry1, cry2 and vip3 genes of potential Bacillus thuringiensis strains. A Evolutionary relationships of taxa. The evolutionary history was inferred using the neighbor-joining method [1]. The optimal tree with the sum of branch length = 0.52363330 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches [2]. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Tamura-Nei method [3] and are in the units of the number of base substitutions per site. The analysis involved 15 nucleotide sequences. Codon positions included were 1st + 2nd + 3rd + noncoding. All positions containing gaps and missing data were eliminated. There were a total of 665 positions in the final dataset. Evolutionary analyses were conducted in MEGA7 [4]. 1. Saitou N. and Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4:406–425. 2. Felsenstein J. (1985). Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39:783–791. 3. Tamura K. and Nei M. (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular Biology and Evolution 10:512–526. 4. Kumar S., Stecher G. and Tamura K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33:1870–1874. Disclaimer: Although utmost care has been taken to ensure the correctness of the caption, the caption text is provided "as is" without any warranty of any kind. Authors advise the user to carefully check the caption prior to its use for any purpose and report any errors or problems to the authors immediately (www.megasoftware.net). In no event shall the authors and their employers be liable for any damages, including but not limited to special, consequential or other damages. Authors specifically disclaim all other warranties expressed or implied, including but not limited to the determination of suitability of this caption text for a specific purpose, use or application. B Evolutionary relationships of taxa. The evolutionary history was inferred using the neighbor-joining method [1]. The optimal tree with the sum of branch length = 0.34599582 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches [2]. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Kimura two-parameter method [3] and are in the units of the number of base substitutions per site. The analysis involved 14 nucleotide sequences. Codon positions included were 1st + 2nd + 3rd + noncoding. All ambiguous positions were removed for each sequence pair. There were a total of 4582 positions in the final dataset. Evolutionary analyses were conducted in MEGA7 [4]. 1. Saitou N. and Nei M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4:406–425. 2. Felsenstein J. (1985). Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39:783–791. 3. Kimura M. (1980). A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution 16:111–120. 4. Kumar S., Stecher G. and Tamura K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33:1870–1874. Disclaimer: Although utmost care has been taken to ensure the correctness of the caption, the caption text is provided "as is" without any warranty of any kind. Authors advise the user to carefully check the caption prior to its use for any purpose and report any errors or problems to the authors immediately (www.megasoftware.net). In no event shall the authors and their employers be liable for any damages, including but not limited to special, consequential or other damages. Authors specifically disclaim all other warranties expressed or implied, including but not limited to the determination of suitability of this caption text for a specific purpose, use or application. C Evolutionary relationships of taxa. The evolutionary history was inferred using the neighbor-joining method [1]. The optimal tree with the sum of branch length = 0.35582526 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches [2]. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Kimura two-parameter method [3] and are in the units of the number of base substitutions per site. The analysis involved 14 nucleotide sequences. All positions containing gaps and missing data were eliminated. There was a total of 858 positions in the final dataset. Evolutionary analyses were conducted in MEGA7 [4]. 1. Saitou N. and Nei M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4:406–425. 2. Felsenstein J. (1985). Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39:783–791. 3. Kimura M. (1980). A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution 16:111–120. 4. Kumar S., Stecher G. and Tamura K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33:1870–1874. Disclaimer: Although utmost care has been taken to ensure the correctness of the caption, the caption text is provided "as is" without any warranty of any kind. Authors advise the user to carefully check the caption prior to its use for any purpose and report any errors or problems to the authors immediately (www.megasoftware.net). In no event shall the authors and their employers be liable for any damages, including but not limited to special, consequential or other damages. Authors specifically disclaim all other warranties expressed or implied, including but not limited to the determination of suitability of this caption text for a specific purpose, use or application
The Vip 3A gene was found to be present in all five Bt strains, viz. Ak2.IIPR (MF143589), F8.IIPR (MF143591), F6.IIPR (MF143590), F1.IIPR (MK124722) and F5.IIPR (MK214423), based on phylogenetic analysis (Fig. 4c).
The partial exochitinase gene chi36 was amplified in F8.IIPR (MN991173), Ak2.IIPR (MN985815), F1.IIPR (MN879415) and F5.IIPR (MN985816) to give a PCR product of 1083 bp when annealed at 50.2 °C.
VIP3 protein profiling
The VIP3 protein from terrific broth was extracted, purified and dialyzed. The SDS–PAGE analysis of the supernatant from F8.IIPR strain reveals the presence of protein with 88 kDa (Fig. 5).
Discussion
The genus Bacillus can be divided into five groups based on the phylogenetic analysis of 16S rRNA, viz. Bacillus cereus, B. megaterium, B. subtilis, B. circulans and B. brevis. The Bt falls in B. cereus group. The B. cereus group has typical colony morphology such as off-white color of colonies, rough waxy growth and irregular spreading margins on nutrient agar as reported by Venkatramanamma et al. (2022). All 12 indigenous Bt strains exhibited the above-said colony morphological characteristics. The present findings were in congruence with those of Hassan et al. (2021), wherein they had reported majority of Bt isolates were off white with wavy margins, dry or moist appearance and flat colony morphology. Apart from recording the colony morphology, the variation in speed of growth in nutrient agar for the 12 Bt strains was also recorded by incubating at 25 °C for 7 days. The colony width ranged from 0.56 to 1.06 cm. The present results contradict the findings of Bautista and Teves (2013) who reported a colony growth of < 1 to 4 mm. The reason for lesser growth was that they had incubated for shorter period (24 h) only at 30 °C but the incubation time was high (7 Days) in the present study. Hi-Bacillus™ Identifications kit (KB013; HiMedia, India) was deployed by Rajasekhar et al. (2017) for biochemical characterization of Bt strains from India and Sudan. The present finding on Voges–Proskauer’s and ONPG test was in congruence with the above report. Thus, all these biochemical tests showed existence of variation among Bt strains as reported by Martin et al. (2010)
Generally, the dipteran toxic strains, i.e., Bt israelensis, have more probability of possessing hemolytic type. Further, they predominantly have β-exotoxin which is responsible for hemolytic activity (Obeidat et al. 2012). The present result is in accordance with those of El-kersh et al. (2012). They also reported the occurrence of non-hemolytic strains (8%) from Saudi Arabia. Further, they had reported only 31% strains were following Bt kurstaki biochemical type, which may be the reason for low occurrence of non-hemolytic strain.
The Bt kurstaki strain, BA 83B from IMTECH, Chandigarh, produced bigger endospores (length 2.1 ± 0.16 µm and diameter 1.63 ± 0.16 µm) in soybean-supplemented medium than in control (length 1.1 ± 0.13 µm and diameter 0.63 ± 0.07 µm). Thus, the endospore size varies according to the nutrition available (Jisha and Benjamin 2014). Moreover, Elsayed et al. (2014) reported that nutrient requirement varied according to the type of Bt strains. The present results are also in accordance with the findings of Elsayed et al. (2014) as there is a variation in endospores (1.63 to 2.36 µm length). Further, the variation in endospore production was also due to genetic variations in the Bt strains growth potential as reported by Argolo Filho et al. (2011) who had divided the Bt strains into three groups based on their growth rate as fast growers (attaining log phase ~1 h after inoculation), medium growers (attaining log phase ~7 h) and slow growers (attaining log phase 10–11 h) which was measured through spectrophotometrically based OD at 590 nm.
The present result supports the findings of Hassan et al. (2021) wherein they had visualized four different parasporal protein shapes in their Bt strains. Further, variation in the crystal protein combination and endospore morphometrics indicates the occurrence of diversity among the native Bt strains isolated from Indo-Gangetic plains.
The present findings were in congruence with the reports of Roy et al. (2021) wherein they had reported a LC50 value of 2.49 X 107 cfu/ml against Culex tritaeniorhynchus Giles (Diptera: Culicidae). The present findings, i.e., two Bt strains (F8.IIPR, Ak2.IIPR) were having higher toxicity index than HD1 against H. armigera, were supported by the reports of Lone et al. (2017) wherein they had reported a native Bt strain (JK12) from Himalayas with a higher toxicity index than HD1 against H. armigera. Similarly, in Sri Lanka, Baragamaarachchi et al. (2019) reported that native Bt strain (AB1) was highly toxic to Dipel® resistant Diamond back moth, Plutella xylostella Linnaeus (Lepidoptera: Plutellidae). Thus, native strains were more toxic to locally occurring insect pest.
The present result corroborates with findings of Lee et al. (2015) wherein they also found LC50 of Bt strains varied from 1.9 X 103 to 9.8 X 105 cfu/ml. The present results contradict findings of Murali Krishna et al. (2018) wherein they had reported that HD1 (9.59 X 104 cfu/ml) is the best strain when compared with LC50 of 21 native Bt isolates (9.76 X 104 to 5.67 X 106 cfu/ml) against third-instar S. litura. The present result supports the findings of Mohan et al. (2014) wherein they had reported S. litura as highly tolerant among the three noctuids (H. armigera, S. litura and Sesamia inferens Walker (Lepidoptera: Noctuidae)) screened against 10 Bt strains. In the present study also, S. litura has exhibited the highest tolerance [highest LC50 (F8.IIPR: 2.52 X 103 cfu/ml) than H. armigera (2.22 X 103 cfu/ml) and S. obliqua (1.99 X 103 cfu/ml)] among three polyphagous insects studied (Table 3). The probable reason for high tolerance of S. litura might be over-digestion of cry protoxin by midgut protease, which was shown in Spodoptera littoralis (Boisduval) (Lepidoptera: Noctuidae) by Ogiwara et al. (1992). Similarly, Hernandez (1988) also had reported that among 13 Bt serovars tested against neonates of Spodoptera frugiperda J.E. Smith (Lepidoptera: Noctuidae), only four serovars, viz. Bt kenyae, Bt tolworthi, Bt aizawai and Bt kurstaki, had recorded 80 to 100% mortality by diet contamination method. The variation in toxicity level among the strains for three different polyphagous lepidopteran pest stated above is due to functional and toxicological diversity of cry genes they harbor and differences in the copy numbers of cry genes present, expression pattern and expression levels (Sujayanand and Pandey 2020). Further, the difference in the receptors present in these three insects’ midgut also contributes to the variation in their toxicity. Another important factor that contributed to variation in toxicity among the strains was their spore and toxin concentration. Mohan et al. (2014) had already confirmed that Bt kurstaki endospores act synergistically with the Cry 2A against Bt-resistant population of Plutella xylostella.
Researchers all over the world are trying to find biological control measures for M. incognita management as chemical nematicides are harming the agro-ecosystem by their toxic residues. There are many bacteria with nematicidal activity such as Pseudomonas fluorescens (Flugge 1886) Migula, P. aeruginosa Migula, Bacillus thuringiensis and B. firmus Bredemann and Werner. P. fluorescens kills soybean cyst nematode by producing 2,4-diacetylphloroglucinolc (Siddiqui and Shaukat 2003). Corynebacterium paurometabolu inhibited nematode egg hatching by producing hydrogen sulfide (Meena and Pimentel 2002). In the present study, F8.IIPR and F5.IIPR exhibited the highest nematicidal activity. Mohammad et al. (2008) studied the biocontrol efficacy of Bt against M. incognita and observed a reduction in egg masses and egg number as compared to the control. Bt DB27 was found to produce two novel proteins, namely Cry21Fa1 and Cry21Hai, which were found to be effective against Caenorhabditis elegans Maupas (Iatsenko et al. 2014).
The present result supports the findings of Lee et al. (2015) wherein bipyramidal protein-producing isolates CAB565 and 566 were similar to Bt kurstaki KB098 strain and Bt kurstaki KB099 strain.
DNA gyrase is more reliable in finding the phylogeny of native Bt isolates based on their ecological and evolutionary relationships between Bt isolates (Shikov et al. 2021). The phylogenetic analysis of DNA gyrase confirmed that F8.IIPR, Ak2.IIPR, F1.IIPR, F5.IIPR and F4.IIPR were more close to gyrase gene of Bt kurstaki BGSC 4D20 strain and they form a single clade, while F6.IIPR and J3.IIPR were showing phylogeny with different Bt serovars. The present report confirms the report that cry1A and cry2A genes occurred in high frequency (90.61%) by various workers. The vip3A gene occurred in medium frequency (40.70%) native Bt strains of Thailand (Boonmee et al. 2019). The difference in the toxicities of Bt strains may be related to the difference in the composition of crystals protein. Chak et al. (1994) reported a new Bt strain with cry l (Ab), cry IC and cry ID genes, and its effectiveness was due to variation in genetic composition of cry genes. The combined chitinolytic activity and entomocidal activity of potential Bt strains in the present study imply their utility in deploying them in the integrated pest management as an effective biocontrol agent.
The molecular weight of protein bands that occur in the supernatant corresponds to that of VIP3A as reported by Argolo Filho et al. (2011) wherein they had reported that Bt supernatant possesses various secreted entomotoxins with an expected size of ∼88 kDa and ∼60 kDa corresponding to Vip3A and Cry 1I. Interestingly, the VIP3 protein profiling using supernatant from F8.IIPR also had confirmed the presence of Cry1I as reported by Argolo Filho et al. (2011). Thus, the present work identifies the potential native Bt strains (F8.IIPR & Ak2.IIPR) for managing three polyphagous lepidopteran pests and root knot nematode along with morphological, biochemical and molecular characterizations of pesticidal genes such as cry1, cry2Aa, cry2Ac, vip3A and chi36.
Conclusion
The present study concluded that two indigenous Bt strains, namely F8.IIPR and Ak2.IIPR, as a potent microbial biocontrol agent among the four indigenous Bt strains, were screened against three polyphagous insect pest, viz., S. obliqua, H. armigera and S. litura. The PCR screening and phylogenetic analysis of entomotoxic and nematotoxic genes (Cry1, Cry2Aa, Cry2Ac, VIP3A and chi36) revealed that the two potential Bt strains had multiple insecticidal cum nematicidal genes. Further, this was confirmed by the presence of bipyramidal, cuboidal and spherical crystal protein in SEM micrographs. Further, exochitinases from these strains were found to exhibit toxicity against polyphagous nematode, M. incognita also. The SDS–PAGE profiling of F8.IIPR supernatant reveals the presence of CRY1I, which is a special CRY that is secretory in nature along with VIP3. The two potential Bt strains F8.IIPR and Ak2.IIPR were submitted in the National Agriculturally Important Microorganism Culture Collection [NAIMCC], ICAR-NBAIM, with the accession numbers NAIMCC-SB-065 and NAIMCC-SB-066, respectively. Further, the Bt strains F1.IIPR, F4.IIPR, F5.IIPR and F6.IIPR were submitted with the following accessions, NAIMCC-B-03049, NAIMCC-B-03050, NAIMCC-B-03051 and NAIMCC-B-03052, respectively. These strains have the potential for further downstream processing to develop as biopesticide for polyphagous lepidopteran pest of this region which will help in conserving the agro-ecological diversity and reduces the dependence of harmful insecticides. Studying the metabolomics of these Bt strains will help us in better understanding about CRY1I and VIP3 toxins synergistic activity against polyphagous lepidopteran insects, and it can be extrapolated for genetic engineering of third-generation insect transgenics (i.e., plants with cry1I/VIP3 toxins). This will also assist in development of liquid biopesticides containing both CRY and VIP3 toxins for management of lepidopteran insects. Further investigation is needed on differences in the gut proteases of the three polyphagous lepidopteran insects, which will help in better understanding of evolution of Bt resistance as these three insects had varying degrees of Bt susceptibility.
Availability of data and materials
Not applicable.
Change history
11 September 2023
A Correction to this paper has been published: https://doi.org/10.1186/s41938-023-00734-8
Abbreviations
- Bt:
-
Bacillus thuringiensis
- ICAR:
-
Indian Council of Agricultural Research
- NAIMCC:
-
National Agriculturally Important Microorganism Culture Collection
- NBAIM:
-
National Bureau of Agriculturally Important Microorganism
- IIPR:
-
Indian Institute of Pulses Research
- SAS:
-
Statistical Analysis Software
- BGSC:
-
Bacillus Genetic Stock Centre
- SEM:
-
Scanning electron micrograph
- kv:
-
Kilovolt
- ml:
-
Milliliter
- LC50 :
-
Lethal concentration 50%
- OD:
-
Optical density
- nm:
-
Nanometer
- HCl:
-
Hydrochloric acid
- rpm:
-
Revolutions per minute
- min:
-
Minutes
- mM:
-
Millimolar
- g:
-
Grams
- SCM:
-
Spore crystal mixture
- CFU:
-
Colony-forming units
- BHC:
-
Bihar hairy caterpillar
- TC:
-
Tobacco caterpillar
- PB:
-
Pod borer
References
Agaisse H, Lereclus D (1995) How does Bacillus thuringiensis produce so much insecticidal crystal protein? J Bacteriol 177(21):6027–6032
Ananda MR, Vaiahnav S, Naide PR, Aruna NV (2022) Long term benefits of legume based cropping systems on soil health and productivity. An overview. Int J Environ Clim Chang 12(9):299–315
Anonymous (2022) Directorate of economics and statistics. Department of agriculture and farmers welfare, GOI. https://eands.dacnet.nic.in/Advance_Estimates.htm Accessed on 23rd Oct 2022
Argôlo Filho RC, Gomes RA Jr, Barreto MR, de P Lana UG, Valicente FH, Loguercio LL, (2011) Growth variation among Bacillus thuringiensis strains can affect screening procedures for supernatant-secreted toxins against insect pests. Pest Manag Sci 67(9):1184–1192
Arora N, Ahmad T, Rajagopal R, Bhatnagar RK (2003) A constitutively expressed 36kDa exochitinase from Bacillus thuringiensis HD-1. Biochem Biophys Res Commun 307(3):620–625
Asokan R, Swamy HMM, Arora DK (2012) Screening, diversity and partial sequence comparison of vegetative insecticidal protein (vip3A) genes in the local isolates of Bacillus thuringiensis Berliner. Curr Microbiol 64(4):365–370
Baragamaarachchi RY, Samarasekera JKRR, Weerasena OVDSJ, Lamour K, Jurat-Fuentes JL (2019) Identification of a native Bacillus thuringiensis strain from Sri Lanka active against Dipel-resistant Plutella xylostella. PeerJ 7:e7535
Bautista JR, Teves FG (2013) Antibiotic susceptibility testing of isolated Bacillus thuringiensis from three soil types around Iligan City. Philipp Afr J Microbiol Res 7(8):678–682
Boonmee K, Thammasittirong SNR, Thammasittirong A (2019) Molecular characterization of lepidopteran-specific toxin genes in Bacillus thuringiensis strains from Thailand. Biotech 9(4):1–11
Chak KF, Chao DC, Tseng MY, Kao SS, Tuan SJ, Feng TY (1994) Determination and distribution of cry-type genes of Bacillus thuringiensis isolates from Taiwan. Appl Environ Microbiol 60(7):2415–2420
El-kersh TA, Al-sheikh YA, Al-akeel RA, Alsayed AA (2012) Isolation and characterization of native Bacillus thuringiensis isolates from Saudi Arabia. Afr J Biotechnol 11(8):1924–1938
Elsayed EA, Othman NZ, Malek R, Awad HM, Wu K, Aziz R, Wadaan MA, El Enshasy HA (2014) Bioprocess development for high cell mass and endospore production by Bacillus thuringiensis var israelensis in semi-Industrial Scale. J Pure Appl Microbiol 8(4):2773–2783
Engelbrecht G, Horak I, Jansen van Rensburg PJ, Claassens S (2018) Bacillus-based bionematicides: development, modes of action and commercialization. Biocontrol Sci Techn 28:629–653
Franco-Rivera A, Benintende G, Cozzi J, Baizabal-Aguirre VM, Valdez-Alarcón JJ, López-Meza JE (2004) Molecular characterization of Bacillus thuringiensis strains from Argentina. Antonie Van Leeuwenhoek 86(1):87–92
Hassan AA, Youssef MA, Elashtokhy MMA, Ismail IM, Aldayel M, Afkar E (2021) Isolation and identification of Bacillus thuringiensis strains native of the eastern province of Saudi Arabia. Egypt J Biol Pest Control 31(1):1–11
Hernandez JL (1988) Évaluation de la toxicité de Bacillus thuringiensis sur Spodopterafrugiperda. Entomophaga 33(2):163–171
Hsu SC, Lockwood J (1975) Powdered chitin agar as a selective medium for enumeration of actinomycetes in water and soil. Appl Microbiol 29(3):422–426
Iatsenko I, Boichenko I, Sommer RJ (2014) Bacillus thuringiensis DB27 produces two novel protoxins, Cry21Fa1 and Cry21Ha1, which act synergistically against nematodes. Appl Environ Microbiol 80:3266–3275
Ichikawa M, Uemori A, Yasutake K, Kagoshima K, Mizuki E, Ohba M (2008) Failure to phenotypically discriminate between non-insecticidal Bacillus thuringiensis strains with anticancer parasporins (PS2, PS3, and PS4) and Bacillus thuringiensis strains that produce insecticidal Cry proteins. Appl Entomol Zool 43(3):421–426
Jisha VN, Benjamin S (2014) Solid-state fermentation for the concomitant production of δ-endotoxin and endospore from Bacillus thuringiensis subsp. kurstaki. Adv Biosci Biotech 5(10):797
Jones JT, Haegeman A, Danchin EG, Gaur HS, Helder J, Jones MG, Kikuchi T, Manzanilla-López R, Palomares-Rius JE, Wesemael WM, Perry RN (2013) Top 10 plant-parasitic nematodes in molecular plantpathology. Mol Plant Pathol 14:946–961
Lee YK, Jin NY, Lee BR, Seo MJ, Youn YN, Gill SS, Yu YM (2015) Isolation and characterization of new Bacillus thuringiensis strains with insecticidal activity to difficult to control lepidopteran pests. J Fac Agric Kyushu Univ 60:103–112
Li H, Bouwer G (2012) Toxicity of Bacillus thuringiensis Cry proteins to Helicoverpa armigera (Lepidoptera: Noctuidae) in South Africa. J Invertebr Pathol 109(1):110–116
Logan NA, Berge O, Bishop AH, Bussse HJ, de Vos P, Fritz D, Kampfer HM, P, Rabinovitch L, Salkinoja-Salonen MS, Seldin L, Ventosa A, (2009) Proposed minimal standards for describing new taxa of aerobic, endospore forming bacteria. Int J Syst Evol Microbiol 59:2114–2121
Lone SA, Malik A, Padaria JC (2017) Selection and characterization of Bacillus thuringiensis strains from north western Himalayas toxic against Helicoverpa armigera. Microbiology Open 6(6):e00484
Martin PA, Gundersen-Rindal DE, Blackburn MB (2010) Distribution of phenotypes among Bacillus thuringiensis strains. Syst Appl Microbiol 33(4):204–208
Masson L, Erlandson M, Puzstai-Carey M, Brousseau R, Juarez-Perez V, Frutos R (1998) A holistic approach for determining the entomopathogenic potential of Bacillus thuringiensis strains. Appl Environ Microbiol 64(12):4782–4788
Meena J, Pimentel E (2002) Mechanism of action of Corynebacterium pauronetabolum strain C-924 on nematodes. J Nematol 4(2):287
Mendoza G, Portillo A, Arias E, Ribas RM, Olmos J (2012) New combinations of cry genes from Bacillus thuringiensis strains isolated from northwestern Mexico. Int Microbiol 15(4):209–216
Mohammed S, El Saedy MA, Enan MR, Ibrahim NE, Ghareeb A, Moustafa SA (2008) Biocontrol efficiency of Bacillus thuringiensis toxins against root-knot nematode, Meloidogyne incognita. J Mol Cell Biol 7:57–66
Mohan M, Rangeshwaran R, Sivakumar G, Verghese, (2014) Relative toxicity of subspecies of Bacillus thuringiensisagainst lepidopterous insect pests of agriculturalimportance. J Biol Control 28(4):197–203
Murali Krishna T, Devaki K, Swarna B, Abhijit N, Naresh M (2018) Computation of LC50 against Spodoptera litura (Fab.) for Bacillus thuringiensis isolates from native soil samples of Andhra Pradesh. J pharmacogn phytochem 7(4):174–178
Nair R, Schreinemachers P (2020) Global status and economic importance of mungbean. Mungbean Genome 1–8
Obeidat M, Khyami-Horani H, Al-Momani F (2012) Toxicity of Bacillus thuringiensis β-exotoxins and δ-endotoxins to Drosophila melanogaster, Ephestiakuehniella and human erythrocytes. Afr J Biotechnol 11(46):10504–10512
Ogiwara K, Indrasith LS, Asano S, Hori H (1992) Processing of -endotoxin from Bacillus thuringiensis subsp. kurstaki HD-1 and HD-73 by gut juices of various insect larvae. J Invertebr Pathol 60:121–126
Rajashekhar M, Shahanaz E, Vinay K (2017) Biochemical and molecular characterization of Bacillus spp. isolated from insects. J Entomol Zool Stud 5(5):581–588
Ramanujam B, Rangeshwaran R, Sivakmar G, Mohan M, Yandigeri MS (2014) Management of insect pests by microorganisms. Proc Indian Natl Sci Acad 80(2):455–471
Roy M, Chatterjee S, Dangar TK (2021) Characterization and mosquitocidal potency of a Bacillus thuringiensis strain of rice field soil of Burdwan, West Bengal. India Microb Pathog 158:105093
Sattar S, Biswas PK, Hossain MA, Maiti MK, Sen SK, Basu A (2008) Search for vegetative insecticidal proteins (VIPs) from local isolates of Bacillus thuringiensis effective against lepidopteran and homopteran insect pests. J Biopestic 1(2):216–222
Sequeros T, Ochieng J, Schreinemachers P, Binagwa PH, Huelgas ZM, Hapsari RT, Juma MO, Kangile JR, Karimi R, Khaririyatun N, Mbeyagala EK (2021) Mungbean in Southeast Asia and East Africa: varieties, practices and constraints. Agric Food Secur 10(1):1–3
Shikov AE, Malovichko YV, Lobov AA, Belousova ME, Nizhnikov AA, Antonets KS (2021) The distribution of several genomic virulence determinants does not corroborate the established serotyping classification of Bacillus thuringiensis. Int J Mol Sci 22(5):2244
Siddiqui IA, Shaukat SS (2003) Suppression of root-knot disease by Pseudomonas fluorescens CHA0 in tomato: importance of bacterial secondary metabolite 2,4-diacetylphloroglucinol. Soil Biol Biochem 35:1615–1623
Singh B, Das A, Parihar AK, Bhagawati B, Singh D, Pathak KN, Gupta S (2020) Delineation of genotype-by-Environment interactions for identification and validation of resistant genotypes in mungbean to root-knot nematode (Meloidogyne incognita) using GGE biplot. Sci Rep 10(1):1–14
Subbanna ARNS, Khan MS, Srivastava RM, Mishra PK, Babu BK, Venkateswarlu V (2018) Interspecies diversity of Bacillus thuringiensis isolates native from North Western Indian Himalayas. J Environ Biol 39(3):306–313
Sujayanand GK, Pandey S, Jagadeeswaran R, Dubey J (2019) Chitinase: a novel biopesticide for insect and nematode pest management. Indian Farmer 6(9):599–603
Sujayanand GK, Akram M, Konda A, Nigam A, Bhat S, Dubey J, Kumar K, Muthusamy SK (2021) Distribution and toxicity of Bacillus thuringiensis (Berliner) strains from different crop rhizosphere in Indo-Gangetic plains against polyphagous lepidopteran pests. Int J Trop Insect Sci 1:1–9
Sujayanand GK, Pandey S (2020) Structural and functional diversity of cry genes and their utilization in pest management of grain legumes. In: Microbial mitigation of stress response of food legumes (pp 257–269). CRC Press
Sun YP, Johnson ER (1960) Analysis of joint action of insecticides against houseflies. J Econ Entomol 53:887–892
Tailor R, Tippett J, Gibb G, Pells S, Jordan L, Ely S (1992) Identification and characterization of a novel Bacillus thuringiensis delta-endotoxin entomocidal to coleopteran and lepidopteran larvae. Mol Microbiol Berkshire 6(9):1211–1217
Venkataramanamma K, Reddy BV, Jayalakshmi RS, Jayalakshmi V, Rajendran L (2022) Isolation, in vitro evaluation of Bacillus spp against Fusarium oxysporum f. sp. ciceris and their growth promotion activity. Egypt J Biol Pest Control 32(1):1–8
Vessey JC, Pegg GF (1973) Autolysis and chitinase production in cultures of Verticillium albo-atrum. Trans Br Mycol Soc 60(1):133–143
Vilas-Bôas GT, Lemos MVF (2004) Diversity of cry genes and genetic characterization of Bacillus thuringiensis isolated from Brazil. Can J Microbiol 50(8):605–613
Zothansanga R, Senthilkumar N, Gurusubramanian G (2016) Diversity and toxicity of Bacillus thuringiensis from shifting cultivation (jhum) habitat. Biocontrol Sci 21(2):99–111
Acknowledgements
The authors were thankful to Drs. Soren K.R. and Senthilkumar M. for providing PCR facilities and ultracentrifugation facilities. The authors also express their gratitude to Director, ICAR-NBAIM, Mau, Dr K. Pandiyan, Dr M. Kumar and Mr. Manish Roy for facilitating SEM micrographs. The authors were thankful to Director, ICAR-IIPR, for co-operation and constant support for executing this work.
Funding
The authors did not receive any specific grant from any funding agency for accomplishing this work. The work was accomplished under the institute project entitled “Identification and evaluation of potential Bt strains for managing lepidopteran pest of pulse crops” [code: CRSCIIPRSIL201700900148].
Author information
Authors and Affiliations
Contributions
SGK conceived, designed and executed the experiments. SP performed various PCRs and trimmed the gene sequences. SGK, SP, SD and JD purified endospore crystal mixture, performed insect bioassay and chitinolytic activity and recorded and digitalized data. SGK and SP prepared SEM samples and biochemical characterization of Bt strains. RJ performed nematode bioassay. RJ, SP and JD prepared colloidal chitin. SGK, AC, RJ and SP wrote the manuscript and prepared the graph. SGK, AC and RJ analyzed the experimental data. VK performed protein quantification, protein profiling and interpretation of results.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All the authors gave their consent to publish the submitted manuscript as “Original paper” in EJBPC.
Competing interests
The authors declare that the research was conducted in the absence of commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sujayanand, G.K., Pandey, S., Jagadeeswaran, R. et al. Characterization of entomotoxic and nematotoxic genes from indigenous Bacillus thuringiensis strains and their biocontrol potential. Egypt J Biol Pest Control 33, 76 (2023). https://doi.org/10.1186/s41938-023-00722-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41938-023-00722-y