Abstract
Background
Bacillus thuringiensis (Bt) Berliner is an omnipresent soil bacterium used as world’s leading biopesticide to combat agriculturally important insect pests. This study was aimed at protein and gene profiling of an indigenous Bt isolate RM11, which was toxic to the larvae of diamondback moth, Plutella xylostella (L.) in laboratory bioassays.
Results
Indigenous Bt isolate RM11 was characterized along with the standard checks B. thuringiensis subsp. kurstaki (Btk) HD1 and 78/11, based on colony characters, protein profile and PCR screening. All three Bt colonies were fried egg type, white in color with flat elevation and undulated margin. PCR screening revealed the presence of cry1Ac and vip3A genes, which encode lepidopteran toxic proteins in RM11. SDS-PAGE results showed the presence of a prominent protein band of cry1Ac, vip3A with molecular weights 135 kDa, 88 kDa and other bands at 70, 50, 32 and 10 kDa. In leaf disk bioassay with spore crystal mixture, RM11 exhibited toxicity with LC50 of 4.51 µg/ml as against 0.07 µg/ml in positive standard HD1, based on mortality at 72 h after treatment. At LC50 of 4.51 µg/ml, solubilized and insolubilized protein of RM11 was found to produce 56 and 70% mortality.
Conclusions
The present study revealed that RM11 could be a viable alternative for consideration in developing a native Bt formulation and for inclusion in the integrated management of P. xylostella with other native isolates producing different toxins. Furthermore, these findings imply that RM11 could be a source of new cry toxin, which can be confirmed through whole-genome sequencing analysis.
Similar content being viewed by others

Background
Bacillus thuringiensis (Berliner) (Bacillales: Bacillaceae), commonly known as Bt, is an entomopathogenic, gram-positive, rod-shaped bacterium that produces insecticidal crystal (Cry) proteins known as δ-endotoxins during the sporulation phase (Jalapathi et al. 2020). This entomopathogenic bacterium is the most extensively used biopesticide against insect pests, mites, nematodes and protozoa (Gupta et al. 2021). Upon ingestion of crystalline proteins (toxins) by susceptible insects, they cause starvation, pore formation in midgut and septicemia, with consequent death (Schnepf et al. 1998). Several Bt isolates have been identified by specific toxicity toward specific insect orders based on the type of endotoxins they produce. More than 400 Bt-based biopesticide formulations are being used in pest management in the form of foliar sprays apart from the development and use of transgenic plants (Babin et al. 2020). Due to its important role in agriculture and medicine, researchers are interested in finding novel Bt isolates with broad-spectrum insecticidal activity and in identifying novel functional genes of B. thuringiensis (Sajid et al. 2018). On the other hand, long-term usage of Bt toxins leads to a decrease in pest population susceptibility. Bt is extensively used against lepidopteran pests of vegetables and forestry (Navon 2000). Historically, Bt has been highly effective against the diamondback moth, Plutella xylostella (Linnaeus) (Lepidoptera: Plutellidae), a worldwide problem in crucifers. It is the most notorious pest of Cole crops, with an estimated annual cost of US$ ~ 4.5 billion (Zalucki et al. 2012). The widespread use of Btk against P. xylostella has resulted in the emergence of field resistance. Bt Cry toxin-based transgenic Cole crops have been developed to control P. xylostella (Bhattacharya et al. 2002). The present work aimed to find novel Bt isolates with cry and vip genes that code for proteins with increased toxicity, which might be exploited to develop new products and manage resistance in various agriculturally important insect pests.
Methods
Insect rearing
Plutella xylostella populations used in the bioassays were initially obtained from the National Bureau of Agricultural Insect Resources, Bangalore, India, and were reared on cauliflower leaves (CFL-1522, Syngenta) grown in greenhouse conditions without the use of insecticides. The larvae were reared at 25 ± 1 °C, 75 ± 5% RH, and 16:8 (L/D) h photoperiod in a (30 × 30 × 30 cm) larval rearing cage in the laboratory.
Bt isolates
Native Bt isolate, RM11, standard strain Bacillus thuringiensis subsp. kurstaki (Btk) HD1 and acrystalliferous Bt strain 78/11 were obtained from the Department of Plant Biotechnology, Centre for Plant Molecular Biology and Biotechnology, TNAU, Coimbatore, India, and maintained on T3 media for the preparation of spore crystal mixtures and DNA extraction. RM11 was selected from 60 Bt isolates, through preliminary screening bioassay with 5 µg/ml of spore crystal mixture suspension (Navya et al. 2021).
Morphological characterization of Bt isolates
The colony color, surface and margin of all bacterial colonies were observed from the T3 culture plate grown at 30 °C for 24 h. To study the morphology of the parasporal crystalline inclusions, culture smears were made on a glass slide, heat-fixed and stained with Coomassie brilliant blue stain (0.133% Coomassie Brilliant Blue G250 in 50% acetic acid) for about 1 min. Finally, smears were gently rinsed under running water, blot-dried and viewed under a phase-contrast microscope (iScope, Euromex).
Culture conditions and protein isolation for toxicity analysis
The spore crystal mixtures from RM11, HD1 and 78/11 were isolated, following standard protocols (Ramalakshmi and Udayasuriyan 2010). In brief, from each Bt isolate, a single colony was picked and inoculated into tubes with 5 ml of T3 broth and incubated overnight at 30 °C and 200 rpm in a shaking incubator (Orbitek). From overnight grown cultures, 250 µl (1.0% inoculum) was added to 25 ml of T3 broth in a 250-ml conical flask and grown in a shaking incubator kept at 30 °C, 200 rpm for 48–60 h. After 48 h, using a phase-contrast microscope bacterial cell growth and lysis was examined. When more than 90% of the cells were found lysed, the culture was centrifuged at 4 °C for 10 min at 10,000 rpm, and the pellet was resuspended in 25 ml of ice-cold Tris–EDTA buffer [Tris 10 mM, EDTA 1 mM, pH 8.0 with 1 mM phenylmethylsulfonyl fluoride (PMSF)] and washed once with 25 ml of ice-cold 0.5 M, NaCl, and centrifuged for 10 min, followed by two washes with 25 ml Tris–EDTA buffer with 0.5 mM PMSF at the same speed and time (Navya et al. 2021). Finally, 500 µl of sterile distilled water and 10 µl of 1 mM PMSF were added to the resulting pellet, and aliquots of 50 µl were made and stored at − 20 °C for further use.
Spore crystal purification
The crude containing spore crystal mixtures was centrifuged at 10,000 rpm for 10 min at 4 °C. To the 25 mg crude pellet, 300 µl of NaOH buffer (50 mM NaOH, complete mini protease inhibitor (Merck),100 mM PMSF in the ratio 91:8:1) pH 10 was added and incubated at 37 °C for 11/2 h at 175 rpm and centrifuged at 10,000 rpm for 10 min at 4 °C. The supernatant was collected and stored at − 20 °C for use in SDS-PAGE. SDS-PAGE was used to monitor the solubilization of crystals.
Protein profiling
Protein profiling was carried out with spore crystal lysate prepared from RM11, HD1 and the acrystalliferous strain 78/11 using sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) (Laemmli 1970), with 10% separating gel and 4% stacking gel. The spore crystal lysate was mixed at 4:1 with loading buffer (4x) (0.25 M Tris HCl (pH 6.8), 8% SDS, 40% glycerol and 0.5% bromophenol blue). Samples were boiled for 2 min before being loaded into wells. The molecular mass of the proteins was assessed using a three-color protein ladder (Puregene, Genetix Biotech Asia Pvt. Ltd.) that comprised a molecular weight range of 10–315 kDa.
PCR screening and sequencing of cry and vip genes
Total genomic DNA was extracted from broth culture grown from a single bacterial colony and utilized as a template for amplifying the cry1, cry1Ac, cry2, and vip3A genes (Sambrook and Russell 2001). NanoDrop spectrophotometer (Genova Nano, Jenway) was used to quantify the isolated DNA, and agarose gel electrophoresis (1%) was employed to assess its integrity. Each PCR mixture contained 20–50 ng template DNA, 1 µl of each primer and 10 µl of 2 × PCR Master Mix (Smart prime) including dNTPs, Taq polymerase and PCR buffer, with the final volume made to 20 µl using sterile double distilled water (Table 1). PCR amplification was performed in a thermal cycler (ProFlex PCR system) using temperature cycles as given in Table 1, and 1 kb DNA ladder was used to check the PCR amplicon in a 1% agarose gel. The amplified products were visualized under a UV transilluminator (Bio-Rad). PCR-amplified products were sent for sequencing (Biokart India Pvt. Ltd.) with the corresponding primers. The forward and reverse sequences were edited using BioEdit program (Hall 1999), and BLAST was performed using BLASTn. The final aligned sequences were submitted to GenBank for accession numbers.
Bioassays with P. xylostella
The pathogenic activity of RM11 toward P. xylostella was determined by leaf disk bioassay against 3-day-old larvae. Cauliflower leaves of 2.5 cm diameter were thoroughly washed with sterile distilled water containing 0.1% Triton X-100 and air-dried. Before conducting the bioassay, the protein concentration in the isolate was determined (Bradford 1976). For each Bt isolate, 20 µl of spore crystal mixture (concentrations ranging from 5 to 0.125 µg ml−1) was applied gently on cauliflower leaf disks, smeared with a glass rod and allowed to dry. To avoid desiccation, all the leaf disks were placed on the wet surface of Whatman filter paper disks in plastic cups (30 mm diameter). The standard strains HD1 and 78/11 were used as positive and negative control, respectively. The assays were performed in 3 replicates with 30 larvae for each concentration. Larval mortality was observed at 24-h intervals for 3 days, and cumulative mortality at 72 h was taken to work out median lethal concentrations (LC50). To check any difference in toxicity between insolubilized and solubilized spore crystal mixtures of RM11, leaf disk bioassay was conducted with 100 larvae (10 larvae each for 10 replications), and mortality was recorded at 24-h intervals and cumulative mortality at 72 h.
Statistical analysis
Statistical analysis for calculating each treatment LC50 and LC95 and standard error values were performed using the EPA probit analysis program (Version 1.5) in DOSBox background. Bioassay with solubilized spore crystal mixtures was laid out in a completely randomized design (CRD) and statistically analyzed by one-way analysis of variance (ANOVA) using AGRES statistical software version 7.01. Significant differences between means were determined by Duncan’s multiple range test (p = 0.05).
Results
Morphological characterization of Bt isolates
Bt isolate, RM11 and standards HD1 and 78/11 were characterized based on colony’s color, surface, shape, elevation and margin. The isolate RM11 and standards HD1 and 78/11 were off-white in color (Fig. 1). The colony surface was determined to be fried egg type in all 3 cultures, with a flat elevation and undulate edge. Phase-contrast microscopic observations disclosed the presence of bipyramidal-, spherical- and cuboidal-shaped crystals in HD1, whereas bipyramidal-, spherical- and cuboidal-shaped crystals in RM11.
Protein profiling and alkali solubilization
SDS-PAGE analysis of spore crystal mixtures from RM11 and HD1 showed the presence of a wide range of banding patterns with molecular weights at 230 kDa, 88 kDa, 70 kDa, 50 kDa, 40 kDa and 26 kDa (Fig. 2a). In RM11, after exposure to alkaline conditions (pH10) with NaOH buffer, the crystalline protein of 230 kDa size was solubilized to 130 kDa as shown in SDS-PAGE. However, in HD1, 130 kDa protein remained intact even after solubilization (Fig. 2b).
PCR screening and sequencing analysis
PCR for three lepidopteran-specific genes (cry1, cry2 and vip3A) confirmed the presence of only cry1 and vip3A genes in RM11, but not cry2 genes (Fig. 3a). Existence of all three genes, viz. both cry1, cry2 and vip3A genes, was confirmed in HD1 alone. PCR analysis with gene specific primer confirmed the presence of cry1Ac (Fig. 3b). PCR products were sequenced to check the presence of cry1 and vip3A genes, and sequences were analyzed with BLASTn, and sequences of partial genes were deposited in the NCBI genetic sequence database under GenBank accession numbers OM305004.1 (cry1) and OM315207.1 (vip3A) (Fig. 4), respectively. Partial cry1 and vip3A sequences amplified from RM11 using screening primers (Table 1) were found to align with cry1Ac of HD73 and vip3A of HD1 at positions 3758 to 4036 bp (Fig. 5) and 415 to 1194 bp (Fig. 6), respectively.
Insecticidal activity against P. xylostella larvae
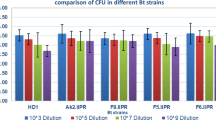
RM11 exhibited LC50 of 4.51 µg/ml with fiducial limits ranging from 2.261 to 15.708 µg/ml, whereas the reference strain HD1 showed LC50 of 0.07 µg/ml with fiducial limits ranging from 0.052 to 0.102 µg/ml. LC50 values were considered significantly dissimilar when fiducial limits did not overlap. The mortality data were found to be significant and confirmed by the chi-square goodness-of-fit test (Table 2, Fig. 7). Solubilized and insolubilized protein of RM11 was found to produce 56 and 70% mortality at LC50 of 4.51 µg/ml (Table 3).
Discussion
Bt have been reported to have white to off-white colony with uneven colony margin and flat to raised colony elevation (Padole et al. 2017). Bipyramidal crystals, cuboidal crystals and spherical crystals were found in the present study, as previously described by Rampersad and Ammons (2005). Similarly, Ramalakshmi and Udayasuriyan (2010) observed the occurrence of cuboidal and bipyramidal crystals among the Bt isolates tested. This wide range of protein banding patterns in these isolates suggested the variable biological activity and specificity for different insect pests. Ramalakshmi and Udayasuriyan (2010) reported protein bands of 135, 95, 65, 43 and 30 kDa. Arrieta and Espinoza (2006), Liu et al. (2009) and others have previously documented diverse electrophoretic patterns of Cry proteins with molecular weights ranging from 20 to 160 kDa. In the present study, protein band at 130 kDa was observed in SDS-PAGE after solubilization of crystalline protein (230 kDa), as previously reported in Bt strain HD73 by Rivers et al. (1988). In the present study, solubilized spore crystal mixture was found to be more toxic (70% mortality), when compared to insolubilized protein (56% mortality). Toxicity was found to be significantly affected by crystalline solubilization (Aronson et al. 1991).
Occurrence of cry1 and cry2 genes together was frequently found (Hernandez-Rodriguez and Ferre 2009). Bt isolates with intergroup crystalline protein-producing genes are excellent prospects for developing broad-spectrum biopesticides (Hernandez-Rodriguez et al. 2009). The isolate RM11 and the positive control (HD1) were found to have the combination of genes cry1 and vip3A. These findings are similar to those of Maheesha et al. (2021). Based on sequence results of RM11, cry1 and vip3A sequences were found to be very similar to the already reported standard strains HD73 and HD1, respectively. Similar results were obtained in a previous study (Sahin et al. 2018). Higuchi et al. (2000) concluded that HD1 was highly effective against P. xylostella, with an LC50 of 0.212 g/ml as against the toxic test isolate with 6.52 g/ml, similar to isolate RM11 which was 64 times less toxic than HD1. The insecticidal activity of spore crystals can be used to find native Bt isolates with insecticidal properties (Sahin et al. 2018). Kahrizeh et al. (2017) reported that Bt isolates are highly toxic to P. xylostella larvae exhibiting 78.3 to 100% mortality. In a study conducted in Northeastern Brazil, 13 Bt isolates out of a total of 104 revealed a mortality rate of 30–100% against P. xylostella neonates (Silva et al. 2012). In a previous study, out of 60 isolates tested, 27 isolates produced 50 to 100% mortality in P. xylostella (Navya et al. 2021). According to Apaydin et al. (2008), 80% of Bt isolates showed varying degree of toxicity against Ephestia kuehniella, with only one strain showing 84% mortality. According to Figueiredo et al. (2019), Vip3 + Cry1 exhibited the highest toxicity than Vip3 + Cry2 protein combinations and thus increased the toxicity against Spodoptera frugiperda. Individual Cry proteins have lower larvicidal activity as compared to the toxicity of Cry proteins when administered in combination (El-Kersh et al. 2016).
Conclusions
The higher level of toxicity of HD1 to P. xylostella, over RM11, could be due to the presence of cry2 genes in addition to cry1 and vip3 which contributed great toxicity to this pest. The whole-genome sequencing of RM11 and identification and cloning of novel cry genes will be helpful in developing a biopesticide formulation for application in integrated pest management or in development of newer transgenic crops. Despite the fact that several Bt toxins have been isolated and proved to be useful in pest management, none of them were able to replace the Cry1Ac-based formulations from Btk strains including HD1 and HD12. Research on the discovery of Bt isolates with novel cry genes is continued in different laboratories throughout the world, anticipating discovery of potential isolates which could be superior to Cry1Ac in toxicity with wide spectrum of activity, so that it could be useful in overcoming insect resistance to already reported Bt toxins. This could be achieved by screening the native Bt isolates in large numbers and against wide spectrum of insect species.
Availability of data and material
Not applicable.
Abbreviations
- Bt :
-
Bacillus thuringiensis
- Cry:
-
Crystal proteins
- Cyt:
-
Cytolytic proteins
- Vip:
-
Vegetative insecticidal proteins
References
Alberola TM, Aptosoglou S, Arsenakis M, Bel Y, Delrio G, Ellar D, Ferre J, Granero F, Guttmann D, Koliais S (1999) Insecticidal activity of strains of Bacillus thuringiensis on larvae and adults of Bactrocera oleae Gmelin (Dipt. Tephritidae). J Invertebr Pathol 74(2):127–136
Apaydin O, Cinar C, Turanli F, Harsa S, Gunes H (2008) Identification and bioactivity of native strains of Bacillus thuringiensis from grain-related habitats in Turkey. Biol Control 45:21–28. https://doi.org/10.1016/j.biocontrol.2008.01.011
Aronson AI, Han ES, McGaughey W, Johnson D (1991) The solubility of inclusion proteins from Bacillus thuringiensis is dependent upon protoxin composition and is a factor in toxicity to insects. Appl Environ Microbiol 57(4):981–986. https://doi.org/10.1128/aem.57.4.981-986.1991
Arrieta G, Espinoza AM (2006) Characterization of a Bacillus thuringiensis strain collection isolated from diverse Costa Rican natural ecosystems. Rev Biol Trop 54:13–27
Babin A, Nawrot E, Marie P, Gallet A, Gatti JL, Poiriac M (2020) Differential side-effects of Bacillus thuringiensis bioinsecticide on non-target Drosophila flies. Sci Rep 10(1):1–18. https://doi.org/10.1038/s41598-020-73145-6
Ben-Dov E, Zaritsky A, Dahan E, Barak ZE, Sinai R, Manasherob R, Margalith Y (1997) Extended screening by PCR for seven cry group genes from field collected strains of Bacillus thuringiensis. Appl Environ Microbiol 63(12):4883–4890. https://doi.org/10.1128/aem.63.12.4883-4890.1997
Bhattacharya RC, Viswakarma N, Bhat SR, Kirti PB, Chopra VL (2002) Development of insect resistant transgenic cabbage plants expressing a synthetic Cry1Ab gene from Bacillus thuringiensis. Curr Sci 83:146–150
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal Biochem 72:248–254. https://doi.org/10.1006/abio.1976.9999
El-kersh TA, Al-sheikh YA, Al-akeel RA, Alsayed AA (2016) Isolation and characterization of native Bacillus thuringiensis isolates from Saudi Arabia. Afr J Biotechnol 11:1924–1938. https://doi.org/10.5897/AJB11.2717
Figueiredo CS, Lemes ARN, Sebastiao I, Desiderio JA (2019) Synergism of the Bacillus thuringiensis Cry1, Cry2, and Vip3 proteins in Spodoptera frugiperda control. Appl Biochem Biotechnol 188(3):798–809. https://doi.org/10.1007/s12010-019-02952-z
Gupta M, Kumar H, Kaur S (2021) Vegetative insecticidal protein (Vip): potential contender from Bacillus thuringiensis for efficient management of various detrimental agricultural pests. Front Microbiol 12:1139. https://doi.org/10.3389/fmicb.2021.659736
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hernandez Rodriguez CS, Ferre J (2009) Ecological distribution and characterization of four collections of Bacillus thuringiensis strains. J Basic Microbiol 49:152–157. https://doi.org/10.1002/jobm.200800121
Hernandez Rodriguez CS, Boets A, Van Rie J, Ferre J (2009) Screening and identification of vip genes in Bacillus thuringiensis strains. J Appl Microbiol 107:219–225. https://doi.org/10.1111/j.1365-2672.2009.04199.x
Higuchi K, Saitoh H, Mizuki E, Ichimatsu T, Ohba M (2000) Larval susceptibility of the diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae), to Bacillus thuringiensis H serovars isolated in Japan. Microbiol Res 155(1):23–29. https://doi.org/10.1016/s0944-5013(00)80018-x
Jalapathi SK, Jayaraj J, Shanthi M, Theradimani M, Venkatasamy B, Irulandi S, Prabhu S (2020) Potential of Cry1Ac from Bacillus thuringiensis against the tomato pinworm, Tutaabsoluta (Meyrick) (Gelechiidae: Lepidoptera). Egypt J Biol Pest Control 30(1):1–4. https://doi.org/10.1186/s41938-020-00283-4
Jain D, Sunda SD, Sanadhya S, Nath DJ, Khandelwal SK (2017) Molecular characterization and PCR-based screening of cry genes from Bacillus thuringiensis strains. 3 Biotech 7(1):1–8
Kahrizeh AG, Khoramnezhad A, Hassanloui RT (2017) Isolation, characterization and toxicity of native Bacillus thuringiensis isolates from different hosts and habitats in Iran. J Plant Prot Res 57(3):212–218
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. https://doi.org/10.1038/227680a0
Liu T, Guo W, Sun W, Sun Y (2009) Biological characteristics of Bacillus thuringiensis strain Bt11 and identification of its cry type genes. Front Agric China 3:159–163. https://doi.org/10.1007/s11703-009-0040-z
Maheesha M, Balasubramani V, Murugan M, Raveendran M, Rajadurai G, Tamilnayagan T, Kokiladevi E, Sathiah N (2021) Characterization of indigenous Bacillus thuringiensis isolate T350 toxic to fall armyworm, Spodoptera frugiperda. J Pharm Innov 10(12):1809–1812
Navya R, Balasubramani V, Raveendran M, Murugan M, Lakshmanan A (2021) Diversity of indigenous Bacillus thuringiensis isolates toxic to the diamondback moth, Plutella xylostella (L.) (Plutellidae: Lepidoptera). Egypt J Biol Pest Control 31(1):1–7. https://doi.org/10.1186/s41938-021-00495-2
Navon A (2000) Bacillus thuringiensis insecticides in crop protection—reality and prospects. Crop Prot 19:669–676. https://doi.org/10.1016/S0261-2194(00)00089-2
Padole DA, Moharil MP, Ingle KP, Munje S (2017) Isolation and characterization of native isolates of Bacillus thuringiensis from Vidarbha Region. Int J Curr Microbiol Appl Sci 6:798–806. https://doi.org/10.20546/ijcmas.2017.601.094
Ramalakshmi A, Udayasuriyan V (2010) Diversity of Bacillus thuringiensis isolated from Western Ghats of Tamil Nadu State, India. Curr Microbiol 61:13–18. https://doi.org/10.1007/s00284-009-9569-6
Rampersad J, Ammons D (2005) A Bacillus thuringiensis isolation method utilizing a novel stain, low selection and high throughput produced a typical result. BMC Microbiol 5(1):1–11. https://doi.org/10.1186/1471-2180-5-52
Rivers DB, Vann CN, Zimmack HL (1988) Trypsin activation of the Bacillus thuringiensis delta-endotoxin in Ostrinia nubilalis larvae. Proc Indiana Acad Sci 98:207–214
Sahin B, Gomis-Cebolla J, Gunes H, Ferre J (2018) Characterization of Bacillus thuringiensis isolates by their insecticidal activity and their production of Cry and Vip3 proteins. PLoS ONE 13(11):1–18
Sambrook J, Russell DW (2001) Preparation and analysis of eukaryotic genomic DNA. In: Argentine J (ed) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, New York, pp 547–576
Sajid M, Geng C, Li M, Wang Y, Liu H, Zheng J, Peng D, Sun M (2018) Whole genomic analysis of Bacillus thuringiensis revealing partial genes as a source of novel Cry toxins. Appl Environ Microbiol 84(14):1–15. https://doi.org/10.1128/AEM.00277-18
Schnepf E, Crickmore N, Van Rie J, Lereclus D, Baum J, Feitelson J, Zeigler DR, Dean DH (1998) Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol Mol Biol Rev 62:775–806
Silva MC, Siqueiraa HAA, Marques EJ, Silva LM, Barros R, Lima Filho JVM, Silva SMFA (2012) Bacillus thuringiensis isolates from north eastern Brazil and their activities against Plutella xylostella (Lepidoptera: Plutellidae) and Spodoptera frugiperda (Lepidoptera: Noctuidae). Biocontrol Sci Technol 22:583–599
Zalucki MP, Shabbir A, Silva R, Adamson D, Shu-Sheng L, Furlong MJ (2012) Estimating the economic cost of one of the world’s major insect pests, Plutella xylostella (Lepidoptera: Plutellidae): Just how long is a piece of string? J Econ Entomol 105(4):1115–1129
Acknowledgements
Technical support received from the Department of Plant Biotechnology, Department of Agricultural Entomology and Department of Nano Science & Technology, Tamil Nadu Agricultural University, is gratefully acknowledged.
Funding
This work was funded by the Department of Science & Technology, Government of India, in the form of INSPIRE fellowship-IF180443 to R. Naga Sri Navya.
Author information
Authors and Affiliations
Contributions
RNSN carried out the experiment for her doctoral research. VB conceptualized the study. RNSN and VB wrote the paper. VB, MR and AL provided research material and helped in conducting the experiments and analyzed the data. VB, MR, MM and AL helped in reviewing and editing the manuscript. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All the authors give their consent to publish the submitted manuscript as “Original paper” in EJBPC.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Navya, R.N.S., Balasubramani, V., Raveendran, M. et al. Characterization of indigenous Bacillus thuringiensis isolate RM11 toxic to the diamondback moth, Plutella xylostella (L.) (Lepidoptera: Plutellidae). Egypt J Biol Pest Control 32, 56 (2022). https://doi.org/10.1186/s41938-022-00553-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41938-022-00553-3