Abstract
Background
The European corn borer (ECB), Ostrinia nubilalis (Hübner, 1796) (Lepidoptera: Crambidae), is the major pest of maize (Zea mays Linnaeus, 1753) in Serbia. One potential method for managing this pest is the augmentative release of naturally occuring egg parasitoids of the genus Trichogramma. The first step in this process is accurately identifying the naturally occuring species and estimating their natural distribution and abundance. Molecular identification, based on differences in DNA sequences, has commonly been employed for the identification of Trichogramma species. A simple, quick, and accurate molecular assay is urgently required for the identification of two common Trichogramma species, associated with ECB in Serbia: T. brassicae Bezdenko, 1968 and Trichogramma evanescens Westwood, 1833. Such an assay will facilitate an expansive survey of resident populations of Trichogramma associated with ECB across agricultural growing regions of Vojvodina province.
Results
A species-specific multiplex PCR assay for the 2 species was developed and validated that assay using a sample of 79 parasitoid wasps reared from ECB egg masses collected from sample sites across Vojvodina province. Trichogramma brassicae was confirmed as the dominant egg parasitoid of ECB in this region, accounting for 77 of the 79 wasps (97.47%). The remaining 2 were confirmed as T. evanescens. Trichogramma brassicae was detected at all 12 sample sites, while T. evanescens was detected at only 2 plots, Mokrin and Nakovo.
Conclusions
The species-specific multiplex PCR assay presented herein can provide the basis of a quick, cheap, and reliable means for identifying the species of Trichogramma that parasitize ECB egg masses in Serbia. Two currently documented species, T. brassicae and T. evanescens, are readily diagnosed by the size of the PCR product they produce in the assay. Any additional species are expected to not produce a band of a diagnostic size. Such species would subsequently be identified by sequencing, which may also allow them to be promptly incorporated into a revised assay.
Similar content being viewed by others
Background
Maize (Zea mays Linnaeus, 1753) is the number one crop in Serbia. The largest areas of maize production are located in the northern province of Vojvodina, where it is grown on about 620,000 ha, with a total production of over 4 million tons (Latković et al., 2018). Although the production of maize is affected by several harmful insects, the most important maize pest in Serbia is the European corn borer (ECB), Ostrinia nubilalis (Hübner, 1796) (Lepidoptera, Crambidae) (Čamprag et al., 1983). Management techniques commonly used to reduce ECB infestations include changes to cultural practices, the planting of resistant cultivars, and the application of chemical insecticides (Maissle et al., 2010), the last being the most commonly employed approach. Chemical insecticides are not entirely effective against ECB because of the relatively short period the insect spends outside the plant prior to boring into it. Furthermore, there are general concerns about the effects of insecticides on beneficial insects and the wider environment, and specific concerns among the general public about insecticide residues on agricultural products (Phoofolo, 1997). Due to such growing awareness, biological control methods are becoming more favored and increasingly important (Ivezić et al., 2020).
One particular biological control method that has attracted a lot of attention is the augmentative release of naturally occurring species of the genus Trichogramma Westwood, 1833 (Hymentoptera: Trichogrammatidae) (Grushevaya, 2020). Trichogramma spp. are tiny (<1mm) wasps that parasitize the eggs of many insect species and are widely used in inundative and inoculative biological control programs against several pests in the EU (Golbaghi et al., 2020). Several species of Trichogramma have been identified as promising biological control agents of ECB, including T. brassicae and T. evanescens (Van Schelt and Ravensberg, 1991). Both species appear to be widespread across Europe and are reared in commercial facilities for release as biological control agents.
Serbia is one of the countries, at the southast border of Europe, that does not actively use Trichogramma species in its agricultural practices, despite evidence suggesting high activity of native populations of these insects (Ivezić et al., 2020). Not all Trichogramma species (or populations) perform equally well in terms of mass rearing or field dispersal and parasitism rates (Borba et al., 2005). Morphological identification of Trichogramma requires expert taxonomic knowledge, due to their minute size (<1 mm long) and a general lack of reliable characters (Stouthamer et al., 1999). Consequently, alternative identification methods have been developed. Sequences of the internal transcribed spacer 2 (ITS2) region of the nuclear ribosomal DNA are one accepted means of identifying Trichogramma species (Fahriye et al., 2009). An intial survey of the Kikinda region identified two species of Trichogramma (T. brassicae and T. evanescens) parasitizing ECB eggs, based on sequences of the ITS2 gene (Ivezić et al., 2018). A second, more geographically expansive study identified the same two species, again based on DNA sequences, but this time of the mitochondrial cytochrome oxidase subunit 1 (COI) gene (Ivezić et al., 2020). In both studies, T. brassicae was found to be far more abundant in maize than was T. evanescens.
The utility of ITS2 and COI sequences as diagnostic tools for Trichogramma (and many other taxa) is widely accepted, but obtaining a DNA sequence incurs significant costs (time and money). One way that can be achieved is through the development of species-specific PCR primers that can be run alone, or better still, in a multiplex reaction. Species-specific multiplex PCR incorporates several PCR primers that bind to the same DNA strand, each of which is specific to only one “target” species (Rugman-Jones et al., 2020). When combined in a single reaction with a single complementary PCR primer that is “universal” to all target species, the multiplex PCR results in the production of a PCR product of a unique size for each of the target species, allowing specimens to be identified directly following gel electrophoresis of the PCR product (Rugman-Jones et al., 2020).
The aims of this study were (1) to develop a diagnostic species-specific multiplex PCR assay for T. brassicae and T. evanescens in Serbia and (2) to use that assay to survey resident populations of Trichogramma associated with ECB across agricultural growing regions of Vojvodina province.
Methods
Multiplex PCR design
Previous studies of the egg parasitoids of ECB in Serbian maize resulted in the identification of just two species, T. brassicae and T. evanescens (Ivezić et al., 2020). Identification in these studies was based on DNA sequences of the ITS2 and/or COI genes. An assay based on the former gene that was chosen to develop ribosomal sequences typically show lower levels of intraspecific variation than mitochondrial sequences. Ivezić et al. (2018) resulted in the acquisition of ITS2 sequences from 26 specimens, 25 of which were T. brassicae and the other T. evanescens (GenBank accessions MW377755-MW377780). These were combined with a further 31 sequences of T. brassicae and 50 sequences of T. evanescens retrieved from GenBank and aligned in MAFFT version 7.050 (http://mafft.cbrc.jp/alignment/software/) using the Q-INS-I iterative strategy (Katoh and Standley, 2013). This alignment (available in the supplementary material) was used as the basis for designing species-specific PCR primers using the Primer3 (v.0.4.0) software (Rozen and Skaletsky, 2000). Criteria for the design of species-specific primers were as follows: (1) they should lie in stretches of the ITS2 that display minimal levels of intraspecific variation particularly at the 3′ end of the primer, (2) they should work in tandem with the “universal” forward primer ITS2 for (Stouthamer et al. 1999) which is located in the highly conserved 5.8S adjacent to ITS2, (3) at least one nucleotide at the 3′ end of the primer must be unique to its target, (4) complementarity between primers should be minimal to avoid the production of primer dimers, (5) they should all work at similar annealing temperatures, and (6) they should produce species-specific amplicons that differ sufficiently in size to allow their diagnosis using standard agarose gel electrophoresis. The specificity of several primers for each species was further assessed by conducting BLAST searches of the primer sequences against the ITS2 sequences of eight further Trichogramma species that are common in the Mediterannean region: T. bourarachae, T. cacoeciae, T. cordubensis, T. dendrolimi, T. euproctidis, T. nerudai, T. oleae, and T. pintoi (GenBank accessions listed in Sumer et al. 2009 (Table 2)). Any primers with significant similarity to these additional species were immediately rejected. Several primers were subsequently tested individually and in multiplex PCR against DNA from the original 26 specimens (Ivezić et al. 2018), resulting in the identification of a set of primers, which gave the best combination of the desired characteristics (Table 1 and Fig. 1). The resulting assay was performed in individual 25 μl reactions containing 1X DreamTaq PCR mastermix (ThermoScientific), 0.2 μM of each primer (Table 1), and 2.0 μl template DNA. Cycling was done on a Mastercycler® ep gradient S thermocycler (Eppendorf North America Inc., New York, NY, USA) programmed for an initial 3-min denaturation step at 95 °C, followed by 38 cycles of 20 s at 95°C, 20 s at 55°C, 1 min at 68°C, and a final extension of 3 min at 68°C. Amplification was visualized by electrophoresis of 5 μl of each PCR product on a 1.5% agarose gel stained with ethidium bromide. To estimate the size of the PCR products, a 100 bp molecular weight marker was used (Fermentas). Verfied Trichogramma evanescens DNA was used as a positive control and the PCR reagent mix without DNA as a negative control.
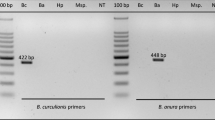
Visualization of the species-specific multiplex PCR products identifying two species of Trichogramma that parasitize the eggs of the European corn borer, in Serbia. Lane legend, from right to left by ordered numbers: 1 T. evanescens; lanes 2–10 T. brassicae; lane 11 no template control; lane 12 positive control T. evanescens; lane 13 100bp DNA size standard (Fermentas Inc, USA). Products were run on a 1.5% agarose gel, stained with ethidium bromide
Identification of native populations of Trichogramma
Trichogramma wasps were reared from parasitized egg clusters of ECB collected from the leaves of maize sampled at 12 different corn fields/plots in Vojvodina (Fig. 2). Sampling took place between late July and late September 2017 in correspondence with the activity of II and III generations of ECB (Čamprag et al., 1983). At each site, 100 randomly selected plants of corn were inspected for the presence of ECB egg clusters using protocols detailed in Reid et al. (1996). Four of the sites were sampled on a single occasion (Čoka, Senta, Vrbas, and Gakovo), three sites were sampled on 2 different dates (Bašaid, Banatska Topola, and Stankić), 4 sites were sampled on 3 different dates (Mokrin, Iđoš, Nakovo, and Despotovo), and only one site was sampled on 4 different dates (Njegoševo) (Table 2). Across all these sampling efforts, a total of 40 parasitized egg clusters were collected and transported to the laboratory, where they were subsequently stored in individual glass tubes (25±1 °C, 65±10% RH, 16:8 h [L:D] photoperiod), until the emergence of the adult parasitoids. Following emergence, the wasps from each parasitized egg cluster were preserved in labeled vials with 95% ethanol and stored at −20 °C.
Locations in Vojvodina where corn crops were sampled in search of parasitized ECB egg masses. Samples were collected in 2017. Map was extracted from Google Maps (earth.google.com/web/) with modifications in Adobe photoshop CC
Genomic DNA was isolated from the ethanol-preserved females, using a HotSHOT method (Truett et al. 2000), following the protocol described by Ivezić et al. (2018). Where possible, 2 female Trichogramma were randomly selected from those that emerged from each of the 40 laboratory-reared ECB egg clusters. Specimens were then identified to species, using the multiplex-PCR assay detailed above.
Results
The species-specific mutiplex PCR assay yielded amplicons (bands) of a diagnostic size for each of the target species, T. brassicae (199bp) and T. evanescens (480bp; Fig. 1). Validation of the assay was sought by aiming to identify 2 female parasitoids emerging from each of 40 collected ECB egg clusters. This was possible for all but one egg cluster (JL40; Table 2), which only yielded a single female wasp. Thus, a total of 79 females were identified, using the species-specific multiplex PCR assay (e.g., Fig. 1). Seventy-seven out of 79 wasps (97.47%) were identified as T. brassicae, and the remaining 2 were confirmed as T. evanescens. No specimens failed to amplify. Trichogramma brassicae was detected at all 12 sample sites, while T. evanescens was detected at only 2 plots, Mokrin and Nakovo (Table 3). In both instances, the T. evanescens individual emerged from an egg cluster that also yielded a T. brassicae individual, indicating multi-parasitism of the cluster by both species. One of those egg clusters (JL14) yielded further 5 females (7 total); therefore, the molecular assay was used to identify each of them. Including the original 2 wasps, 5 of the females emerging from that cluster were T. evanescens, and 2 were T. brassicae.
Discussion
The species-specific multiplex PCR assay described herein relies on inter- and intraspecific variations in the DNA sequences of the locus that were targeted, ITS2. It worked well in the particular agricultural system that was chosen, Serbian maize production. As such, it provided an excellent diagnostic tool for monitoring the Trichogramma species present in that specific system. Whether the assay, in its current format, would be applicable to further agricultural and natural ecosystems, in Serbia or further afield, or for identifying Trichogramma from the eggs of different insect pests, is unknown. The ITS2 region was targeted because it is typically well-conserved within species. However, it is clear from the alignment of ITS2 sequences retrieved from GenBank that the locus was not 100% homogenous across all specimens/populations from different studies (supplementary file). Indeed, the species-specific primer designed for T. evanescens actually spanned a variable stretch which was subject to a 2–4 bp insertion in 10 of the 50 (20%) existing GenBank accessions. Without access to that DNA, there was no way to certain that the assay would result in the amplification of DNA from those specimens, although there was some hope in the fact that the first 9 bases from the 3′ end of the primer were identical across all 50 accessions. It cannot be certain exactly how the introduction of further species into the Serbian maize system might affect the assay, but non-significant similarity was found between the species-specific primers and the ITS2 sequences of 8 further Trichogramma species that are common in the Mediterannean region (Sumer et al. 2009). It was more likely that specimens of additional species would simply fail to produce any PCR product. As such, the integrity of the extracted DNA of any specimen that did not yield an amplicon should be confirmed and followed by the amplification and sequening of the complete ITS2 gene. If a “new” species was detected, it was then likely that it could be included in a redesigned assay.
The identification of arthropod pests and natural enemies using traditional morphological keys and/or DNA sequences of appropriate genes is critical, if expensive and time-consuming part of surveying any new area/ecosytem. Compared with traditional morphological identification, which requires lengthy processing and slide mounting of specimens, coupled with expert knowledge of the genus, the multiplex PCR assay allowed the accurate diagnosis of a large number of Trichogramma specimens in just a few hours. Importantly, such a diagnosis can be completed by a person that has training only in relatively straightforward laboratory techniques (DNA extraction, PCR, and gel electrophoresis), thus helping to overcome the taxonomic impediment that may be associated with identifying large numbers of very small parasitoid wasps. A further advantage of the assay was that it allows the identification of females, which, in Trichogramma, can often only be identified by their association with males (Pinto, 1997). This is particulalry important since Trichogramma populations often display heavily female-biased sex-ratios (Stouthamer et al., 1999).
ECB control in maize, in Serbia, is a very complex task, primarily due to the difficulties associated with the application of insecticides in advanced stages of corn development and because of the long oviposition period of ECB females. Given the negative side effects of chemical insecticides, the use of alternative strategies for ECB control should be considered by relevant decision-making and biological control stakeholders. The use of the multiplex assay in this study again demonstrated the widespread occurrence of 2 resident species of Trichogramma parasitizing the eggs of ECB. Coupled with the findings of surveys conducted in 2016 and 2018 (Ivezić et al. 2018, 2020), it is clear that Trichogramma species, particularly T. brassicae, are important, naturally occuring egg parasitoids of ECB in Kikinda and other areas of Serbia. Indeed, natural rates of parasitism by Trichogramma spp. have been reported to be as high as 70% in north-west Serbia, although that rate fluctuates year to year (Tancik, 2017). Collectively, these studies strongly suggest the potential for using Trichogramma in a biological control program. While the multiplex PCR assay described herein provided a useful tool for surveying naturally occuring Trichogramma populations, should future management plans for ECB in Serbia adopt an approach that does incorporate biological control, this assay (or something similar) may also provide an efficient means of monitoring the immediate success of field-released wasps and/or longer term genetic trends within parasitoid populations (Roltsch et al. 2021). Furthermore, this assay has a practical application for simply assisting with quality control in any future mass-rearing facilities, ensuring that rearing stocks are not invaded by another strain or species.
Conclusions
It is clear that the naturally occuring egg parasitoid T. brassicae and to a much lesser extent T. evanecens parasitize a significant number of ECB egg masses in Serbian maize. As such, Serbian agriculture should seriously consider developing a biological control plan against ECB that focuses on the augementative release of mass-reared T. brassicae. Species-specific multiplex PCR can provide a quick, cheap, and reliable tool to assist the planning, implemantation, and future monitoring of such augmentative releases.
Availability of data and materials
All data generated or analyzed during this study are included in this published manuscript and its supplementary information files.
Abbreviations
- ECB:
-
European corn borer
- ITS2:
-
Internal transcribed spacer 2
- COI:
-
Mitochondrial cytochrome oxidase subunit 1 gene
References
Borba RS, Garcia MS, Kovaleski A, Oliveira AC, Zimmer PD, Castelo Branco JS, Malone G (2005) Dissimilaridade genética de linhagens de Trichogramma Westwood (Hymenoptera: Trichogrammatidae) através de marcadores moleculares ISSR. Neotropical Entomology 34(4):565–569. https://doi.org/10.1590/S1519-566X2005000400005
Čamprag D, Krnjaić Đ, Maceljski M, Maček J, Marić A, Vrabl S (1983) Priručnik izveštajne i prognozne službe zaštite poljoprivrednih kultura. Savez društava za zaštitu bilja Jugoslavije, Beograd, Jugoslavija
Fahriye S., Tuncibilek AS, Oztemiz S, Pintureau B, Rugman-Jones P, Stouthamer R (2009) Molecular key to the common species of Trichogramma of the Mediterranean region. BioControl 54:(5)617-624.
Golbaghi FH, Goldansaz SH, Rahmani S, Attaran MR (2020) Preference and performance of Trichogramma embryophagum when parasitizing Cydia pomonella and two stored-product moths. Bulletin of Insectology 73(1):79–86
Grushevaya I (2020) Prevalence rates of Trichogramma evanescens (Westwood) in Ostrinia nubilalis (Hübner) population in the south of Russia in 2013-2018. IV All-Russian Plant Protection Congress. 18. https://doi.org/10.1051/bioconf/20201800012
Ivezić A, Rugman-Jones PF, Stouthamer R, Ignjatović-Ćupina A (2018) Molecular identification of Trichogramma egg parasitoids of Ostrinia nubilalis in the north eastern Serbia. Archives of Biological Sciences 70(3):425–432. https://doi.org/10.2298/ABS171103002I
Ivezić A, Rugman-Jones PF, Thibaut M, Ris N, Ignjatović-Ćupina A (2020) Molecular identification of Trichogramma species parasitizing Ostrinia nubilalis in corn and pepper in south-east border of Europe. International Journal of Pest Management. https://doi.org/10.1080/09670874.2020.1779383:1–12
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30(4):772–780. https://doi.org/10.1093/molbev/mst010
Latković D, Crnobarac J, Jaćimović G, Visković J (2018) Production of corn in Serbia in the light of climate change. Acta Agraria Debreceniensis 305-322 DOI:https://doi.org/10.34101/actaagrar/150/1726.
Maissle M, Mouron P, Musa T, Bigler F, Pons X, Vasileiadis VP et al (2010) Pests, pesticides use and alternative options in Eropean maize production: current status and future prospects. J Appl Entomol 134:357–375
Phoofolo MW (1997) Evaluation of natural enemies of the European corn borer, Ostrinia nubilalis (Lepidoptera: Pyralidae). PhD Thesis, Iowa State University p. 171.
Pinto JD (1997) Chapter 22. Trichogrammatidae. In: Gibons, G.A.P., Hubner, J.T., J.B., Woolley (Eds.), Annotated keys to the genera of Neartic Chalcidoidea (Hymenoptera). NRC research press, Ottawa, Canada, pp. 726-752.
Reid, LM, Hamilton, RI, Mather DE (1996) Screening maize for resistance to Gibberella ear rot. Agriculture and Agri-Food Canada technical bulletin 40 p.
Roltsch WJ, Bürgi LP, Tomic-Carruthers N, Rugman-Jones PF, Stouthamer R, Mills NJ (2021) Mortality of light brown apple moth egg masses in coastal California: impact of resident Trichogramma parasitism and predation. Biological Control 152:104465 https://doi.org/10.1016/j.biocontrol.2020.104465
Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In: Krawet S, Misener S (Eds.), Bioinformatics methods and protocols: methods in molecular biology. Humana Press.pp. 365–386.
Rugman-Jones PF, Roltsch WJ, Stouthamer R (2020) Species-specific multiplex PCR for the rapid diagnosis of egg parasitoids of light brown apple moth, Epiphyas postvittana, in northern California. Biocontrol Science and Technology 30(6):548–558. https://doi.org/10.1080/09583157.2020.1743817
Stouthamer R, Hu J, Van Kan FJPM, Platner GR, Pinto JD (1999) The utility of internally transcribed spacer 2 DNA sequences of the nuclear ribosomal gene for distinguishing sibling species of Trichogramma. BioControl, 43(4):421–440. https://doi.org/10.1023/A:1009937108715
Sumer F, Tuncbilek AS, Oztemiz S, Pintureau B, Rugman-Jones P, Stouthamer R (2009) A molecular key to the common species of Trichogramma of the Mediterranean region. BioControl 54(5):617–624. https://doi.org/10.1007/s10526-009-9219-8
Tancik J (2017) Natural parasitism of the second generation European Corn Borer Eggs Ostrinia nubilalis (Hubner) (Lepidoptera, Pyralidae) by Trichogramma spp. in sweet corn fields in Vojvodina, Serbia. Plant protection science 53(1):50–54
Truett GE, Heeger P, Mynatt RL, Truett AA, Walker JA, Warman ML (2000) Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). BioTechniques 29(1):52–54. https://doi.org/10.2144/00291bm09
Van Schelt J, Ravensberg WJ (1991) Some aspects on the storage and application of Trichogramma maidis in corn. In: Wajnberg E, Vinson SB (eds) The Third International Symposium on Trichogramma and other Egg Parasitoids. San Antonio, USA, pp 239–242
Acknowledgements
The authors are grateful to Milena Marčić, Jelena Petrović, Melita Dejanović, Dragan Kerkez, Dragana Dragomirov, and Mladen Đuran from the Forecasting and Warning Service in Plant Protection of Serbia for the help in the field and laboratory work.
Funding
The study was supported by the Forecasting and Warning Service of Plant Protection in Serbia and Department of Entomology, University of California Riverside, USA.
Author information
Authors and Affiliations
Contributions
AI conducted field and laboratory work and participated in the interpretation of the obtained results as well as in writing of the manuscript. PRJ designed species-specific primers for multiplex PCR and was an important contributor in data analysis and writing of the manuscript. BT revised the manuscript and made a significant contribution to the final version of submitted paper. All authors contributed equally to this work. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1..
All ITS2 for alignment and multiplex primer design
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ivezić, A., Rugman-Jones, P.F. & Trudić, B. Rapid molecular identification of Trichogramma (Hymenoptera: Trichogrammatidae) parasitizing the eggs of the European corn borer, Ostrinia nubilalis (Lepidoptera: Crambidae) in Serbia. Egypt J Biol Pest Control 31, 74 (2021). https://doi.org/10.1186/s41938-021-00414-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41938-021-00414-5