Abstract
Transgenic Bt cotton with genes from soil inhabiting spore forming bacterium Bacillus thuringiensis Berliner produces δ-endotoxin for the control of lepidopteran insects. The prey-mediated effects of Cry protein on the third trophic level is the most realistic exposure pathway that needs to be addressed as an important component of environment risk assessment. The green lacewing, Chrysoperla zastrowi sillemi (Esben-Petersen) (Neuroptera: Chrysopidae) is the most important generalist predator in the cotton ecosystem in India. The tri-trophic interactions involving Bt cotton expressing single (Cry1Ac) and dual toxins (Cry1Ac and Cry2Ab) fed herbivores, i.e. mealybug, Phenacoccus solenopsis Tinsley, whitefly Bemisia tabaci (Gennadius) and leafhopper, Amrasca biguttula biguttulla (Ishida) on the fitness of C. zastrowi sillemi, were studied. The development, survival and body weight of C. zastrowi sillemi had no deleterious effect as there were insignificant differences in any of the studied fitness parameters regardless of having consumed prey fed on Bollgard, Bollgard II and non-Bt cotton plants. The feeding potential of C. zastrowi sillemi on mealybug was also not different on Bt or non-Bt cotton plants. ELISA studies confirmed the presence of Cry proteins in Bt cotton leaves; however, no Cry1Ac or Cry2Ab protein was detected in prey herbivores (P. solenopsis, B. tabaci and A. biguttula biguttula) or in the predator C. zastrowi sillemi. It could be concluded that transgenic cotton that expresses single (Cry1Ac) or dual (Cry1Ac and Cry2Ab) toxins had no apparent effect on the fitness of the predator through its preys P. solenopsis, B. tabaci and A. biguttula biguttulla.
Similar content being viewed by others
Background
Genetically modified cotton with genes from soil inhabiting spore-forming bacterium Bacillus thuringiensis Berliner (Bt) produces δ-endotoxin, which is lethal to many lepidopteran pests. The area planted under biotech upland cotton globally in 2017 was 24.1 million hectares, India, having the largest area. In India, the area under Bt cotton has increased from 50,000 ha in 2002 to 11.4 million hectares in 2017, representing an unprecedented 227-fold jump in 16 years (James 2017). Initially, the cotton scenario was dominated by Bt cotton varieties/hybrids producing a single Cry protein (Cry1Ac), but these have now been replaced by a second generation of cotton producing two Cry proteins (Cry1Ac and Cry2Ab) (Kumar et al. 2014). The adoption of transgenic Bt cotton in countries such as India has changed the entire pest scenario in the cotton ecosystem. The pest status of bollworms and leaf-feeding insects has declined, but sap feeders, including whitefly, Bemisia tabaci (Gennadiaus); leafhopper, Amrasca biguttula biguttula (Ishida); mealybug, Phenacoccus solenopsis Tinsley; thrips, Thrips tabaci (Lindemann); aphid, Aphis gossypii (Glover); and mirid, Creontiades biseratense (Distant), have emerged as serious pests (Kumar et al. 2015).
Studies conducted in Australia, USA, China and India have indicated that the Cry proteins in transgenic Bt cotton are available throughout the cropping season with temporal and spatial variations (Kranthi et al. 2005 and Shera and Arora 2016). So the target and non-target arthropods are continuously exposed directly or indirectly to Cry proteins, expressed in Bt cotton plants. At the third trophic level, predators and parasitoids may get exposed to Cry proteins, when they feed on herbivores that have consumed plant tissues having Bt protein (Torres et al. 2006; Mota et al. 2012; and Kumar et al. 2014). Exposure on the third trophic level through prey is the most realistic exposure pathway that needs to be addressed as an important component of environment risk assessment.
The chrysopids (Neuroptera) commonly known as lacewings or aphid lions are among the most beneficial predators of agricultural ecosystems and a potent arsenal of biological control. Chrysoperla zastrowi sillemi (Esben-Petersen), previously known as Chrysoperla carnea (Stephen), is the most important generalist predator in the cotton ecosystem in India. Adults are free living while larvae feed on soft-bodied insects like aphids, whitefly, leafhoppers, thrips, mites, and eggs and larvae of many lepidopteran pests (Takalloozadeh 2015). Thus, this predator can be exposed to Cry toxins by feeding on preys, which may affect its fitness parameters and thus hamper its performance as a biocontrol agent.
Most studies on the ecological impact of transgenic Bt cotton (Cry1Ac) or Bt corn (Cry1Ab) on the chrysopids (C. carnea, C. rufilabris, C. externa and Chrysopa pallens), using non-target preys, have focused only on aphids or mites (Torres et al. 2006 and Mota et al. 2012). Studies on the interactions involving Bt cotton, non-target herbivores (P. solenopsis, B. tabaci and A. biguttula biguttula) and their predator C. zastrowi sillemi have not been previously reported.
The objectives of the study were (1) to quantify the impact of cotton cultivars expressing single toxin (Bollgard; Cry1Ac), dual toxins (Bollgard II; Cry1Ac + Cry2Ab) and non-toxin on the survival, development, and body weight of the predator C. zastrowi sillemi through non-target sucking insect pests (mealybug, whitefly and leafhopper); (2) to study the predatory potential of the predator when fed on cotton mealybug P. solenopsis reared on Bt cotton; and (3) to study whether Cry1Ac and Cry2Ab proteins can pass via the food chain to the third trophic level.
Materials and methods
Plant material
Two transgenic Bt cotton hybrids, MRC 6301 Bt (event Mon 831) and Ankur 3028 BG-II (event 15895), approved by Genetic Engineering Approval Committee (GEAC) in India and one non-Bt variety, LH 2108, recommended by Punjab Agricultural University, Ludhiana, were used for the experiments. MRC 6301 Bt plants express Cry1Ac gene from B. thuringiensis targeting bollworms, while Ankur 3028 BG-II plants express Cry1Ac and Cry2Ab genes targeting bollworms and leaf-feeding insects (Navarro and Hautea 2014). Seeds were individually sown in earthen pots (12 l), filled with humus rich soil and plants were raised for rearing of herbivores and further experimentation.
Insect materials (preys)
Plants raised in pots were used for rearing of sucking insect pests P. solenopsis, B. tabaci and A. biguttula biguttula in separate insect proof screen cages (1.5 × 1.5 × 1.5 m) under field conditions. The insects were initially collected from field-grown eggplant, Solanum melongena L., and okra, Abelmoschus esculentus (L.) Moench, crops. Different screen cages were also used for Bollgard, Bollgard II and non-Bt cultivars to prevent insects from moving between cultivars. The plants of the respective cotton cultivar/variety were changed from time to time for continuous supply of fresh food to the insects and availability of their culture throughout the study period. The cultures of all insects used as preys in bioassays were maintained for multiple generations.
Predator
The culture of C. zastrowi sillemi was maintained on eggs of Corcyra cephalonica (Stainton) (Lepidoptera: Pyralidae), as a factitious host, in the Biocontrol Laboratory at Entomological Research Farm, Punjab Agricultural University, Ludhiana. Field-collected C. zastrowi sillemi adults were released in specially designed wooden oviposition cage (50 × 30 × 20 cm; Amar Chand & Company, India) with a sliding roof plank, covered with black muslin cloth. Standard adult diet (honey 1 g, sucrose 5 mg, protein 5 g, yeast 1 g and distilled water 40 ml) was provided twice daily as droplets on Perspex sheet strips by the help of a fine camel hair brush (Sattar and Abro 2011). The stalked eggs laid by the females on the roof plank were destalked after 24 h by the help of a sterilised razor blade. These eggs were transferred to individual plastic vials (4 × 3 cm) by the help of a soft camel hair brush. After hatching, fresh eggs of C. cephalonica were provided daily in each individual vial to the Chrysoperla larva till pupation. The adults emerging from the cocoons were collected individually and transferred again to the oviposition cages. The newly emerged adults were provided by a nutritional diet as described earlier. The culture of C. zastrowi sillemi was used for further experimentations.
Tri-trophic bioassay with C. zastrowi sillemi
Development, survival and body weight
All bioassays were conducted in an environmental chamber at 27 ± 2 °C and 65 ± 5% RH (Macro Scientific Works Ltd., India). Leaves from the upper third portion of 70-day-old Bollgard or Bollgard II or non-Bt plants were detached and placed in separate plastic jars (20 × 10 cm) lined with a muslin cloth for aeration. Similar sized mealybug nymphs (n = 50), whitefly adults (n = 50) and leafhopper nymphs (n = 50), from the stock cultures reared on Bollgard or Bollgard II or non-Bt leaves were collected and released in the respective jars. A soft camel hair brush was used for collection and release of mealybug nymphs, while an aspirator (Rescholar Equipment, India) was used to collect whitefly adults and leafhopper nymphs. The collected insects were released in separate plastic containers, with a hole in the screw cap. The cotton leaf was placed in each container before release of insects. The petiole of detached leaves was wrapped with water-soaked cotton swab so as to keep the leaves fresh and turgid for a longer period of time. A newly hatched single Chrysoperla larva was introduced in each plastic container for feeding. The hole was then plugged by a cotton wool. Each prey treatment (P. solenopsis, B. tabaci and A. biguttula biguttula), reared on Bollgard, Bollgard II or non-Bt leaves, was conducted simultaneously. The experiment was initiated with 30 Chrysoperla larvae for each treatment (one larva/replication). Chrysoperla larvae were inspected twice daily, and life-table parameters (development and mortality) were recorded. Last (3rd) instar Chrysoperla larvae, in each prey-predator combination, were weighed at the end of bioassay, using an electronic balance. The cocoons were collected, weighed and kept in separate glass vials individually to record the pupal period and percentages of adults’ emergence. After emergence from cocoons, adults were provided by an adult diet as described earlier and their longevity was recorded (Additional file 1).
Predatory potential
An experiment on predatory potential was conducted in an environmental chamber set at 27 ± 2 °C and 65 ± 5% RH (Macro Scientific Works Ltd., India). In no-choice tests, 2nd instar mealybug nymphs (n = 30) reared on Bollgard or Bollgard II or non-Bt leaves were released by the help of a soft camel hair brush on respective leaves in separate plastic jars (20 × 10 cm) lined with muslin cloth for aeration (10 replications for each set). A newly hatched single Chrysoperla larva was introduced in each jar for feeding. The number of mealybugs consumed were counted daily and replaced with new sets till pupation. The mean consumption of mealybugs per day was worked out for each larval instar.
Cry toxin expression in tri-trophic pathway (cotton leaves, prey and predator)
To confirm Cry1Ac and Cry2Ab expression in bioassays, leaf samples were collected from the upper third portion of 70-day-old Bollgard (Cry1Ac) or Bollgard II (Cry 1Ac and Cry2Ab) or non-Bt plants. Leaf discs measuring 20 mm per replication were placed in separate 1.5 ml centrifuge tubes, and Cry protein measurements, using enzyme-linked immunosorbent assays (ELISA; see below), were made. To quantify Bt toxin in P. solenopsis nymphs, B. tabaci adults and A. biguttula biguttula nymphs, protein was extracted from each prey herbivore, reared either on Bollgard or Bollgard II or non-Bt leaves. Similarly, to assess the potential transfer of Cry1Ac and Cry2Ab proteins via food chain, C. zastrowi sillemi larvae fed on prey herbivores reared on Bollgard or Bollgard II or non-Bt leaves were collected and assayed, using ELISA. For all ELISA samples, five replications were maintained.
ELISA procedures
Bt protein concentrations in cotton leaves, insect prey and predator were measured, using sandwich ELISA, using quantiplate kits (Wu et al. 2014). The concentration of Cry1Ac protein was measured, using Cry 1Ab/Cry1Ac kit (AP 003 QT V50) and Cry2Ab using Cry2A kit (AP 005 QT BC V50) of ENVIROLOGIX 500 Riverside Industrial Parkway Portland, ME, USA. Before analysis, all insects (preys and predator) were washed in phosphate-buffered saline with Tween-20 buffer to remove any protein from their outer surface. Leaf samples weighing 20 mg were homogenised in 0.5 ml buffer solution and diluted (1: 10 and 1: 50) for Cry1Ac and Cry2Ab, respectively. Quantities (mg/replication) of material and buffer dilutions used from different prey herbivores and predator were ≈20 mg P. solenopsis (in batch) homogenised in 0.5 ml buffer (no dilution), ≈20 mg B. tabaci (in batch) homogenised in 0.5 ml buffer (no dilution), ≈20 mg A. biguttula biguttula (in batch) homogenised in 0.5 ml buffer (no dilution) and 15 ± 2 mg C. zastrowi sillemi homogenised in 0.5 ml buffer (no dilution). All the samples in buffer were ground by hands, using a plastic pestle. For every sample, a fresh pestle was used to avoid any possible cross contamination of the individual sample. After vortexing for 3 h on a vortex shaker (Spinix; Tarson Products Ltd., India), centrifugation for 1 min (at 10,000 rpm in microcentrifuge) (Eppendorf AG 5415D, Germany) and appropriate dilution of supernatants, ELISA was performed according to manufacturer’s protocol. The absorbance of each well solution was recorded at 450 nm by using micro filter plate reader (Thermo Electron Corporation, China). The optical density (OD) value of each calibrator and corresponding concentrations of Cry1Ac and Cry2Ab (standards provided in the kit) were used to prepare the standard curve. The proteins’ concentration of each sample was determined by finding its OD value and the corresponding concentration level in the linear curve, using regression analysis. The results from standard curve were multiplied by the dilution factor incurred during extraction. To determine the dilution factor, the volume in milliliter of extraction solution was divided by weight of samples in grams. For leaf samples, protein concentrations in μg g−1 fresh weight were calculated by multiplying these results by (1:10 or 1:50) dilutions made for Cry1Ac and Cry2Ab, respectively.
Data analyses
The data on the life parameters (larval period, pupal period and adult longevity) and weight (larval and cocoon weight) were subjected to one-way ANOVA and Tukey’s multiple range test. Data are presented as mean ± standard error. Data on Chrysoperla larval and pupal survivals were analysed and presented in the form of Wald Chi-square χ2 test and P values. Data on feeding performance were subjected to two-way ANOVA and Tukey’s multiple range tests. All statistical tests were carried out using IBM SPSS 22.0 for Windows (IBM Corporation, Armonk, New York, USA).
Results and discussion
Development, survival and weight of C. zastrowi sillemi fed on P. solenopsis reared on Bt and non-Bt cotton leaves
Different life parameters, i.e. larval period (F = 0.01; df = 2.70; P = 0.994), pupal period F = 0.08; df = 2.60; P = 0.927) and adult longevity (F = 0.01; df = 2.60; P = 0.987) of C. zastrowi sillemi did not differ when fed on P. solenopsis reared on Bollgard or Bollgard II or non-Bt cotton leaves (Table 1). There was also insignificant difference in larval (χ2 = 0.324; df = 2; P = 0.956) and pupal survival (χ2 = 0.333; df = 2; P = 0.954) among the treatments. The prey mediated effects of Bollgard or Bollgard II on the body weight of last larval instar (F = 0.09; df = 2.57; P = 0.912), cocoon (F = 0.04; df = 2.57; P = 0.958) and adults (F = 0.06; df = 2.27; P = 0.938) of Chrysoperla also showed insignificant difference than the non-Bt cotton (Table 1).
Development, survival and weight of C. zastrowi sillemi fed on B. tabaci reared on Bt and non-Bt cotton leaves
The prey (B. tabaci)-mediated effects of Bt cotton, expressing single (Cry1Ac) or dual (Cry1Ac Cry2Ab) toxin, on the larval (F = 0.01; df = 2.73; P = 0.991) and pupal periods (F = 0.08; df = 2.60; P = 0.926) of C. zastrowi sillemi were not different than the non-Bt cotton. Similarly, the larval survival (χ2 = 0.317; df = 2; P = 0.957) and pupal survival (χ2 = 0.463; df = 2; P = 0.927) survival also were not significantly affected by the treatments. The body weight of larvae (F = 0.03; df = 2.57; P = 0.971), and cocoons (F = 0.07; df = 2.57; P = 0.929) had insignificant difference when Bollgard or Bollgard II or non-Bt cotton leaves fed B. tabaci adults were offered to Chrysoperla as prey (Table 2).
Development, survival and weight of C. zastrowi sillemi fed on A. biguttula biguttula reared on Bt and non-Bt cotton leaves
The developmental period of C. zastrowi sillemi immature stages, i.e. larval (F = 0.01; df = 2.87; P = 0.980) and pupal periods (F = 0.07; df = 2.54; P = 0.930), had insignificant difference, when fed on the prey (A. biguttula biguttula), reared on cotton cultivars expressing single (Cry1Ac) or dual (Cry1Ac and Cry2Ab) or no toxin. Further, there were also insignificant differences in larval (χ2 = 0.342; df = 2; P = 0.952) and pupal survivals (χ2 = 0.375; df = 2; P = 0.945) of the predator among different treatments. Larval (F = 0.03; df = 2.57; P = 0.957) and cocoon weights (F = 0.02; df = 2.54; P = 0.979) of Chrysoperla were also not affected, regardless to the prey that had fed on cotton cultivars with or without toxin (Table 3).
Bt proteins in genetically modified crops may pose a risk to non-target beneficial arthropods providing biological control, pollination and decomposition services in the ecosystem. The possible lethal effects posed by insecticidal proteins in biological control agents can be determined under laboratory studies in different ways (Tian et al. 2013): (1) directly, the organism can be exposed to the protein incorporated in an artificial diet or mixed with non-genetically and genetically modified plant materials; and (2) indirectly, predatory species can be offered target or non-target prey herbivores fed on genetically modified plants. The latter way involving prey-mediated effects at higher trophic level provides a very realistic exposure pathway and has been experimented in numerous tri-trophic interaction studies with genetically modified Bt crops. Target prey herbivores susceptible to Bt proteins may result in prey quality-mediated effects at the third trophic level. One way to avoid this impact is to use non-target herbivores, which are not susceptible to proteins expressed in transgenic plants (Romeis et al. 2011).
Obtained results revealed that the larval period and survival, pupal period and survival, and adult longevity of C. zastrowi sillemi as well the body weight (larval, cocoon or adult) of the predator were not different when the predator was offered P. solenopsis or B. tabaci or A. biguttula biguttula as food that had consumed cotton leaves expressing single toxin (Cry1Ac) or two toxins (Cry1Ac and Cry2Ab) or no toxin. Non-significant differences might be due to the fact that Cry1Ac and Cry2Ab proteins were not detected in any of the herbivore (lack of exposure) and consequently not reflected in the third trophic level. The results agree with Guo et al. (2008). Similar findings have been reported in tri-trophic interactions between Bt cotton (Cry1Ac), A. gossypii and C. carnea (Magar et al. 2012) and C. externa (Mota et al. 2012). In a comprehensive review, Romeis et al. (2014) concluded that no direct impact on survival and developmental parameters has also been noticed in C. carnea feeding on Bt maize pollen containing Cry 1Ab or Cry 3Bb1 (Li et al. 2008) and Cry 1Ab protein mixed in sucrose diet (Romeis et al. 2014). Simon et al. (2006) reported that Cry1Ab or Cry2Ab toxins did not have detrimental effects on C. carnea when ingested either directly or through prey as the larval midgut lacks specific receptors for these proteins. No prey-mediated effects of Cry proteins have also been reported in other predatory species including the coccinellids (Wu et al. 2014), the rove beetles (Garcia et al. 2010) and the wolf spiders (Niu et al. 2017).
Predatory potential of C. zastrowi sillemi fed on P. solenopsis reared on Bt and non-Bt cotton leaves
Daily consumption rate of P. solenopsis by C. zastrowi sillemi on non-Bt cotton was not different than the Bt cotton expressing single (Cry1Ac) or dual (Cry1Ac Cry2Ab) toxin (F = 0.04; df = 2.81; P = 0.961) (Table 4). However, consumption by different larval instars were significantly different from each other (F = 1803.73; df = 2.81; P < 0.001). Third instar Chrysoperla larvae consumed more preys, followed by the 2nd and 1st instar larvae. The interaction between the factors, cotton type and Chrysoperla larval stages, was also insignificant and indicating that differential consumption rate by larval stages was not a function of the cotton type (Bt or non-Bt cotton) (F = 0.15; df = 4.81; P = 0.861).
Quantification of Cry proteins in tri-trophic pathway
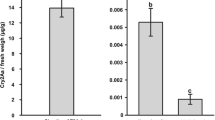
The concentration of Cry1Ac protein in MRC 6301 Bt cotton leaves averaged (2.52 ± 0.02 μg/g fresh weight). The expressed levels of Cry1Ac and Cry2Ab proteins in Ankur 3028 BG-II leaves averaged (1.95 ± 0.02 μg/g) and (21.64 ± 0.32 μg/g) fresh weight, respectively. ELISA studies revealed that prey herbivores (P. solenopsis, B. tabaci and A. biguttula biguttula) reared on Bollgard and Bollgard II leaves did not contain detectable amounts of the Cry1Ac or Cry2Ab protein. Similarly no Cry1Ac or Cry2Ab protein was detected in C. zastrowi sillemi larvae fed on any of the three prey herbivore. None of non-Bt cotton leaves or prey herbivores or C. zastrowi sillemi samples was found to contain any Cry1Ac or Cry2Ab protein in respective tri-trophic pathway.
The expression of Cry proteins in Bollgard (Kranthi et al. 2005) and Bollgard II (Kranthi et al. 2009) cotton have been known to be high in leaves as compared to other plants parts. In the present study, high concentrations of both Cry1Ac and Cry2Ab proteins in plant leaves were recorded. Moreover, Cry2Ab protein was expressed at higher level in Bollgard II than Cry1Ac which is consistent with earlier studies (Kranthi et al. 2009 and Shera and Arora 2016).
The majority of earlier studies that aimed to quantify the Cry protein concentration in non-target herbivores that have fed on Bt transgenic plants have shown Cry proteins, either absent or detected at very low levels. Highly varying concentrations of Cry proteins in herbivores depend on their feeding mode and feeding location on the plant (Eisenring et al. 2017). Phloem feeders (mealybugs, whiteflies, aphids) do not ingest the Cry proteins, but cell content feeders (spider mites), herbivores that do not target the phloem (thrips, plant bugs), or tissue feeders (beetles, caterpillars) ingest relatively high concentrations of Cry proteins (Eisenring et al. 2017; Meissle and Romeis 2018). Raps et al. (2001) reported that Bt toxin is not present in the phloem sap, so aphids do not ingest the toxin feeding on Bt maize plants, thereby excluding its direct effects on aphids or on other phloem feeding arthropods. Similarly, the toxin content (Cry1Ab) from Bt maize was found to be negligible in leafhoppers (Obrist et al. 2006). In contrast, the leafhoppers Empoasca pteridis (Dahlbom), Eupteryx atropunctata (Goeze) and Zyginidia scutellaris (Herrich-Schäffer) also feed on mesophyll cells and thus ingest certain amount of Cry protein. This has been shown previously for Z. scutellaris, which was found to acquire certain amount of Cry protein (Cry1Ab) from Bt maize (Dutton et al. 2004).
Conclusions
It could be concluded that phloem feeders like P. solenopsis, B. tabaci and A. biguttula biguttula contained no detectable Cry proteins (Cry1Ac or Cry2Ab) despite their high expression levels in Bollgard or Bollgard II cotton leaves. Considering that the green lacewing C. zastrowi sillemi is mostly a generalist predator of phloem feeders in the cotton crop, it is unlikely that these non-target herbivores feeding on Bt cotton pose any risk to this beneficial predator. Thus, this important generalist predator will continue to render its biological control services in the cotton ecosystem dominated by transgenic Bt cotton cultivars.
References
Dutton A, Obrist L, D’ Alessandro M, Diener L, Muller M, Romeis J, Bigler F (2004) Tracking Bt-toxin in transgenic maize to assess the risks on non-target arthropods. IOBC/WPRS Bulletin 27:57–63
Eisenring M, Romeis J, Naranjo SE, Meissle M (2017) Multitrophic Cry-protein flow in a dual-gene Bt-cotton field. Agric Ecosyst Environ 247:283–289
Garcia M, Ortego F, Castanera P, Farinoa GP (2010) Effects of exposure to the toxin Cry1Ab through Bt maize fed prey on the performance and digestive physiology of the predatory rove beetle Atheta coriara. Biol Ctrl 55:225–233
Guo JV, Wan FH, Dong L, Lovei GL, Han AJ (2008) Tri-trophic interactions between Bt cotton, the herbivore Aphis gossypii Glover (Homoptera: Aphididae), and the predator Chrysopa pallens (Rambur) (Neuroptera: Chrysopidae). Environ Ent 37:263–270
James C (2017) Global status of commercialized biotech/GM crops in 2017: biotech crop adoption surges as economic benefits accumulate in 22 years. ISAAA Briefs No. 53. International Service for the Acquisition of Agri-Biotech Applications (ISAAA), Ithaca
Kranthi KR, Naidu S, Dhawad CS, Tatwawadi A, Mate K, Patil E, Bharose AA, Behere GT, Kranthi S (2005) Temporal and intra-plant variability of CrylAc expression in Bt cotton arid its influence on the survival of the cotton bollworm, Helicoverpa armigera (Hubner) (Noctuidae: Lepidoptera). Curr Sci 89:291–298
Kranthi S, Dhawad CS, Naidu S, Bharose AA, Chaudhary A, Sangode V, Nehare SK, Bajaj SR, Kranthi KR (2009) Susceptibility of the cotton bollworm, Helicoverpa armigera (Hubner) (Lepidoptera: Noctuidae) to the Bacillus thuringiensis toxin Cry2Ab before and after the introduction of Bollgard-II. Crop Prot 28:371–375
Kumar R, Tian JC, Naranjo SE, Shelton AM (2014) Effects of Bt cotton on Thrips tabaci (Thysanoptera: Thripidae) and its predator, Orius insidiosus (Hemiptera: Anthocoridae). J Econ Ent 107:927–932
Kumar V, Dhawan AK, Shera PS (2015) Transgenic cotton in India: ten years and beyond. In: Singh B, Arora R, Gosal SS (eds) Biological and molecular approaches in pest management. Scientific Publishers, Jodhpur, pp 202–227
Li Y, Meissle M, Romeis J (2008) Consumption of Bt maize pollen expressing Cry1Ab or Cry3Bb1 does not harm adult green lacewings, Chrysoperla carnea (Neuroptera: Chrysopidae). PLoS One 3:2909
Magar PN, Satpute NS, Madankar SS (2012) Biological parameters of Chrysoperla carnea Stephens as influenced by Bt and non-Bt cotton fed insect pests. J Cotton Res Dev 26:230–233
Meissle M, Romeis J (2018) Transfer of Cry1Ac and Cry2Ab proteins from genetically engineered Bt cotton to herbivores and predators. Insect Sci 25:823–832
Mota TA, Fernandes MC, de Souza MF, da Fionseca PRB, de Quadros JC, Kassab AO (2012) Tritrofic interactions between Bt cotton plants, the aphid Aphis gossypii Glover, 1827 (Hemiptera: Aphididae), and the predator, Chrysoperla externa (Hagen, 1861) (Neuroptera: Chrysopidae). Afr J Agri Res 7:5919–5924
Navarro MJ, Hautea RA (2014) Adoption and up take pathways of GM/biotech crops by small-scale, resource-poor farmers in China, India, and the Philippines. ISAAA Brief No. 48, Ithaca
Niu L, Mannakkara A, Qui L, Hua X, Lei C, Juan LJ, Ma W (2017) Transgenic Bt rice lines producing Cry1Ac, Cry2Aa or Cry1Ca have no detrimental effects on Brown Planthopper and Pond Wolf Spider. Sci Rep 7:1940. https://doi.org/10.1038/s41598-017-02207
Obrist LB, Dutton A, Albajes R, Bigler F (2006) Exposure of arthropod predators to Cry1Ab toxin in Bt maize fields. Ecol Ent 31:143–154
Raps A, Kehr J, Gugerli P, Moar WJ, Bigler F, Hilbeck A (2001) Immunological analysis of phloem sap of Bacillus thuringiensis corn and of the nontarget herbivore Rhopalosiphum padi (Homoptera: Aphididae) for the presence of Cry1Ab. Mol Ecol 10:525–533
Romeis J, Meissle M, Naranjo SE, Li Y, Bigler F (2014) The end of myth – Bt (Cry1Ab) maize does not harm green lacewings. Front Pl Sci 5. https://doi.org/10.3389/fpls.2014.00391
Romeis J, Richard LH, Marco PC, Keri C, Adinda DS, Gatehouse AMR, Herman RA, Huesing JE, Mclean AM, Raybould A, Shelton AM (2011) Recommendations for the design of laboratory studies on non-target arthropods for risk assessment of genetically engineered plants. Transgenic Res 20:1–22
Sattar M, Abro GH (2011) Mass rearing of Chrysoperla carnea (Stephens) (Neuroptera: Chrysopidae) adults for integrated pest management programmes. Pak J Zool 43:483–487
Shera PS, Arora R (2016) Survival and development of spotted bollworm, Earias vittella (Fabricius) (Lepidoptera: Nolidae) on different transgenic Bt and isogenic non-Bt cotton genotypes. Phytoparasitica 44:99–113
Simon AR, deMaagd RA, Avilla C, Bakker PL, Molthoff J, Zamora JEG, Ferre J (2006) Lack of detrimental effects of Bacillus thuringiensis Cry toxins on the insect predator Chrysoperla carnea: a toxicological, histopathological, and biochemical analysis. App Environ Microbiol 72:1595–1603
Takalloozadeh HM (2015) Effect of different prey species on the biological parameters of Chrysoperla carnea (Neuroptera: Chrysopidae) in laboratory conditions. J Crop Prot 4:11–18
Tian JC, Wang XP, Long LP, Romeis J, Naranjo SE, Helmich RL, Wang P, Earle ED, Shelton AM (2013) Bt crops producing Cry1Ac, Cry2Ab and Cry1F do not harm the green lacewing, Chrysoperla rufilabris. PLoS One 8:e60125 https://doi.org/1journal/journal.pone.0060125
Torres JB, Ruberson JR, Adang MJ (2006) Expression of Bacillus thuringiensis Cry1Ac protein in cotton plants, acquisition by pests and predators: a tritrophic analysis. Agric Forest Ent 8:191–202
Wu H, Zhang Y, Liu P, Xie J, He Y, Deng C, Clercq P, Pang H (2014) Effects of transgenic Cry1Ac + CpTi cotton on non-target mealybug pest Ferrisia virgata and its predator Cryptolaemus montrouzieri. PLoS One 9:e95537. https://doi.org/1journal/journal.prone.0095537
Acknowledgements
The authors are thankful to the Professor & Head, Department of Entomology and Dr. Vijay Kumar, Senior Entomologist, Department of Entomology, Punjab Agricultural University, Ludhiana for providing necessary facilities.
Funding
Not applicable
Availability of data and materials
The datasets used during the current study are available from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Contributions
All authors contributed equally in the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1:
Development time, survival and weight of Chrysoperla zastrowi sillemi fed on Phenacoccus solenopsis reared on Bt and non-Bt cotton cultivars. (DOCX 31 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Shera, P.S., Karmakar, P., Sharma, S. et al. Impact of Bt cotton expressing single (Cry1Ac) and dual toxins (Cry1Ac and Cry2Ab) on the fitness of the predator Chrysoperla zastrowi sillemi (Esben-Petersen): prey-mediated tri-trophic analysis. Egypt J Biol Pest Control 28, 98 (2018). https://doi.org/10.1186/s41938-018-0102-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41938-018-0102-8