Abstract
Background
Artificial intelligence (AI) technology has been increasingly developed and studied in cardiac imaging. This systematic review summarizes the latest progress of image segmentation, quantification, and the clinical application of AI in evaluating cardiac adipose tissue.
Methods
We exhaustively searched PubMed and the Web of Science for publications prior to 30 April 2021. The search included eligible studies that used AI for image analysis of epicardial adipose tissue (EAT) or pericoronary adipose tissue (PCAT). The risk of bias and concerns regarding applicability were assessed with the Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2) tool.
Results
Of the 140 initially identified citation records, 19 high-quality studies were eligible for this systematic review, including 15 (79%) on the image segmentation and quantification of EAT or PCAT and 4 (21%) on the clinical application of EAT or PCAT in cardiovascular diseases. All 19 included studies were rated as low risk of bias in terms of flow and timing, reference standards, and the index test and as having low concern of applicability in terms of reference standards and patient selection, but 16 (84%) studies did not conduct external validation.
Conclusion
AI technology can provide accurate and quicker methods to segment and quantify EAT and PCAT images and shows potential value in the diagnosis and risk prediction of cardiovascular diseases. AI is expected to expand the value of cardiac adipose tissue imaging.
Similar content being viewed by others
Background
Artificial intelligence (AI) refers to technology in which computers or other machines simulate human intelligence to enable problem solving (Chartrand et al. 2017). The progress of AI has gradually permeated the medical field, and its fusion with medical imaging is constantly deepening (Yu et al. 2018). A large amount of image data collected in clinical practice is a rich resource for AI, which significantly improves AI models. In radiology, AI excels at identifying complex image patterns and offers more quantitative and objective evaluation methods (Hosny et al. 2018b).
Machine learning and deep learning are AI methods. Machine learning refers to computer-based algorithms that perform tasks without explicitly programming and can conduct classification or regression tasks through learning patterns from engineered features (Chartrand et al. 2017). Radiomics is an emerging field widely used to mine predefined high-throughput features extracted from medical images to support clinical decision-making, and machine learning is the primary tool for feature selection and advanced model construction in radiomics (Lambin et al. 2012; Avanzo et al. 2020). Furthermore, the training of deep learning models obtains features and patterns directly from images rather than through engineered features (Hosny et al. 2018a). Deep learning is an end-to-end learning method that directly maps the entire learning process from the original data to the desired output (Chartrand et al. 2017).
Convolutional neural networks (CNNs) are one of the most popular deep learning architectures, which utilize convolution operations that effectively decrease the number of parameters, thus requiring less training data (Litjens et al. 2019). The application of CNNs in cardiac imaging has achieved remarkable results. For example, Betancur et al. exploited a CNN-based approach to identify obstructive coronary stenosis based on myocardial perfusion imaging (MPI) in single-photon emission computed tomography (SPECT), and the approach achieved an area under the curve (AUC) that was higher than that of MPI alone (per patient 0.80 vs. 0.78; per vessel: 0.76 vs. 0.73) (Betancur et al. 2018). With the growing interest in the investigation of epicardial adipose tissue (EAT) and pericoronary adipose tissue (PCAT), AI has begun to play an active and critical role in image analysis of EAT and PCAT.
EAT is located between the visceral pericardium and the myocardium and is in direct contact with the myocardium and coronary artery; EAT functions as an endocrine organ with metabolic and inflammatory activities (Gaborit et al. 2017). It can secrete bioactive molecules (adipocytokines, proinflammatory cytokines, growth factors, etc.) and affect the myocardium and coronary artery through vasocrine or paracrine (Iacobellis et al. 2005; Cherian et al. 2012). Studies have shown that EAT is associated with cardiovascular diseases (CVDs) such as arrhythmia, coronary artery disease, and heart failure (Topuz and Dogan 2017; Abe et al. 2018; Mancio et al. 2018). In addition, there is evidence that the PCAT around the coronary artery is the source of adipocytokines, which influences the development of coronary artery disease (Wang et al. 2013; Opincariu et al. 2020).
With the increasing potential of AI in disease assessment, the role of AI in the assessment of EAT and PCAT has attracted increasingly more attention. Since this field is novel but rapidly developing, we systematically review the latest progress of image segmentation, quantification, and the clinical application of AI in EAT and PCAT imaging.
Methods
This study was conducted according to the preferred reporting items for systematic reviews and meta-analyses (PRISMA) 2020 statement (Page et al. 2021).
Information sources and search strategy
We conducted a systematic literature search of PubMed and the Web of Science prior to 30 April 2021 using terms related to artificial intelligence and epicardial adipose tissue. The reference lists of relevant studies and reviews were checked to find other possible studies. The language was restricted to English. The detailed search strategies are listed in the supplement.
Selection process and data collection
After removing duplicate articles, two reviewers (L.Z. and BB.J.) independently reviewed the titles, abstracts, and full texts to select eligible articles. Articles that did not use AI or that did not involve EAT or PCAT were excluded. The same two reviewers independently extracted the data from the included articles, including the article characteristics (authors, year of publication, and research purposes), sample sizes, imaging equipment, AI methods, and main results. In the process, the two reviewers resolved their disagreement via consensus with the third senior reviewer (XQ.X.).
Study risk of bias assessment
Two reviewers (L.Z. and BB.J.) independently assessed the risk of bias and concerns regarding applicability of the included studies using the Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2) tool (Whiting et al. 2011) and resolved the disagreement by consultation with the third senior reviewer (XQ.X.). QUADAS-2 consists of four key domains: (1) patient selection, (2) index test, (3) reference standard, and (4) flow and timing. Considering the concerns regarding applicability in the index test domain, the studies with external validation were considered low concern.
Results
Study selection and study characteristics
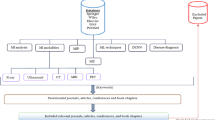
The initial search identified 140 citation records. Of these, 45 duplicate records were deleted, and 56 records were removed by browsing the titles and abstracts. After reading the full texts, 22 studies were excluded. Thereafter, 19 studies were finally included in the systematic review. Figure 1 depicts the flowchart of the selection process.
Figure 2 and Table 1 show the characteristics of the included studies. Of the 19 included studies, 15 (79%) used AI technology to segment and quantify EAT or PCAT, while 4 (21%) explored the clinical value of AI in the diagnosis and risk prediction of cardiovascular diseases. In terms of imaging equipment, 18 studies (95%) used computed tomography (CT), including 12 (63%) applying non-contrast scans and 6 (32%) performing coronary CT angiography (CCTA) scans. Only one study (5%) used cardiac magnetic resonance (CMR) imaging.
Study quality assessment
Figure 3 and Table S1 represent the results of risk of bias and concerns regarding applicability according to the QUADAS-2 tool. For the assessment of bias risk, all 19 studies were rated as low risk in terms of flow and timing, reference standards, and index tests. In terms of patient selection, 5 studies (26%) were rated as low risk while 14 (74%) were rated as unclear risk. For the assessment of the concerns of applicability, all 19 studies were rated as low concern in terms of reference standards and patient selection. However, for the index test, only one study (5%) was rated as low concern because of the existence of external validation; 16 studies (84%) were considered unclear because they only conducted internal validation (cross or leave-one-out validation), and 2 studies (11%) were rated as high concern due to a lack of validation.
Discussion
This systematic review summarized the research progress of AI in EAT and PCAT image analysis. An exhaustive literature search indicated that AI can improve the image segmentation and quantification of EAT or PCAT and can also be used as a tool for the diagnosis and risk prediction of coronary artery diseases based on EAT or PCAT. However, most of the studies have not applied external validation, which suggests that the generalizability of these studies remains to be improved.
Segmentation of EAT and PCAT
Given the potential clinical value of EAT, the segmentation of EAT images is the crucial step for further quantitative analysis. In the traditional analysis mode, radiologists have to delineate the boundary of EAT manually. This procedure is user-dependent, time-consuming, and poorly reproducible, so it is necessary to develop an accurate, quicker, and reproducible method for EAT segmentation (Norlén et al. 2016; Commandeur et al. 2018). Researchers have developed machine learning algorithms to segment EAT. Rodrigues et al. utilized a genetic algorithm to optimize the parameters of an ellipse that was used to simulate the pericardium contour (Rodrigues et al. 2017b). After 10, 100, and 200 generations of genetic algorithm iteration, the percentage of epicardial fat engulfed by the ellipse was 97.3%, 98.8%, and 99.5%, respectively. Norlén et al. proposed a method to detect the pericardium using the random forest classification algorithm (Norlén et al. 2016). This method used feature-based multi-atlas regulations for spatial initiation in CCTA images. Finally, the segmentation was completed in global optimization through graph cuts, and the Dice similarity coefficient (DSC) between the proposed method and experts was 0.91. The average segmentation time was 51.9 s. The DSC is a commonly used measurement to evaluate the similarity between two structures, and its value ranges from 0 to 1. A higher DSC represents a higher similarity of image segmentation (Andrews and Hamarneh 2015).
Researchers have also explored the utilization of deep learning in EAT segmentation. Zhang et al. developed a dual U-Net CNN for the automatic segmentation and quantification of EAT (Zhang et al. 2020). Compared with a single U-Net (DSC = 0.766) and Seg-Net (DSC = 0.767), this method segmented the EAT of 20 patients with a mean DSC of 0.912.
In addition to tracking PCAT manually on axial images, PCAT can also be automatically segmented in 3D space. PCAT is considered adipose tissue whose radial distance from the outer wall of the coronary artery is equal to the mean diameter of the artery. A well-developed software package (Aquarius Workstation, TeraRecon GmbH) automatically segments PCAT radially outwards from the outer wall of coronary vessels in 3D space (Antonopoulos et al. 2017). PCAT is then defined by the CT value range (typically from − 190 HU to − 30 HU) of the adipose tissue within the segmented volume of interest.
In short, researchers have developed several AI-based methods for the automatic segmentation of EAT and PCAT. The studies applying AI technology to evaluate EAT (Hasebe et al. 2020) and PCAT (Oikonomou et al. 2019) showed good interobserver agreement, and the correlation coefficients were 0.93 and 0.94, respectively. These AI-based methods have the advantages of high accuracy, short processing time, and good reproducibility, which lay the foundation for further research.
Quantification of EAT and PCAT
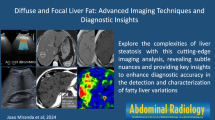
Quantification follows image segmentation. The quantitative parameters for EAT or PCAT are generally volume, area, and thickness. Commandeur et al. proposed a new multi-task framework using deep CNNs to quantify epicardial and thoracic adipose tissues (Commandeur et al. 2018). This method showed that the automatic volume quantification of epicardial and thoracic adipose tissue was strongly consistent with the manual method, and the median DSC was 0.823 and 0.905, respectively. The correlation coefficient between automatic and manual EAT volume quantification was 0.924. The automatic method for quantifying EAT took 25.63 ± 3.72 s per case, much shorter than the 10–11 min of the manual method. Figures 4 and 5 represent the image segmentation and quantification of EAT and PCAT, respectively.
Schematic diagram of pericoronary adipose tissue and FAI. (left) 3D volume rendering image of pericoronary adipose tissue (orange color); (right) the numbers in the lower right corner represent the values of fat attenuation index. RCA = right coronary artery; LAD = left anterior descending artery; CX = circumflex artery; HU = Hounsfield unit
Clinical applications of machine learning in EAT
EAT plays an important role in the development of CVD through pathophysiological mechanisms (Mazurek et al. 2003; Cheng et al. 2008). Evidence suggests that abundant EAT is a predictor of adverse cardiovascular events (Mahabadi et al. 2013; Goeller et al. 2018). Since machine learning algorithms can maximize the data mining of image features and clinical factors, researchers have used them to analyze EAT. In a prospective study, Commandeur et al. developed a machine learning-derived risk score based on the extreme gradient boosting (XGBoost) algorithm, which combined clinical risk factors with the coronary artery calcium (CAC) score and automatically quantified EAT to predict the long-term risk of myocardial infarction (MI) and cardiac death (Commandeur et al. 2020). The machine learning-derived risk score (AUC = 0.82) performed better than the risk factors for atherosclerotic CVD (AUC = 0.77) and CAC score (AUC = 0.77). Otaki et al. applied the same algorithm (XGBoost) and found that the epicardial fat volume was an independent predictor of impaired myocardial flow reserve (Otaki et al. 2015). Compared with the CAC score (AUC = 0.66) and epicardial fat volume (AUC = 0.67), the AUC of the combined risk score integrating the CAC score and epicardial fat volume was 0.73, which improved the prediction ability of impaired myocardial flow reserve. In general, the existing evidence suggests that EAT, with the support of machine learning, can improve the diagnosis and prognostic prediction of CVD.
Clinical applications of machine learning in PCAT
PCAT is the adipose tissue enveloping the coronary artery, which involves the evolution of atherosclerosis (Dey et al. 2010). The pericoronary fat attenuation index (FAI) is a novel imaging biomarker that maps the spatial changes in PCAT attenuation in CCTA images. The FAI has been used to identify subclinical coronary artery disease (Antonopoulos et al. 2017) and improve the risk prediction of all-cause cardiac mortality (Oikonomou et al. 2018). In addition to the FAI that simply represents the CT value, radiomics can mine the high-throughput quantitative information of the PCAT. Oikonomou et al. reported that the radiomics signature of the PCAT was associated with the fibrosis and vascularity of the PCAT, and the PCAT represented the adipose tissue remodelling caused by coronary inflammation (Oikonomou et al. 2019). In addition, they proposed a random forest method to analyze 1391 radiomic features of the PCAT extracted from 101 patients who had major adverse cardiac events (MACEs) within 5 years after undergoing CCTA and 101 matched controls. Their machine learning-based model performed better than the existing clinical risk factor model (Δ[C-statistic] = 0.126, p < 0.001) in predicting the outcome of MACEs. Lin et al. explored the ability of the radiomics signature of the PCAT in CCTA images to discriminate patients with MI from those with stable or no coronary artery disease (Lin et al. 2020). Using an XGBoost that combined clinical factors, PCAT attenuation, and radiomics features, their method significantly improved the discrimination ability of acute MI (AUC = 0.87) compared with a model with clinical factors and PCAT attenuation (AUC = 0.77) or clinical factors alone (AUC = 0.76). In brief, the implementation of machine learning can maximize the information extracted from PCAT and play an important role in the diagnosis and risk prediction of coronary artery disease.
Multi-modal imaging of cardiac adipose tissue
Multiple types of equipment can be used to evaluate EAT, including CT, CMR, and echocardiography. CT offers volumetric visualization and quantification of cardiac adipose tissue with high spatial resolution. However, CT has the disadvantage of unavoidable radiation exposure. CMR is considered to be the best imaging method for adipose tissue because of its superb fat signal display (Wong et al. 2017). According to the results of Mahajan et al., there was a significant correlation between CMR and autopsy for measuring the mass of adipose tissue with an intraclass correlation coefficient > 0.8 (Mahajan et al. 2013). The main disadvantages of CMR are its limited availability and high costs. Echocardiography is widely used, but the image quality is poor and not conducive to quantitative analysis. The quantification of EAT by echocardiography was limited to the thickness of adipose tissue on the right ventricular free wall (related to EAT thickness on CMR: R = 0.905) (Iacobellis et al. 2003; Gaborit et al. 2017).
Compared with the CT widely used in EAT analysis, research based on echocardiography and CMR is limited. In the included studies, only one used deep learning to segment EAT in CMR images with a low DSC of 0.56. More studies are expected to investigate EAT using AI and multi-modal imaging.
Future prospects
The segmentation and quantification of EAT is an important basis of EAT research. Researchers have developed AI-based methods to make this process more accurate, quicker, and reproducible. In addition to developments in the experimental environment, more robust and generalizable AI methods are needed to assist the clinical application of EAT analysis.
As a booming field in recent years, radiomics enables the in-depth mining of image information. Based on EAT and PCAT, scholars have begun to use radiomics to diagnose and predict the risk of cardiovascular disease. However, to validate the clinical value of radiomics-based imaging biomarkers, more well-conducted multi-center studies are needed. Deep learning has become an auxiliary tool for image segmentation. On the basis of automatic segmentation of PCAT, an AI algorithm was used to quantitatively analyze and model the FAI, which expands the research of cardiac imaging. In addition to image segmentation, more AI-based research is expected to be launched to further explore the diagnostic and prognostic value of EAT and PCAT in CVD.
Conclusion
With the recent development of AI, researchers have begun to apply AI to analyze EAT and PCAT images. AI-based methods provide accurate and quicker image segmentation and quantification. Although the publications are not numerous, the published studies have suggested the potential clinical application of EAT and PCAT in the diagnosis and risk prediction of CVD. Although AI research on cardiac adipose tissue is still in its infancy, the application of AI technology is expected to expand the clinical value of cardiac adipose tissue in cardiac imaging.
Availability of data and materials
Not applicable.
Abbreviations
- AI:
-
Artificial intelligence
- CNN:
-
Convolutional neural network
- AUC:
-
Area under the curve
- EAT:
-
Epicardial adipose tissue
- PCAT:
-
Pericoronary adipose tissue
- CVD:
-
Cardiovascular disease
- PRISMA:
-
Preferred reporting items for systematic reviews and meta-analyses
- QUADAS-2:
-
Quality Assessment of Diagnostic Accuracy Studies-2
- CT:
-
Computed tomography
- CCTA:
-
Coronary computed tomography angiography
- CMR:
-
Cardiac magnetic resonance
- XGBoost:
-
Extreme gradient boosting
- CAC:
-
Coronary artery calcium
- MI:
-
Myocardial infarction
- FAI:
-
Fat attenuation index
- MACE:
-
Major adverse cardiac events
References
Aarthy D, Priya C, Sudha S (2019) Deep Learning for Quantification of Epicardial Fat from Non-Contrast CT. 2019 IEEE International Conference on Intelligent Techniques in Control, Optimization and Signal Processing (INCOS). https://doi.org/10.1109/INCOS45849.2019.8951387
Abe I, Teshima Y, Kondo H, Kaku H, Kira S, Ikebe Y, Saito S, Fukui A, Shinohara T, Yufu K, Nakagawa M, Hijiya N, Moriyama M, Shimada T, Miyamoto S, Takahashi N (2018) Association of fibrotic remodeling and cytokines/chemokines content in epicardial adipose tissue with atrial myocardial fibrosis in patients with atrial fibrillation. Heart Rhythm 15(11):1717–1727. https://doi.org/10.1016/j.hrthm.2018.06.025
Andrews S, Hamarneh G (2015) Multi-region probabilistic dice similarity coefficient using the aitchison distance and bipartite graph matching. Commun Res Rep (CoRR) arXiv:1509.07244
Antonopoulos AS, Sanna F, Sabharwal N, Thomas S, Oikonomou EK, Herdman L et al (2017) Detecting human coronary inflammation by imaging perivascular fat. Sci Transl Med 9(398):eaal2658
Avanzo M, Wei L, Stancanello J, Vallières M, Rao A, Morin O, Mattonen SA, el Naqa I (2020) Machine and deep learning methods for radiomics. Med Phys 47(5):e185–e202. https://doi.org/10.1002/mp.13678
Betancur J, Commandeur F, Motlagh M, Sharir T, Einstein AJ, Bokhari S, Fish MB, Ruddy TD, Kaufmann P, Sinusas AJ, Miller EJ, Bateman TM, Dorbala S, di Carli M, Germano G, Otaki Y, Tamarappoo BK, Dey D, Berman DS, Slomka PJ (2018) Deep learning for prediction of obstructive disease from fast myocardial perfusion SPECT: a multicenter study. JACC Cardiovasc Imaging 11(11):1654–1663. https://doi.org/10.1016/j.jcmg.2018.01.020
Chartrand G, Cheng PM, Vorontsov E, Drozdzal M, Turcotte S, Pal CJ, Kadoury S, Tang A (2017) Deep learning: a primer for radiologists. RadioGraphics 37(7):2113–2131. https://doi.org/10.1148/rg.2017170077
Cheng KH, Chu CS, Lee KT, Lin TH, Hsieh CC, Chiu CC, Voon WC, Sheu SH, Lai WT (2008) Adipocytokines and proinflammatory mediators from abdominal and epicardial adipose tissue in patients with coronary artery disease. Int J Obes 32(2):268–274. https://doi.org/10.1038/sj.ijo.0803726
Cherian S, Lopaschuk GD, Carvalho E (2012) Cellular cross-talk between epicardial adipose tissue and myocardium in relation to the pathogenesis of cardiovascular disease. Am J Physiol Endocrinol Metab 303(8):E937–E949. https://doi.org/10.1152/ajpendo.00061.2012
Commandeur F, Goeller M, Betancur J, Cadet S, Doris M, Chen X, Berman DS, Slomka PJ, Tamarappoo BK, Dey D (2018) Deep learning for quantification of epicardial and thoracic adipose tissue from non-contrast CT. IEEE Trans Med Imaging 37(8):1835–1846. https://doi.org/10.1109/TMI.2018.2804799
Commandeur F, Goeller M, Razipour A, Cadet S, Hell MM, Kwiecinski J, Chen X, Chang HJ, Marwan M, Achenbach S, Berman DS, Slomka PJ, Tamarappoo BK, Dey D (2019) Fully Automated CT Quantification of Epicardial Adipose Tissue by Deep Learning: A Multicenter Study. Radiol Artif Intell 1(6):e190045. https://doi.org/10.1148/ryai.2019190045
Commandeur F, Slomka PJ, Goeller M, Chen X, Cadet S, Razipour A, McElhinney P, Gransar H, Cantu S, Miller RJH, Rozanski A, Achenbach S, Tamarappoo BK, Berman DS, Dey D (2020) Machine learning to predict the long-term risk of myocardial infarction and cardiac death based on clinical risk, coronary calcium, and epicardial adipose tissue: a prospective study. Cardiovasc Res 116(14):2216–2225. https://doi.org/10.1093/cvr/cvz321
Dey D, Wong ND, Tamarappoo B, Nakazato R, Gransar H, Cheng VY, Ramesh A, Kakadiaris I, Germano G, Slomka PJ, Berman DS (2010) Computer-aided non-contrast CT-based quantification of pericardial and thoracic fat and their associations with coronary calcium and metabolic syndrome. Atherosclerosis 209(1):136–141. https://doi.org/10.1016/j.atherosclerosis.2009.08.032
Fulton MR, Givan AH, Fernandez-del-Valle M, Klingensmith JD (2020) Segmentation of epicardial adipose tissue in cardiac MRI using deep learning. Proceedings of the SPIE - Progress in Biomed Opt Imaging 11317:113170Q(113177 pp.). https://doi.org/10.1117/12.2550013
Gaborit B, Sengenes C, Ancel P, Jacquier A, Dutour A (2017) Role of epicardial adipose tissue in health and disease: a matter of fat? Compr Physiol 7(3):1051–1082. https://doi.org/10.1002/cphy.c160034
Goeller M, Achenbach S, Marwan M, Doris MK, Cadet S, Commandeur F, Chen X, Slomka PJ, Gransar H, Cao JJ, Wong ND, Albrecht MH, Rozanski A, Tamarappoo BK, Berman DS, Dey D (2018) Epicardial adipose tissue density and volume are related to subclinical atherosclerosis, inflammation and major adverse cardiac events in asymptomatic subjects. J Cardiovasc Comput Tomogr 12(1):67–73. https://doi.org/10.1016/j.jcct.2017.11.007
Hasebe H, Yoshida K, Nogami A, Ieda M (2020) Difference in epicardial adipose tissue distribution between paroxysmal atrial fibrillation and coronary artery disease. Heart Vessels 35(8):1070–1078. https://doi.org/10.1007/s00380-020-01575-3
He X, Guo BJ, Lei Y, Wang T, Fu Y, Curran WJ, Zhang LJ, Liu T, Yang X (2020a) Automatic segmentation and quantification of epicardial adipose tissue from coronary computed tomography angiography. Phys Med Biol 65(9):095012. https://doi.org/10.1088/1361-6560/ab8077
He X, Guo BJ, Lei Y, Wang T, Curran WJ, Liu T, Zhang LJ, Yang X (2020b) Automatic quantification of myocardium and pericardial fat from coronary computed tomography angiography: a multicenter study. Eur Radiol 31(6):3826–3836. https://doi.org/10.1007/s00330-020-07482-5
Hosny A, Parmar C, Coroller TP, Grossmann P, Zeleznik R, Kumar A, Bussink J, Gillies RJ, Mak RH, Aerts HJWL (2018a) Deep learning for lung cancer prognostication: a retrospective multi-cohort radiomics study. PLoS Med 15(11):e1002711. https://doi.org/10.1371/journal.pmed.1002711
Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts HJWL (2018b) Artificial intelligence in radiology. Nat Rev Cancer 18(8):500–510. https://doi.org/10.1038/s41568-018-0016-5
Iacobellis G, Assael F, Ribaudo MC, Zappaterreno A, Alessi G, Di Mario U et al (2003) Epicardial fat from echocardiography: a new method for visceral adipose tissue prediction. Obes Res 11(2):304–310. https://doi.org/10.1038/oby.2003.45
Iacobellis G, Corradi D, Sharma AM (2005) Epicardial adipose tissue: anatomic, biomolecular and clinical relationships with the heart. Nat Clin Pract Cardiovasc Med 2(10):536–543. https://doi.org/10.1038/ncpcardio0319
Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RGPM, Granton P, Zegers CML, Gillies R, Boellard R, Dekker A, Aerts HJWL (2012) Radiomics: Extracting more information from medical images using advanced feature analysis. Eur J Cancer 48(4):441–446. https://doi.org/10.1016/j.ejca.2011.11.036
Li Z, Zou L, Yang R (2019). A Neural Network-based Method for Automatic Pericardium Segmentation. Proceedings of the 2nd International Conference on Computer Science and Software Engineering. Xi'an, China, Association for Computing Machinery 45–49. https://doi.org/10.1145/3339363.3339372
Lin A, Kolossváry M, Yuvaraj J, Cadet S, McElhinney PA, Jiang C, Nerlekar N, Nicholls SJ, Slomka PJ, Maurovich-Horvat P, Wong DTL, Dey D (2020) Myocardial Infarction associates with a distinct pericoronary adipose tissue radiomic phenotype: a prospective case-control study. JACC Cardiovasc Imaging 13(11):2371–2383. https://doi.org/10.1016/j.jcmg.2020.06.033
Litjens G, Ciompi F, Wolterink JM, de Vos BD, Leiner T, Teuwen J et al (2019) State-of-the-art deep learning in cardiovascular image analysis. JACC Cardiovasc Imaging 12(8, Part 1):1549–1565
Mahabadi AA, Berg MH, Lehmann N, Kälsch H, Bauer M, Kara K, Dragano N, Moebus S, Jöckel KH, Erbel R, Möhlenkamp S (2013) Association of epicardial fat with cardiovascular risk factors and incident myocardial infarction in the general population: the Heinz Nixdorf Recall Study. J Am Coll Cardiol 61(13):1388–1395. https://doi.org/10.1016/j.jacc.2012.11.062
Mahajan R, Kuklik P, Grover S, Brooks AG, Wong CX, Sanders P, Selvanayagam JB (2013) Cardiovascular magnetic resonance of total and atrial pericardial adipose tissue: a validation study and development of a 3 dimensional pericardial adipose tissue model. J Cardiovasc Magn Reson 15(1):73. https://doi.org/10.1186/1532-429X-15-73
Mancio J, Azevedo D, Saraiva F, Azevedo AI, Pires-Morais G, Leite-Moreira A, Falcao-Pires I, Lunet N, Bettencourt N (2018) Epicardial adipose tissue volume assessed by computed tomography and coronary artery disease: a systematic review and meta-analysis. Eur Heart J Cardiovasc Imaging 19(5):490–497. https://doi.org/10.1093/ehjci/jex314
Mazurek T, Zhang L, Zalewski A, Mannion JD, Diehl JT, Arafat H, Sarov-Blat L, O’Brien S, Keiper EA, Johnson AG, Martin J, Goldstein BJ, Shi Y (2003) Human epicardial adipose tissue is a source of inflammatory mediators. Circulation 108(20):2460–2466. https://doi.org/10.1161/01.CIR.0000099542.57313.C5
Norlén A, Alvén J, Molnar D, Enqvist O, Norrlund RR, Brandberg J, Bergström G, Kahl F (2016) Automatic pericardium segmentation and quantification of epicardial fat from computed tomography angiography. J Med Imaging (Bellingham) 3(3):034003. https://doi.org/10.1117/1.JMI.3.3.034003
Oikonomou EK, Marwan M, Desai MY, Mancio J, Alashi A, Hutt Centeno E, Thomas S, Herdman L, Kotanidis CP, Thomas KE, Griffin BP, Flamm SD, Antonopoulos AS, Shirodaria C, Sabharwal N, Deanfield J, Neubauer S, Hopewell JC, Channon KM, Achenbach S, Antoniades C (2018) Non-invasive detection of coronary inflammation using computed tomography and prediction of residual cardiovascular risk (the CRISP CT study): a post-hoc analysis of prospective outcome data. Lancet 392(10151):929–939. https://doi.org/10.1016/S0140-6736(18)31114-0
Oikonomou EK, Williams MC, Kotanidis CP, Desai MY, Marwan M, Antonopoulos AS, Thomas KE, Thomas S, Akoumianakis I, Fan LM, Kesavan S, Herdman L, Alashi A, Centeno EH, Lyasheva M, Griffin BP, Flamm SD, Shirodaria C, Sabharwal N, Kelion A, Dweck MR, van Beek EJR, Deanfield J, Hopewell JC, Neubauer S, Channon KM, Achenbach S, Newby DE, Antoniades C (2019) A novel machine learning-derived radiotranscriptomic signature of perivascular fat improves cardiac risk prediction using coronary CT angiography. Eur Heart J 40(43):3529–3543. https://doi.org/10.1093/eurheartj/ehz592
Opincariu D, Benedek T, Chițu M, Raț N, Benedek I (2020) From CT to artificial intelligence for complex assessment of plaque-associated risk. Int J Cardiovasc Imaging 36(12):2403–2427. https://doi.org/10.1007/s10554-020-01926-1
Otaki Y, Hell M, Slomka PJ, Schuhbaeck A, Gransar H, Huber B, Nakazato R, Germano G, Hayes SW, Thomson LEJ, Friedman JD, Achenbach S, Berman DS, Dey D (2015) Relationship of epicardial fat volume from noncontrast CT with impaired myocardial flow reserve by positron emission tomography. J Cardiovasc Comput Tomogr 9(4):303–309. https://doi.org/10.1016/j.jcct.2015.03.005
Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD et al (2021) The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ 372:n71
Rodrigues ÉO, Cordeiro de Morais FF, Conci A (2015) On the Automated Segmentation of Epicardial and Mediastinal Cardiac Adipose Tissues Using Classification Algorithms. Stud Health Technol Inform 216:726–730. https://doi.org/10.3233/978-1-61499-564-7-726
Rodrigues ÉO, Morais FF, Morais NA, Conci LS, Neto LV, Conci A (2016) A novel approach for the automated segmentation and volume quantification of cardiac fats on computed tomography. Comput Methods Programs Biomed 123:109–128. https://doi.org/10.1016/j.cmpb.2015.09.017
Rodrigues ÉO, Pinheiro VHA, Liatsis P, Conci A (2017a) Machine learning in the prediction of cardiac epicardial and mediastinal fat volumes. Comput Biol Med 89:520–529. https://doi.org/10.1016/j.compbiomed.2017.02.010
Rodrigues ÉO, Rodrigues LO, Oliveira LSN, Conci A, Liatsis P (2017b) Automated recognition of the pericardium contour on processed CT images using genetic algorithms. Comput Biol Med 87:38–45. https://doi.org/10.1016/j.compbiomed.2017.05.013
Tamarappoo BK, Lin A, Commandeur F, McElhinney PA, Cadet S, Goeller M, Razipour A, Chen X, Gransar H, Cantu S, Miller RJH, Achenbach S, Friedman J, Hayes S, Thomson L, Wong ND, Rozanski A, Slomka PJ, Berman DS, Dey D (2021) Machine learning integration of circulating and imaging biomarkers for explainable patient-specific prediction of cardiac events: A prospective study. Atherosclerosis 318:76–82. https://doi.org/10.1016/j.atherosclerosis.2020.11.008
Topuz M, Dogan A (2017) The effect of epicardial adipose tissue thickness on left ventricular diastolic functions in patients with normal coronary arteries. Kardiol Pol 75(3):196–203. https://doi.org/10.5603/KP.a2016.0139
Wang J, Wang LJ, Peng YP, Zhang LJ, Jiang SS, Gong JB (2013) Association of pericardial adipose tissue volume with presence and severity of coronary atherosclerosis. Clin Invest Med 36(3):E143–E150. https://doi.org/10.25011/cim.v36i3.19725
Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, Leeflang MM, Sterne JA, Bossuyt PM, QUADAS-2 Group (2011) QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med 155(8):529–536. https://doi.org/10.7326/0003-4819-155-8-201110180-00009
Wong CX, Ganesan AN, Selvanayagam JB (2017) Epicardial fat and atrial fibrillation: current evidence, potential mechanisms, clinical implications, and future directions. Eur Heart J 38(17):1294–1302. https://doi.org/10.1093/eurheartj/ehw045
Yu KH, Beam AL, Kohane IS (2018) Artificial intelligence in healthcare. Nat Biomed Eng 2(10):719–731. https://doi.org/10.1038/s41551-018-0305-z
Zhang Q, Zhou J, Zhang B, Jia W, Wu E (2020) Automatic epicardial fat segmentation and quantification of CT scans using dual U-Nets with a morphological processing layer. IEEE Access 8:128032–128041. https://doi.org/10.1109/ACCESS.2020.3008190
Zlokolica V, Krstanović L, Velicki L, Popović B, Janev M, Obradović R, Ralević NM, Jovanov L, Babin D (2017) Semiautomatic epicardial fat segmentation based on fuzzy c-means clustering and geometric ellipse fitting. J Healthc Eng 2017:5817970. https://doi.org/10.1155/2017/5817970
Acknowledgements
Not applicable.
Funding
This study was sponsored by Ministry of Science and Technology of China (2016YFE0103000), National Natural Science Foundation of China (project nos. 81971612 and 81471662), and Shanghai Jiao Tong University (ZH2018ZDB10).
Author information
Authors and Affiliations
Contributions
LZ and XQX designed the concept of this review. LZ and BBJ conducted literature search and data collection, and drafted the manuscript. JQS draw the computer graphics. LZ, JQS, and XQX revised the manuscript. XQX supervised this manuscript. All authors participated in writing and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
Assessment of risk of bias and concerns regarding applicability according to the Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2) tool.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, L., Sun, J., Jiang, B. et al. Development of artificial intelligence in epicardial and pericoronary adipose tissue imaging: a systematic review. European J Hybrid Imaging 5, 14 (2021). https://doi.org/10.1186/s41824-021-00107-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41824-021-00107-0