Abstract
Background
Numerous biomarkers have been proposed for diagnosis, therapeutic, and prognosis in sepsis. Previous evaluations of the value of biomarkers for predicting mortality due to this life-threatening condition fail to address the complexity of this condition and the risk of bias associated with prognostic studies. We evaluate the predictive performance of four of these biomarkers in the prognosis of mortality through a methodologically sound evaluation.
Methods
We conducted a systematic review a systematic review and meta-analysis to determine, in critically ill adults with sepsis, whether procalcitonin (PCT), C-reactive protein (CRP), interleukin-6 (IL-6), and presepsin (sCD14) are independent prognostic factors for mortality. We searched MEDLINE, EMBASE, and the Cochrane Central Register of Controlled Trials up to March 2023. Only Phase-2 confirmatory prognostic factor studies among critically ill septic adults were included. Random effects meta-analyses pooled the prognostic association estimates.
Results
We included 60 studies (15,681 patients) with 99 biomarker assessments. Quality of the statistical analysis and reporting domains using the QUIPS tool showed high risk of bias in > 60% assessments. The biomarker measurement as a continuous variable in models adjusted by key covariates (age and severity score) for predicting mortality at 28–30 days showed a null or near to null association for basal PCT (pooled OR = 0.99, 95% CI = 0.99–1.003), CRP (OR = 1.01, 95% CI = 0.87 to 1.17), and IL-6 (OR = 1.02, 95% CI = 1.01–1.03) and sCD14 (pooled HR = 1.003, 95% CI = 1.000 to 1.006). Additional meta-analyses accounting for other prognostic covariates had similarly null findings.
Conclusion
Baseline, isolated measurement of PCT, CRP, IL-6, and sCD14 has not been shown to help predict mortality in critically ill patients with sepsis. The role of these biomarkers should be evaluated in new studies where the patient selection would be standardized and the measurement of biomarker results.
Trial registration
PROSPERO (CRD42019128790).
Similar content being viewed by others
Introduction
Sepsis, a life-threatening organ dysfunction resulting from a dysregulated host response to an infection [1], occurs in one per 1000 people worldwide and accounts for 10% of intensive care unit (ICU) admissions [2]. Its complications include acute renal failure, polyneuropathy, cardiomyopathy, and multiple organ dysfunction [3, 4]. Mortality in septic shock can be > 40% [1]. About 50–70% of sepsis survivors suffer persistent physical, psychological, mental, and social issues [5, 6]. Management of sepsis is challenging, and it may include early administration of antibiotics, restoring tissue perfusion by resuscitation with crystalloids and vasopressors, and controlling the infection source [3, 7]. Care needs to be individualised, with stratification of patient’s mortality risk [8, 9].
Biomarkers have been proposed as the key to tailoring therapies for specific patients and monitoring their effects [10]. Currently, more than 150 potential sepsis biomarkers have been described [11]; however, many of them correspond to the measurement of substances derived from the complex process that sepsis represents, in its microorganism-host interaction, without its clinical utility being determined to date. Four of these biomarkers: procalcitonin (PCT), C-reactive protein (CRP), interleukin (IL)-6, and presepsin (sCD14) are commonly assessed biomarkers, and they have been evaluated for diagnosis and prognosis purposes [8, 12,13,14,15]. With respect to sepsis prognosis, single biomarkers or their various combinations may be useful in estimating the risk of death, re-hospitalisation, or long-term complications as they can help detect endothelial damage, intestinal permeability and organ failure early [15, 16]. Biomarker results may be added to clinical scores to improve prediction. The timing of measurement and the kinetics of their clearance are key points in the prognostic investigation [17]. However, conclusions about their value are not firm. Although the Surviving Sepsis Campaign guidelines for the management of sepsis consider biomarkers as an add-on of clinical evaluation, in the Sepsis-3 definition consensus their role remains still undefined [1].
Beyond the well-established role of some biomarkers for the diagnosis and management of septic patients, a formal assessment of the prognostic role of biomarkers in the prediction of critical sepsis outcomes using a methodologically sound critical appraisal is lacking [18]. The objective of this systematic review was to determine whether PCT, CRP, IL-6, and sCD14 are independent prognostic factors for predicting mortality in critically ill adults with sepsis.
Materials and methods
Search strategy and selection criteria
We conducted this systematic review and meta-analysis following recommended methods [18, 19]. The review was prospectively registered (PROSPERO, CRD42019128790) and reported following the PRISMA (Preferred Reporting Items for Systematic reviews and Meta-Analysis) statement [20]. We searched MEDLINE-Ovid, EMBASE-Ovid, and the Cochrane Central Register of Controlled Trials (CENTRAL) using a sensitive search strategy without language restrictions, from inception date to March 24 2023 (Additional file S1). We also hand-searched the reference lists of included studies.
Pairs of reviewers independently screened titles, abstracts, and then full-text articles. Disagreements were resolved by discussion with a third reviewer. We selected for this review phase 2 confirmatory prognostic factor studies [21], which measure a factor’s independent prognostic effect while controlling for known covariates. Studies considered phase 1 exploratory studies (i.e., those identifying potential associations or differences without an adequately adjusted analysis), as well as case series, diagnostic accuracy studies, and other studies not focussed on prognosis were excluded.
Studies were eligible for inclusion if they (a) included at least ten patients and at least 5 events; (b) included adults (males and females aged ≥ 18 years) reported as being critically ill and with a confirmed diagnosis of sepsis following valid clinical criteria; (c) measured at least one of the four biomarkers under assessment within the first 24 h of sepsis diagnosis or admission to the intensive care unit, emergency department or hospital ward; (d) reported any measure of patient’s survival (such as mortality or survival), ideally at 28–30 days following hospitalisation. If a study reported similar patients from the same institution in more than one publication, we included the paper with the largest sample in our analysis to avoid spurious precision due to duplication.
Data analysis
Pairs of reviewers independently extracted the following data from each eligible study, using a standardised form: study general characteristics, including number and name of centres involved, study funding and declaration of conflict of interests; population characteristics, including criteria for sepsis diagnosis, age and sex, severity scores (e.g., SOFA, APACHE, SAPS) and hospital setting (e.g., ICU, emergency room); biomarkers characteristics, including the timing and unit of measurement; and mortality reports, including the number of survivors/deaths and the corresponding proportions. Study authors were contacted for clarifications when necessary.
The primary outcome of this systematic review was mortality. This outcome included all-cause mortality, sepsis-related mortality, ICU or hospital mortality, ideally measured at 28–30 days, as defined by the study authors. For statistical analysis, we classified all measurements performed at 28–30 days as “mortality at 28–30 days” (i.e., hospital mortality at 28–30 days, ICU mortality at 28–30 days, survival at 28–30 days and sepsis-related mortality at 28–30 days); other reports without a clear statement about the follow-up duration (i.e., hospital mortality, ICU mortality, survival/non-survival) were classified as “mortality-no details”.
We assessed the methodological quality of the included studies using the Quality In Prognosis Studies (QUIPS) tool, which contains six domains [22] that have been tailored for this review (see Additional file S2). Clinical experts participating in these reviews selected a list of prognostic covariates to be considered in the QUIPS assessment. These covariates were age, a severity score (e.g., SOFA, SAPS, or APACHE), use of vasopressors, key comorbidities (i.e., immunosuppression, pulmonary disease, cancer, alcohol dependence, lymphocytopenia), inappropriate empirical antibiotic regimen, late antibiotic coverage and control of septic focus. Age and severity score were regarded as the key covariates that predictive models should include for a minimal adequacy of the adjustment (QUIPS Domain 5). Pairs of reviewers independently assessed the risk of bias. Clarifications were requested from the study authors when necessary. We made a consensus judgement for each of the 6-QUIPS domains, using one of the three risk of bias categories (i.e., low, moderate or high) suggested by the tool guidelines [18].
We extracted information regarding the predictive value of each biomarker, including the baseline measurement of survival and no-survival groups (e.g., medians and interquartile ranges, IQRs), the corresponding effect measure derived from a multivariable model (e.g., adjusted odds ratio, OR, with 95% confidence interval, CI). If the authors had categorized the underlying continuous variables of the biomarkers, we extracted the proposed positivity threshold (i.e., the biomarker cut-off proposed by the authors to predict mortality).
Due to the biomarkers being measured in different units across studies (e.g., mg/L, g/mL, µg/L), we transformed the numerical information to the most frequent unit reported for the biomarker, as follow: PCT: ng/ml; CRP: mg/L; IL-6: pg/mL; and sCD14: pg/mL. Studies in which the biomarker was transformed into a logarithmic scale were analysed separately because a transformation to a common unit was not possible. We cross-checked the accuracy of the numerical findings using all the additional numerical information provided in the reports, such as the p-values, the CIs and Wald’s test results, and clarifications were requested from the study authors when necessary. In cases where the study authors did not evaluate the biomarker in their final model but provided information from univariable analysis (e.g., an initial univariable logistic model), we extracted this information as a proxy for the performance of the biomarker in a multivariable analysis as a last resource. In cases where only narrative information about the biomarker was available, we extracted the reported “statistically significance” of the association (commonly based on p-values) and added it narratively to our synthesis. We prioritised as main analysis the association of the biomarker measured as a continuous variable with “mortality at 28–30 days” in models adjusted for the covariates age and severity score based on the consensus of our expert panel.
We aggregated data within clinical and statistical relevant groups by each biomarker using a random effects restricted maximum likelihood (REML) meta-analysis model, providing pooled estimates (hazard ratio or OR) according to the Sidik-Jonkman method, along with the corresponding Hartung-Knapp 95% CI and between-study variance estimates. To account for the heterogeneity in the pooled effects, we calculated also the 95% prediction interval. All statistical analyses were performed using Stata 17.
Results
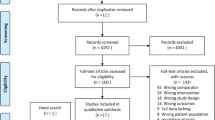
After removing duplicates, we identified 61 432 records in electronic database searches (Fig. 1). After excluding 51,189 records at the title and abstract level, 208 full texts were screened. Forty-nine publications were excluded due to ineligible participant sample (undefined mix of septic and no-septic patients), prognostic factor, outcome, or study design (Additional file S3). Another 99 studies were excluded as they were exploratory studies, i.e., those reporting differences between survivors and non-survivors. Finally, 60 studies that met our inclusion criteria and provided 99 biomarker assessments (Fig. 1) evaluating the prognostic role of PCT (43 assessments), CRP (27 assessments), IL-6 (22 assessments), and sCD14 (7 assessments).
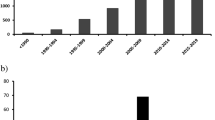
Sixty studies including 15,681 critically ill septic patients evaluated the role of at least one of our selected biomarkers for mortality prediction [23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62]. We contacted 11 study teams for additional information, and we received replies from three [27, 33, 56]. Fifty-five (91.7%) studies were published in the last 10 years (2012–2023), and 30 (50.0%) were published in the previous 3 years. Studies were primarily performed in China (28 studies), South Korea (6) and Spain (4). Most of the studies involved patients from a single centre (49 studies, 81.7%), intensive care unit (ICU) being the most frequent hospital setting (41 studies, 68.3%). Only one study comprised information from two study centres (two ICUs from Spain and France). Fifty-three studies (88.3%) were cohorts, three analysing data from a randomised controlled trial. One study team reported information about a similar set of patients from the same institution in two different publications [32, 63]; we included the data form the report with the largest sample size (187 patients versus 160). Study funding was mainly public (22 studies, 36.7%), with 20 studies not reporting this information. Authors of 44 studies (73.3%) declared no conflict of interest (Additional file S4).
Regarding the population under analysis, the sample size ranged from 29 [25] to 1984 [64] septic patients, with a median of 145 (25th–75th percentiles 89–267). The percentage of women ranged from 23.7% to 67%. Studies that reported mean age for septic patients found the range from 47.5 to 84.3 years (39). Diagnosis of sepsis was primarily performed using the 2015 SCCM/ESICM Sepsis-3 (1) (30 studies, 50.0%) and the 2001 SCCM/ESICM/ACCP/ATS/SIS [7] (16 studies, 26.7%). In some cases, authors used a specific definition of a septic condition (i.e., community-acquired infection) or local guidelines about sepsis. Sepsis origin was mixed in 71.7% of populations (i.e., combining respiratory, urogenital, intra-abdominal, blood and skin-soft tissues, among others). One study excluded patients under mechanical ventilation, [55] and two excluded patients using vasopressors [32, 55]. Only three studies reported the use of empirical antibiotics, being in both cases used in more than 90% of patients [48, 54, 65].
Regarding the assessment of biomarkers, 41/60 studies (68.3%) evaluated a single biomarker in their analysis. PCT was the most frequently assessed biomarker (40 studies and 43 assessments). Mortality rate across the studies ranged from 7 to 65%. Eighty-two assessments addressed the relationship between one biomarker and the mortality at 28–30 days (87.2%), while the remaining assessments focused on mortality without further details, including hospital and ICU mortality (Table 1).
Across all biomarkers, the QUIPS domain most affected by risk of bias was the statistical analysis and reporting domain (Domain 6), with > 60% of assessments at high risk of bias, mostly due to inadequate reporting of the final model (i.e., report of statistically significant values only), optimal cut-off used to estimate the final effect measure and impossibility to reproduce the confidence intervals using the standard errors and coefficients provided in the report. The study participation domain (Domain 1) was judged as low risk of bias for three of the four biomarkers, with more than 50% of assessments considered with an adequate participation of the eligible participants, a clear reporting of inclusion/exclusion criteria, and in some cases, a provision of a flow diagram. Study confounding domain (Domain 5) was commonly affected because important key prognostic covariates were not accounted in the analysis. Due to the poor reporting of confounders in the included studies, we considered only age and severity score (i.e., SOFA, SAPS or APACHE) as the minimal set of prognostic factors for the multivariable analysis. We found 31 assessments with these prognostic factors being considered in the final statistical models and so, they were considered at low risk of bias (31%). The remaining domains were considered at moderate risk of bias, mostly due to unclear reporting of missing participants (i.e., losses at follow-up), missing information (i.e., missing biomarker information), and unclear methods of measurement (i.e., mortality measurement). Only two study providing information about PCT, IL-6 and CRP was considered at low risk of bias in all QUIPS-domains (Additional file S5) [62, 66].
We found a heterogeneity in the analysis and reporting of the association between biomarkers and mortality from the included studies. Baseline biomarker levels were analysed as a continuous variable in 64 assessments (65%), 13 of them being analysed after a logarithmic transformation. In 23 assessments, authors omitted the report of the effect measure, mostly due the analyses were not significant either in the univariable or the multivariable analysis. In the remaining assessments, the study authors chose a threshold, mostly derived as the optimal cut-off, and analysed the biomarker as a categorical variable. We report here the findings of biomarkers as a continuous variable in prognostic models accounting for age and severity score as confounding factors in relation to mortality at 28–30 days. Detailed information regarding the remaining assessments from included studies (e.g., categorical, log- transformed and narrative reports), as well as the information for other mortality outcomes (i.e., mortality-general, no details), is shown in the Additional files S5–S8 and Additional figs. S1–S5.
Regarding basal PCT (ng/ml), forty out of 94 assessments focused on baseline PCT levels and its prognostic value for mortality, mortality at 28–30 days being the outcome most reported (35/43, 81.4%). Analysis focus on studies accounting by age and a severity score as a confounding factors, showed a null association between basal PCT and mortality at 28–30 days (pooled OR = 0.99, 95% CI = 0.99 to 1.00, 95% prediction interval = 0.97 to 1.03; I2 = 4.9%; 3 studies, 1129 patients) (Fig. 2, Additional Table S6, Additional Fig. S1). Only one study accounted by age and a severity in a multivariable cox regression model, and it showed a significant association (HR = 1.04, 95% CI = 1.01 to 1.07, 1 study, 2015). Additional pooled analysis without this minimal adjustment showed similar findings, either using OR (pooled OR = 1.02, 95% CI = 0.85 to 1.24, 95% prediction interval = 0.60 to 1.73; I2 = 99.9%; 11 studies, 3565 patients) or HR measures of effect (pooled HR = 1.16, 95% CI = 0.95 to 1.40, 95% prediction interval = 0.99 to 1.01; I2 = 99.9%; 9 studies, > 2344 patients) (Fig. 2, Additional Table S6, Additional Fig. S1). Most of the remaining assessments derived from categorical, log-transformed variables and narrative reports showed no role for PCT in the prognosis of mortality (Additional file S6).
Regarding Basal CRP (mg/L), twenty-five out of 94 assessments focused on baseline CRP levels and its association with mortality, mortality at 28–30 days being the most frequent outcome (21/25, 84.0%). Analysis focus on studies accounting for age and a severity score as a confounding factors, showed a null association between basal CRP and mortality at 28–30 days in multivariable logistic and Cox regression models (pooled OR = 1.01, 95% CI = 0.87 to 1.17, I2 = 61.8%, 2 studies, 303 patients; pooled HR = 1.01, 95% CI = 0.92 to 1.11, I2 = 57.7%, 2 studies, 631 patients) (Fig. 2, Additional Fig. S2, Additional Table S7). Pooled analysis without this minimal adjustment showed similar findings, either using OR (pooled OR = 1.01, 95% CI = 0.99 to 1.03, 95% prediction interval = 0.95 to 1.06, I2 = 95.7%; 5 studies, 2287 patients) or HR measures of effect (pooled HR = 1.03, 95% CI = 0.96 to1.10, 95% prediction interval = 0.99 to1.02; I2 = 98.8%; 6 studies, > 1741 patients) (Fig. 2, Additional Fig. S2). Most of the remaining assessments derived from categorical, log-transformed variables and narrative reports showed no role for CRP in the prognosis of mortality (Additional Table S7).
Regarding basal IL-6 (pg/ml), 22 out of 94 assessments were focused on baseline IL-6 levels and its association with mortality, mortality at 28–30 days being the most frequent outcome (20/22, 90.9%). Only one study accounted by age and a severity score as a confounding factors, and it showed a near to null association between basal IL-6 and mortality at 28–30 days (OR = 1.02, 95% CI = 1.01 to1.03) (Fig. 2, Additional Fig. S3, Additional Table S8). Pooled analysis without this minimal adjustment showed similar findings (pooled OR = 1.00, 95% CI = 0.99 to 1.01, 95% prediction interval = 0.98 to 1.02; I-square = 99.5%; 7 studies, > 963 patients; HR = 1.00, 95% CI = 1.00 to 1.00, 1 study, 97 patients) (Fig. 2, Additional Fig. S3). Most of the remaining assessments derived from categorical, log-transformed variables and narrative reports showed no role for IL-6 in the prognosis of mortality (Additional Table S8).
Regarding basal sCD14 (pg/ml), only six assessment focused on baseline sCD14 levels and prediction of mortality (Additional Table S9). Only two assessments accounting by key prognostic factors and analysing the biomarker as a continuous variable without minimal adjustment. This analysis shows a significant relationship between sCD14 levels, although the effect size is small (pooled HR = 1.00, 95% CI = 1.00 to 1.01; I2 = 0.4%; 2 studies, > 278 patients. Information from categorical and log-transformed analysis is contradictory for the association between sCD14 and mortality, including 28–30 days (Additional Table S9).
A sensitivity analysis combining information of mortality categories only affect selected pooled estimations of PCT and CRP levels. However, the measures of effect estimated combining these assessments, as well as the corresponding 95% confidence intervals, did not show an increased risk of mortality with any of these biomarkers, similar to the findings using separate mortality categories.
Discussion
This systematic review, comprising of studies mostly published in the last decade (2011–2023), found a lack of association between the baseline values of PCT, CRP, IL-6, and sCD14 and the risk of mortality, especially at 28–30 days, in critically ill septic patients diagnosed using current criteria. This evidence covered information from several countries and teams worldwide and recruited a large number of septic patients. Quality of the evidence was affected by several sources of bias; the QUIPS domain most affected being the statistical analysis and reporting (Domain 6) across all biomarkers. This deficiency limited the strength of the inferences drawn.
In general, the number of assessments showing no association between the baseline biomarker values and mortality was greater than those showing a positive association. When we focussed on assessments of the biomarker as a continuous variable and adjusted by key covariates for the optimal prognostic evaluation, we found a null association of basal PCT and CRP with mortality at 28–30 days (For PCT = pooled OR = 0.99, 95% CI = 0.99–1.003, 95% prediction interval (PI) = 0.97–1.03; I2 = 4.9%; 3 studies, 1129 patients. For CRP = pooled OR = 1.01, 95% CI = 0.87 to 1.17, I2 = 61.8%, 2 studies, 303 patients). For IL-6, we only found individual assessment accounting for these key covariates and showed also null or close to null association for mortality 28–30 days prediction (For IL-6: OR = 1.02, 95% CI = 1.01–1.03). However, additional pooled analysis accounting for other prognostic covariates are in agreement with these findings (see Additional Tables S6–S9). Regarding SCD14 assessment, we only found information derived from six assessments with contradictory findings. These assessments are mostly from log-transformed data, which prevented their aggregation with other untransformed estimates to avoid compromising the clinical interpretability of the pooled estimates [67].
Our study has several strengths. Firstly, our systematic review involved a comprehensive search of the literature, with more than 60,000 hits identified from electronic searches. We identified more evidence regarding the prediction of mortality at 28–30 days, which is a clinically important outcome for managing patients adequately according to their levels of basal risk. We made extra efforts to confirm the accuracy of the numerical data and standardised the units of measurement to able a pooled data analysis. Although we made efforts to unify the information to synthesize the data, the variability of reporting was the main constraint. In addition, our review follows the current recommended methodology to conduct systematic reviews to evaluate prognostic factors, selecting information from phase 2 confirmatory prognostic studies using multivariable models and adjustment for confounding [18, 68]. Other non-systematic reviews regarding the prognosis of mortality in sepsis have focused on exploratory data with no control for confounding (i.e., the merely compute differences in means between survivors and non-survivors) and claim these biomarkers’ role in the clinical setting without a discussion about the limitations [15, 16].
Limitations of our findings include the risk of bias affecting the included studies, primarily due to the lack of report of critical statistical and clinical information (e.g., the full results of the multivariable model). Although we asked for clarifications from the study authors, this information was dismissed without proper registration in some cases. We also noticed that control of confounding was not optimal in several analyses, and models failed to incorporate the key set of prognostic confounders, such as age and a severity score. The sample size of the studies was also a concern, as in some studies it led to unreliable estimates from the multivariable models. Future studies, to be considered confirmatory (i.e., phase 2) should include a minimal set of covariates while assessing the prognostic value of factors in the sepsis scenario. Regrettably, the limited availability of valid information for analysis hindered our ability to thoroughly evaluate the influence of the clinical setting and mortality definitions on the results. Finally, we believe that the publication of non-significant findings might been affected by reporting biases. Studies that found no association between the biomarker values and mortality probably have fewer chances to be published in indexed journals or in other publicly available sources [69]. Besides, we could not analyse the results of several publications due to their incomplete reporting of results, which increases the risk of selective outcome reporting. Researchers and journals in this field are encouraged to publish studies with null results and carefully report properly their results, as this information is crucial for the assessment of the actual value of any biomarker to guide the management of critically ill patients. Our study has not assessed the added predictive value of the biomarkers. Future investigations should specifically evaluate the biomarkers’ added predictive value in the context of other prognostic information.
Conclusion
The use of resources and related costs for treating sepsis are considerable, so all additional information proposed to guide clinicians in the adequate treatment for each patient need to be supported with the maximum level of evidence to be helpful in the daily clinical setting [2, 8, 70,71,72,73,74]. Biomarkers have the potential to predict or detect sepsis complications, as well as improve prognostication and therefore support clinical management decisions. However, the results of this review showed that unique basal and isolated measurement of PCT, CRP, IL-6, and sCD14 biomarkers are not useful for determining the mortality prognosis among critically ill sepsis patients during their hospital stay. Biomarkers are valuable tools for understanding the biology of sepsis and can guide clinical care by identifying different molecular phenotypes involved in critical illness precision medicine. Before using a biomarker to determine prognosis in clinical practice, we should consider better standardization of clinical studies, which include a group of patients with a specific sepsis phenotype, as well as uniformity in the measurement methods of these biomarkers. Future assessments that integrate these biomarkers with clinical severity scores, individual patient variables, and complex sepsis pathways such as inflammation, clotting or endothelial damage improved patient selection in clinical trials, allowing for the recruitment of more homogenous patient cohorts in objectively predicting adverse outcomes in septic patients [16].
Availability of data and materials
All data generated or analysed during this study are included in this published article [and its supplementary information files].
References
Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA. 2016;315(8):801–10.
Angus DC, Linde-Zwirble WT, Lidicker J, Clermont G, Carcillo J, Pinsky MR. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit Care Med. 2001;29(7):1303–10.
Dellinger RP, Levy MM, Rhodes A, Annane D, Gerlach H, Opal SM, et al. Surviving sepsis campaign: international guidelines for management of severe sepsis and septic shock: 2012. Crit Care Med. 2013;41(2):580–637.
Rhodes A, Evans LE, Alhazzani W, Levy MM, Antonelli M, Ferrer R, et al. Surviving sepsis campaign: international guidelines for management of sepsis and septic shock: 2016. Intensive Care Med. 2017;43(3):304–77.
Whittaker SA, Fuchs BD, Gaieski DF, Christie JD, Goyal M, Meyer NJ, et al. Epidemiology and outcomes in patients with severe sepsis admitted to the hospital wards. J Crit Care. 2015;30(1):78–84.
Yende S, Linde-Zwirble W, Mayr F, Weissfeld LA, Reis S, Angus DC. Risk of cardiovascular events in survivors of severe sepsis. Am J Respir Crit Care Med. 2014;189(9):1065–74.
Levy MM, Fink MP, Marshall JC, Abraham E, Angus D, Cook D, et al. 2001 SCCM/ESICM/ACCP/ATS/SIS international sepsis definitions conference. Crit Care Med. 2003;31(4):1250–6.
Evans L, Rhodes A, Alhazzani W, Antonelli M, Coopersmith CM, French C, et al. Surviving sepsis campaign: international guidelines for management of sepsis and septic shock 2021. Crit Care Med. 2021;49(11):e1063–143.
Ely EW. The ABCDEF bundle: science and philosophy of how ICU liberation serves patients and families. Crit Care Med. 2017;45(2):321–30.
Rello J, van Engelen TSR, Alp E, Calandra T, Cattoir V, Kern WV, et al. Towards precision medicine in sepsis: a position paper from the European Society of Clinical Microbiology and Infectious Diseases. Clin Microbiol Infect. 2018;24(12):1264–72.
Sankar V, Webster NR. Clinical application of sepsis biomarkers. J Anesth. 2013;27(2):269–83.
van Engelen TSR, Wiersinga WJ, Scicluna BP, van der Poll T. Biomarkers in sepsis. Crit Care Clin. 2018;34(1):139–52.
Kibe S, Adams K, Barlow G. Diagnostic and prognostic biomarkers of sepsis in critical care. J Antimicrob Chemother. 2011;66 Suppl 2:ii33-40.
Pierrakos C, Velissaris D, Bisdorff M, Marshall JC, Vincent J-L. Biomarkers of sepsis: time for a reappraisal. Crit Care (London, England). 2020;24(1):287.
Barichello T, Generoso JS, Singer M, Dal-Pizzol F. Biomarkers for sepsis: more than just fever and leukocytosis—a narrative review. Crit Care. 2022;26(1):14.
Giannakopoulos K, Hoffmann U, Ansari U, Bertsch T, Borggrefe M, Akin I, et al. The use of biomarkers in sepsis: a systematic review. Curr Pharm Biotechnol. 2017;18(6):499–507.
Meng FS, Su L, Tang YQ, Wen Q, Liu YS, Liu ZF. Serum procalcitonin at the time of admission to the ICU as a predictor of short-term mortality. Clin Biochem. 2009;42(10–11):1025–31.
Riley RD, Moons KGM, Snell KIE, Ensor J, Hooft L, Altman DG, et al. A guide to systematic review and meta-analysis of prognostic factor studies. BMJ. 2019;364:k4597.
Skoetz N, Collins G, Moons K, Estcourt LJ, Engert A, Kobe C, et al. Interim PET for prognosis in adults with Hodgkin lymphoma: a prognostic factor exemplar review. Cochrane Database Syst Rev. 2017;(4):CD012643. https://doi.org/10.1002/14651858.CD012643. Accessed 25 July 2023.
Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ. 2009;339:b2535.
Hayden JA, Côté P, Steenstra IA, Bombardier C; QUIPS-LBP Working Group. Identifying phases of investigation helps planning, appraising, and applying the results of explanatory prognosis studies. J Clin Epidemiol. 2008;61(6):552–60. https://doi.org/10.1016/j.jclinepi.2007.08.005.
Hayden JA, van der Windt DA, Cartwright JL, Cote P, Bombardier C. Assessing bias in studies of prognostic factors. Ann Intern Med. 2013;158(4):280–6.
Aalto H, Takala A, Kautiainen H, Siitonen S, Repo H. Monocyte CD14 and soluble CD14 in predicting mortality of patients with severe community acquired infection. Scand J Infect Dis. 2007;39(6–7):596–603.
Amancio RT, Japiassu AM, Gomes RN, Mesquita EC, Assis EF, Medeiros DM, et al. The innate immune response in HIV/AIDS septic shock patients: a comparative study. PLoS One. 2013;8(7):e68730.
Andaluz-Ojeda D, Bobillo F, Iglesias V, Almansa R, Rico L, Gandia F, et al. A combined score of pro- and anti-inflammatory interleukins improves mortality prediction in severe sepsis. Cytokine. 2012;57(3):332–6.
Andaluz-Ojeda D, Nguyen HB, Meunier-Beillard N, Cicuéndez R, Quenot JP, Calvo D, et al. Superior accuracy of mid-regional proadrenomedullin for mortality prediction in sepsis with varying levels of illness severity. Ann Intensive Care. 2017;7(1):15.
Carbonell R, Moreno G, Martin-Loeches I, Gomez-Bertomeu F, Sarvise C, Gomez J, et al. Prognostic value of procalcitonin and C-reactive protein in 1608 critically ill patients with severe influenza pneumonia. Antibiotics (Basel, Switzerland). 2021;10(4):350.
Chen J, He Y, Zhou L, Deng Y, Si L. Long non-coding RNA MALAT1 serves as an independent predictive biomarker for the diagnosis, severity and prognosis of patients with sepsis. Mol Med Rep. 2020;21(3):1365–73.
De La Torre-Prados MV, Garcia-De La Torre A, Enguix A, Mayor-Reyes M, Nieto-Gonzalez M, Garcia-Alcantara A. Mid-regional pro-adrenomedullin as prognostic biomarker in septic shock. Minerva Anestesiol. 2016;82(7):760–6.
Eidt MV, Nunes FB, Pedrazza L, Caeran G, Pellegrin G, Melo DAS, et al. Biochemical and inflammatory aspects in patients with severe sepsis and septic shock: the predictive role of IL-18 in mortality. Clin Chim Acta. 2016;453:100–6.
Elke G, Bloos F, Wilson DC, Brunkhorst FM, Briegel J, Reinhart K, et al. The use of mid-regional proadrenomedullin to identify disease severity and treatment response to sepsis - a secondary analysis of a large randomised controlled trial. Crit Care (london, england). 2018;22(1):79.
Erdogan M, Findikli HA, Okuducu TI. A novel biomarker for predicting sepsis mortality: SCUBE-1. Medicine. 2021;100(6):e24671.
Guo Y, Yang H, Gao W, Ma C, Li T. Combination of biomarkers in predicting 28-day mortality for septic patients. J Coll Physicians Surg Pak. 2018;28(9):672–6.
Hu C, Zhou Y, Liu C, Kang Y. Pentraxin-3, procalcitonin and lactate as prognostic markers in patients with sepsis and septic shock. Oncotarget. 2018;9(4):5125–36.
Jain S, Sinha S, Sharma SK, Samantaray JC, Aggrawal P, Vikram NK, et al. Procalcitonin as a prognostic marker for sepsis: a prospective observational study. BMC Res Notes. 2014;7:458.
Jekarl DW, Lee SY, Lee J, Park YJ, Kim Y, Park JH, et al. Procalcitonin as a diagnostic marker and IL-6 as a prognostic marker for sepsis. Diagn Microbiol Infect Dis. 2013;75(4):342–7.
Jiang Y, Jiang F-Q, Kong F, An M-M, Jin B-B, Cao D, et al. Inflammatory anemia-associated parameters are related to 28-day mortality in patients with sepsis admitted to the ICU: a preliminary observational study. Ann Intensive Care. 2019;9(1):67.
Li H, Shan-Shan Z, Jian-Qiang K, Ling Y, Fang L. Predictive value of C-reactive protein and NT-pro-BNP levels in sepsis patients older than 75 years: a prospective, observational study. Aging Clin Exp Res. 2020;32(3):389–97.
Liu S, Wang X, She F, Zhang W, Liu H, Zhao X. Effects of neutrophil-to-lymphocyte ratio combined with interleukin-6 in predicting 28-day mortality in patients with sepsis. Front Immunol. 2021;12:639735.
Masson S, Caironi P, Spanuth E, Thomae R, Fumagalli R, Pesenti A, et al. Presepsin (SCD14-ST) is an early predictor of outcome in patients with severe sepsis or septic shock: preliminary data from the albumin Italian outcome sepsis (ALBIOS) study. Biochim Clin. 2013;37:S44.
Oberholzer A, Souza SM, Tschoeke SK, Oberholzer C, Abouhamze A, Pribble JP, et al. Plasma cytokine measurements augment prognostic scores as indicators of outcome in patients with severe sepsis. Shock (Augusta, Ga). 2005;23(6):488–93.
Phua J, Koay ESC, Lee KH. Lactate, procalcitonin, and amino-terminal pro-B-type natriuretic peptide versus cytokine measurements and clinical severity scores for prognostication in septic shock. Shock (Augusta, Ga). 2008;29(3):328–33.
Ryoo SM, Han KS, Ahn S, Shin TG, Hwang SY, Chung SP, et al. The usefulness of C-reactive protein and procalcitonin to predict prognosis in septic shock patients: a multicenter prospective registry-based observational study. Sci Rep. 2019;9(1):6579.
Shao R, Li C-S, Fang Y, Zhao L, Hang C. Low B and T lymphocyte attenuator expression on CD4+ T cells in the early stage of sepsis is associated with the severity and mortality of septic patients: a prospective cohort study. Crit Care (London, England). 2015;19:308.
Siddiqui S, Gurung RL, Liu S, Ping Seet EC, Lim SC. Genetic polymorphisms and cytokine profile of different ethnicities inseptic shock patients and their association with mortality. Indian J Crit Care Med. 2019;23(3):135–8.
Song J, Park DW, Moon S, Cho HJ, Park JH, Seok H,et al. Diagnostic and prognostic value of interleukin-6, pentraxin 3, and procalcitonin levels among sepsis and septic shock patients: a prospective controlled study according to the Sepsis-3 definitions. BMC Infect Dis. 2019;19(1):968. https://doi.org/10.1186/s12879-019-4618-7.
Su L, Feng L, Song Q, Kang H, Zhang X, Liang Z, et al. Diagnostic value of dynamics serum sCD163, sTREM-1, PCT, and CRP in differentiating sepsis, severity assessment, and prognostic prediction. Mediators Inflamm. 2013;2013:969875.
Suberviola B, Castellanos-Ortega A, Ruiz Ruiz A, Lopez-Hoyos M, Santibanez M. Hospital mortality prognostication in sepsis using the new biomarkers suPAR and proADM in a single determination on ICU admission. Intensive Care Med. 2013;39(11):1945–52.
Sun J, Song SD, Zhao HJ. Expression of soluble triggering receptor expressed on myeloid cells-1 in septic patients and its relation with prognosis. Chin Crit Care Med. 2011;23(5):305–8.
Viallon A, Guyomarc’h S, Marjollet O, Berger C, Carricajo A, Robert F, et al. Can emergency physicians identify a high mortality subgroup of patients with sepsis: role of procalcitonin. Eur J Emerg Med. 2008;15(1):26–33.
Wang M, Zhang Q, Zhao X, Dong G, Li C. Diagnostic and prognostic value of neutrophil gelatinase-associated lipocalin, matrix metalloproteinase-9, and tissue inhibitor of matrix metalloproteinases-1 for sepsis in the Emergency Department: an observational study. Crit Care (London, England). 2014;18(6):634.
Webb AL, Kramer N, Stead TG, Mangal R, Lebowitz D, Dub L, et al. Serum procalcitonin level is associated with positive blood cultures, in-hospital mortality, and septic shock in emergency department sepsis patients. Cureus. 2020;12(4):e7812.
Wu C, Ma J, Yang H, Zhang J, Sun C, Lei Y, et al. Interleukin-37 as a biomarker of mortality risk in patients with sepsis. J Infect. 2021;82(3):346–54.
Wu HP, Chen CK, Chung K, Jiang BY, Yu TJ, Chuang DY. Plasma transforming growth factor-β1 level in patients with severe community-acquired pneumonia and association with disease severity. J Formos Med Assoc. 2009;108(1):20–7.
Xia Y, Zou L, Li D, Qin Q, Hu H, Zhou Y, et al. The ability of an improved qSOFA score to predict acute sepsis severity and prognosis among adult patients. Medicine. 2020;99(5):e18942.
Xie Y, Li B, Lin Y, Shi F, Chen W, Wu W, et al. Combining blood-based biomarkers to predict mortality of sepsis at arrival at the emergency department. Med Sci Monit. 2021;27:e929527.
Yang B, Wang J, Tao X, Wang D. Clinical investigation on the risk factors for prognosis in patients with septic shock. Zhonghua Wei Zhong Bing Ji Jiu Yi Xue. 2019;31(9):1078–82.
Yin Q, Liu B, Chen Y, Zhao Y, Li C. The role of soluble thrombomodulin in the risk stratification and prognosis evaluation of septic patients in the emergency department. Thromb Res. 2013;132(4):471–6.
Zhang Y, Khalid S, Jiang L. Diagnostic and predictive performance of biomarkers in patients with sepsis in an intensive care unit. J Int Med Res. 2019;47(1):44–58.
Zhao J, He Y, Xu P, Liu J, Ye S, Cao Y. Serum ammonia levels on admission for predicting sepsis patient mortality at D28 in the emergency department: a 2-center retrospective study. Medicine. 2020;99(11):e19477.
Zhao M, Duan M. Lactic acid, lactate clearance and procalcitonin in assessing the severity and predicting prognosis in sepsis. Zhonghua Wei Zhong Bing Ji Jiu Yi Xue. 2020;32(4):449–53.
Zhao Y, Li C, Jia Y. Evaluation of the mortality in emergency department sepsis score combined with procalcitonin in septic patients. Am J Emerg Med. 2013;31(7):1086–91.
Findikli HA, ErdoGan M. Serum G protein-coupled estrogen receptor-1 levels and its relation with death in patients with sepsis: a prospective study. Minerva Anestesiol. 2021;87(5):549–55.
Koozi H, Lidestam A, Lengquist M, Johnsson P, Frigyesi A. A simple mortality prediction model for sepsis patients in intensive care. J Intensive Care Soc. 2023;(0). https://doi.org/10.1177/17511437221149572.
Suranadi IW, Sinardja CD, Suryadi IA. Role of procalcitonin in predicting mortality and organ dysfunction at intensive care admission. Int J Gen Med. 2022;15:4917–23.
Yu B, Chen M, Zhang Y, Cao Y, Yang J, Wei B, et al. Diagnostic and prognostic value of interleukin-6 in emergency department sepsis patients. Infect Drug Resist. 2022;15:5557–66.
Nieboer D, Steyerberg EW, Soedamah-Muthu S, Vergouwe Y. Log transformation in biomedical research: (mis)use for covariates. Stat Med. 2013;32(21):3770–1.
Hayden JA, Côté P, Steenstra IA, Bombardier C. Identifying phases of investigation helps planning, appraising, and applying the results of explanatory prognosis studies. J Clin Epidemiol. 2008;61(6):552–60.
Hwang TJ, Carpenter D, Lauffenburger JC, Wang B, Franklin JM, Kesselheim AS. Failure of investigational drugs in late-stage clinical development and publication of trial results. JAMA Intern Med. 2016;176(12):1826–33.
Danna DM. Hospital costs associated with sepsis compared with other medical conditions. Crit Care Nurs Clin North Am. 2018;30(3):389–98.
Sandquist M, Wong HR. Biomarkers of sepsis and their potential value in diagnosis, prognosis and treatment. Expert Rev Clin Immunol. 2014;10(10):1349–56.
Angus DC, Bindman AB. Achieving diagnostic excellence for sepsis. JAMA. 2022;327(2):117–18. https://doi.org/10.1001/jama.2021.23916.
Morden NE, Colla CH, Sequist TD, Rosenthal MB. Choosing wisely–the politics and economics of labeling low-value services. N Engl J Med. 2014;370(7):589–92.
Wammes JJ, van den Akker-van Marle ME, Verkerk EW, van Dulmen SA, Westert GP, van Asselt AD, et al. Identifying and prioritizing lower value services from Dutch specialist guidelines and a comparison with the UK do-not-do list. BMC Med. 2016;14(1):196.
Acknowledgements
SEPSIS BIOMARKERS Collaborators: Alvaro Estupiñan, Luis Franco, Jorge Cardenas, Ivan Robayo, Mario Villabon, Mario Gomez, Elena Stalling, Noelia Alvarez
Funding
Instituto de Salud Carlos III, Spain and European Union (“Fondo Europeo de Desarrollo Regional, Una manera de hacer Europa”), grant number [PI 19/0048].
Author information
Authors and Affiliations
Consortia
Contributions
DMF, IAR, AM, GS, BFF, JLA, XN, RF, and JZ conceived the study. DMF, IAR, AM, SFG, GS, BFF, JLA, XN, RF, and JZ developed the research protocol and search strategy. DMF, IAR, LDCA, SFG, AAM, DSR, AV, GS, BFF, and JLA screened the titles, abstracts, and full texts, and did the data abstraction. AM and JZ verified the study data. AM, LDCA, SFG, BFF, and JZ did the statistical analysis. DMF, IAR, AM, BFF, XN, RF, and JZ co-wrote the first draft of the manuscript, and GS, DSR, IS, DM and JLA reviewed subsequent drafts. XN, RF, and KK provided major revisions to the first draft and reviewed subsequent drafts. All authors had full access to all of the study data throughout the entire review process, and DMF, IAR, AM, and JZ verify the study data. All authors contributed to final edits and agreed to submission of the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
Dr. Ferrer reports personal fees from Shionogi, personal fees from Pfizer, personal fees from MSD, personal fees from Gilead, personal fees from Menarini, personal fees from GSK during the conduct of the study. The remaining authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Additional file S1.
MEDLINE and EMBASE literature search strategies. Additional file S2. QUIPS domains-items considering for assessment. Additional file S3. Excluded studies and reason for exclusion. Additional file S4. Characteristics of Included Studies. Additional file S5. Methodological quality of included studies using the QUIPS tool. Additional Table S6. Baseline PCT values and mortality: measure effects. Additional Table S7. Baseline CRP values and mortality: measure effects. Additional Table S8. Baseline IL-6 values and mortality: measure effects. Additional Table S9. Baseline sCD14 values and mortality: measure effects. Additional Figure S1. Procalcitonin and prediction of mortality at 28-30 days in critically-ill septic patients. Additional Figure S2. C-reactive protein and prediction of mortality at 28-30 days in critically-ill septic patients. Additional Figure S3. Interleukin-6 and prediction of mortality at 28-30 days in critically-ill septic patients. Additional Figure S4. Summary of mortality at 28-30 days and baseline biomarkers measures. Additional Figure S5. Summary of mortality, no details provided and baseline biomarkers measures.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Molano-Franco, D., Arevalo-Rodriguez, I., Muriel, A. et al. Basal procalcitonin, C-reactive protein, interleukin-6, and presepsin for prediction of mortality in critically ill septic patients: a systematic review and meta-analysis. Diagn Progn Res 7, 15 (2023). https://doi.org/10.1186/s41512-023-00152-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41512-023-00152-2