Abstract
Background
Clinical prediction models are often constructed using multicenter databases. Such a data structure poses additional challenges for statistical analysis (clustered data) but offers opportunities for model generalizability to a broad range of centers. The purpose of this study was to describe properties, analysis, and reporting of multicenter studies in the Tufts PACE Clinical Prediction Model Registry and to illustrate consequences of common design and analyses choices.
Methods
Fifty randomly selected studies that are included in the Tufts registry as multicenter and published after 2000 underwent full-text screening. Simulated examples illustrate some key concepts relevant to multicenter prediction research.
Results
Multicenter studies differed widely in the number of participating centers (range 2 to 5473). Thirty-nine of 50 studies ignored the multicenter nature of data in the statistical analysis. In the others, clustering was resolved by developing the model on only one center, using mixed effects or stratified regression, or by using center-level characteristics as predictors. Twenty-three of 50 studies did not describe the clinical settings or type of centers from which data was obtained. Four of 50 studies discussed neither generalizability nor external validity of the developed model.
Conclusions
Regression methods and validation strategies tailored to multicenter studies are underutilized. Reporting on generalizability and potential external validity of the model lacks transparency. Hence, multicenter prediction research has untapped potential.
Registration
This review was not registered.
Similar content being viewed by others
Introduction
Clinical predictive models (CPMs) are clinically useable mathematical equations that relate multiple predictors for a particular individual to the probability of risk for the presence (diagnosis) or future occurrence (prognosis) of a particular outcome [1]. They are an increasingly common and important methodological tool for patient-centered outcomes research and for clinical care. By providing evidence-based estimates of the patient’s probability of health outcomes, CPMs enable clinicians and patients to make decisions that are more rational and consistent with a patient’s own risks, values, and preferences.
It is no surprise that many researchers use multicenter datasets as a substrate to develop clinical prediction models. To be useful, clinical prediction models must be reliable in new patients, potentially including patients from different hospitals, countries, or care settings. Recruitment at multiple sites makes it easier to collect sufficient data to estimate model parameters reliably, especially when the outcome of interest is a rare event.
Currently, the TRIPOD guidelines do not mention any requirements that are specific to multicenter studies, other than reporting the number and location of centers [1, 2]. Nonetheless, multicenter studies pose particular challenges to statistical data analysis and offer new opportunities. On the one hand, a key assumption underlying common regression techniques is violated. Observations are not independent as they are clustered within hospitals; patients within a hospital may be more alike than patients from different hospitals. On the other hand, the fact that hospital populations differ from one another may shed light on the generalizability of the model.
Studies have shown that mixed (i.e., random intercept) and fixed (i.e., center variables) effects regression allow to study differences between centers in event rates and predictor effects and may even provide better predictions [3,4,5,6,7,8]. Leave-center-out cross-validation has been proposed to efficiently assess the generalizability of the model to centers not included in the development set [9, 10]. For example, small center effects and successful cross-validation of a model developed in multiple tertiary care centers in one country strongly indicate generalizability to other tertiary centers in that country. However, transportability of the model to clinical care settings or distinct populations not represented in the data can never be guaranteed [11,12,13]. In the example above, predictive performance in primary or secondary care or in foreign centers (“transportability”) remains to be assessed in external validation studies.
Hence, multicenter studies are extremely interesting if they are representative of the settings in which the model is intended to be used. This is uncertain if centers that participate in studies differ from those who do not, for example, because they have an academic interest or specialization in the disease under study (selection bias) [14]. Cases identified from specialist centers may not be representative of all cases in the general population, and patients without the condition may have been referred there because they presented with many risk factors, distorting regression estimates (referral bias). Moreover, the predictive performance of a model may differ between subgroups in a population (spectrum bias), which has led to the recommendation to use the prevalence in the studied setting as a guide when evaluating whether the reported predictive performance is applicable to a particular clinical setting [15,16,17,18].
The purpose of this research is to investigate the properties, analysis, and reporting of multicenter studies in the Tufts PACE Clinical Prediction Model Registry, a comprehensive database of clinical prediction models for cardiovascular disease [19, 20]. In addition, we provide simulated illustrations of consequences of common design and analysis choices.
Methods
We searched the Tufts PACE Clinical Prediction Model Registry for multicenter studies. This registry contains published clinical prediction models for patients at risk for and with known cardiovascular disease. The inclusion and exclusion criteria and electronic search strategy are published in detail elsewhere [19, 20]. Briefly, the registry was constructed from a PubMed search for English-language articles containing newly developed prediction models published from January 1990 to March 2015. It includes prognostic and diagnostic models to predict binary outcomes (e.g., myocardial infarction or death). Only articles that show the model in a format that allows readers to make individual predictions were included (e.g., an equation, a point score, an online calculator).
We considered studies labeled in the registry as multicenter and published between January 2000 and March 2015. From these, a randomly selected subset of 50 papers underwent full-text screening. When a paper presented multiple prediction models, we selected the primary model as identified by the authors. Where no primary model was identified, we selected the one with the smallest number of events per variable (EPV). We extracted sample size and other dataset characteristics for the development dataset. In two studies, the development dataset was single center (due to a geographical train-test split of multicenter data). In these cases, we described the complete multicenter dataset.
One researcher (LW) extracted the following and entered it in an excel spreadsheet:
-
The total sample size of the development dataset;
-
The number of events of interest in the development dataset;
-
The ratio of the number of patients with events divided by the total sample size (ignoring censoring in time-to-event data);
-
The number of centers;
-
The ratio of the sample size in the largest and smallest center;
-
Center sizes, or any descriptive information related to center size;
-
A description of the setting in which the model was developed (e.g., all tertiary/academic centers, a community cohort, mixed settings);
-
The type of study data used for model development (registry/cohort, trial, individual patient data meta-analysis (IPD-MA));
-
The regression technique used for model development;
-
Whether an external validation was included in the study (a random train-test split is not considered external validation);
-
How many external validation datasets were used;
-
Whether external validation happened in the same center (or community) as model development;
-
Whether external validation happened in the same country as model development;
-
Whether external validation happened in the same care setting as model development;
-
Whether external validation used data from the same time period as model development;
-
Statements regarding the generalizability of the prediction model (including selection, referral, and spectrum bias).
When a publication did not report on these items, but the authors referred to earlier publications or a study website when describing the methodology, we screened these sources. The retrieved information was cross-checked with recorded information in the Tufts PACE database, and conflicts were resolved [19, 20]. We summarized all extracted data as frequencies, medians, ranges, and interquartile ranges.
We did not register this review, nor did we publish a review protocol. We used the PRISMA checklist to prepare this manuscript [21].
To highlight potential issues with common design and analysis choices for multicenter prediction models, we supplemented the review with illustrations, the detailed methods of which are provided in the Appendix. Briefly, in the first illustration, we compare marginal predicted probabilities (obtained by using standard logistic regression ignoring clustering) to conditional predicted probabilities from a mixed effects model. We simulated a dataset with a binary outcome with event rate 0.33 and a single standard normal predictor (with the same distribution across centers) with an effect of 0.8 on the logit scale. We generated data for 500 centers, with 1000 observations each, and let the intercepts vary per center with a standard deviation of 1 or 0.5. We fitted a standard logistic regression model and a mixed effects logistic regression model and evaluated predictions in terms of discrimination and calibration.
In the second illustration, we exemplify non-transportability with a published example of a risk score to predict recurrent cardiovascular events [22] and by using a real multicenter clinical dataset that was collected to develop preoperative prediction models for ovarian cancer diagnosis [23]. The dataset contains information on 3439 patients from 12 oncology referral centers and 1664 patients from 8 general hospitals. We consider a binary outcome Y (ovarian malignancy) and two predictors: age and the log-transformed CA125 biomarker value. The observed relations (from logistic regression) between the outcome and predictors in tertiary care and secondary care are taken to be the true models in each setting. Using resampling techniques, we generated a typical multicenter model development dataset in tertiary care, a large external validation set in tertiary care, and a large external validation set in secondary care. We made calibration plots and computed C-statistics to assess the generalizability and transportability of the model.
Results
The number of clinical prediction models published per year showed a strong increase over time. The proportion of models built using multicenter data increased slightly over time. Sixty-four percent (602/944 models) of models published after 2000 used multicenter data (see Fig. 1).
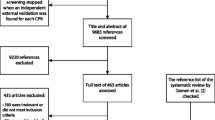
Of all studies included in the Tufts PACE Clinical Prediction Model Registry, 52% (390/747 studies) were multicenter studies published after 2000 (see flowchart in Fig. 2). We discuss the details of a random subset of 50 studies (Table 1) [24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73]. Forty of them analyzed observational multicenter data. Six created a prediction model making secondary use of multicenter trial data. For one study, the reporting on the data source was equivocal. Three studies were individual patient data meta-analyses (IPD-MA), of which two combined datasets from multiple community cohorts and one combined data from multiple trials (some of them are multicenter trials themselves). In IPD-MA of existing study data, the primary source of heterogeneity is at the study level and not at the center level. Hence, from these three studies, study-level information was extracted and analyzed instead of center-level information (e.g., number of studies in the IDP-MA, n in each of the original studies, statistical account of clustering within studies).
The sample size for model development in the 50 selected studies varied from 155 to 834,696 with a median of 2263 (IQR 667 to 13,705). Sixteen studies had between 100 and 1000 observations, 18 studies had between 1000 and 10,000 observations, 13 studies had between 10,000 and 100,000 observations, and 3 studies had more than 100,000 observations.
Multicenter: a catch-all term
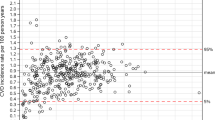
The number of centers contributing data to a multicenter study varied widely from just two centers to 5473 centers, with a median of 10 (IQR 3 to 76) (Fig. 3). As such, “multicenter” is a term covering anything from two researchers joining forces to large international registries. There was a moderate positive correlation between the total sample size and the number of contributing centers (Fig. 3; Spearman ρ 0.4). Four studies did not report the number of participating centers. Two of them were large studies (N = 562,791 and 9556) using data from a quality of care improvement program mentioning more than a hundred centers participated, one was a secondary analysis of a multicenter trial (N = 1692), and one was a cohort study (N = 747).
The median number of patients per center was 133 (range 1 to 16,295; IQR 62 to 530), but imbalance in center contributions was very common. Sample sizes for each included center were reported in only 9/50 studies. The ratio of largest to smallest center size could be computed in 10 studies. Among these, the median sample size ratio of the largest center to the smallest was 4 (range 1 to 34, IQR 3 to 7). Twelve additional studies contained summary statements on sample sizes (e.g., interquartile ranges) or hospital volume (e.g., number of beds, surgical volume, catchment population sizes). These indicated imbalance in center sizes in nine studies, were equivocal in one study, and, interestingly, described active control to limit imbalance in two studies. These active controls were including only the first 10 to 20 consecutive patients in each center and setting an upper inclusion limit for centers. The majority (28/50) did not report on center size imbalance. These tended to be smaller studies (median N 1654 versus 4294).
Multicenter collaborations were not exclusively used to study rare outcomes. The median ratio of the number of events to the total sample size was 0.07 (range 0.001 to 0.60, IQR 0.03 to 0.16, not reported in 1), and the median number of events 177 (range 6 to 48,412; IQR 98 to 497; not reported in 1).
Clustered data is commonly ignored during analysis
The vast majority of studies (39/50) completely ignored the clustered data structure that is typical for multicenter studies during analysis (Fig. 4; 24 used logistic regression models, 13 Cox models, 2 Fine and Gray models). The other studies took clustering into account when building the prediction model in a variety of ways: standard Cox or logistic regression with a center-level covariate (e.g., a fixed center effect, a region effect, a physician experience effect), mixed effects logistic regression, or stratified regression. Two studies split their dataset into a single-center model development set and used the other centers for geographical validation.
Validation of the model
In leave-center out-cross-validation, each cluster is left out of the dataset as a validation set once, while the model is fitted on all other clusters [9]. Only one study used this technique, an IPD-MA that used leave-one-study-out cross-validation.
Ten studies included an external validation in the model development study, two of which used two separate external validation datasets. In 3/10 studies, validation was merely temporal. In 6/10 studies, validation was a mixture of temporal and geographic validation (in a different center or community, four studies used validation data from a different country). Validation in data collected at the same time as model development, but from another center, occurred in 1/10. All validations took place in hospitals or care settings similar to the ones in which the model was developed.
Multicenter data does not guarantee generalizability
To evaluate in what type of settings a prediction model could be applied, it is crucial that researchers describe the settings from which study data was obtained. In our sample, 23/50 studies failed to do so.
Among the studies that did report on the settings from which data was obtained, 16/27 were single-setting studies: 13 collected all data in tertiary/academic centers, 2 collected all data in community hospitals, and 1 collected all data in veteran affair hospitals. Data from multiple settings (8/27; e.g., a mix of academic and community hospitals, a mix of teaching and non-teaching hospitals) and use of population or community cohort data (3/27) were rare.
Most studies used data from only one country; 11/50 were international studies, and 3/50 did not report where data was collected.
Only 17/50 of studies included a direct or indirect statement on the generalizability of their findings to the target population or the potential applicability in other centers. Eleven studies addressed potential selection bias (including referral bias and spectrum bias) in their study. Five studies critically reflected on generalizability in light of a high disease prevalence in their study population. One study questioned generalizability due to the use of specific devices in the participating centers. More than half the studies (27/50) did not discuss representativeness of the study sample or selection bias, but simply stated that external validation is needed. A few studies (4/50) discussed neither generalizability nor external validity. Only two studies (2/50) claimed generalizability of their model to the target population. The first did so on the basis of using data from a large, multinational registry (N = 63,118; 126 centers) covering a heterogeneous population, accrued using consecutive enrolment with a limit on the number of inclusions per month per site to ensure representativeness, with standardized definitions, quality control efforts, and audits. The second (N = 1048, 288 centers, a mix of teaching and non-teaching hospitals) did so based on their use of registry data rather than clinical trial data, a claimed lack of selection, recall, and respondent bias, and successful external validation in an independent registry with a lower proportion of severely ill patients than the development dataset.
Discussion
This review of the Tufts PACE Clinical Prediction Model Registry indicated that 64% of clinical prediction models published after 2000 were derived from multicenter data. Our survey of published studies resulted in a number of important observations.
Firstly, “multicenter” is a broad term, covering small-scale studies conducted at only two collaborating centers as well as very large international registries. Secondly, all studies had in common that the collected patient data cluster within centers, yet this data structure is most often ignored during analysis. In the minority of studies that did acknowledge the data structure, a broad array of analysis techniques were used, indicating there is no commonly accepted way to build prediction models in clustered data. Thirdly, even though multicenter studies hold the promise of better generalizability to the clinical target population than single-center studies, reporting on the potential generalizability and external validity is not transparent. Nearly one in two studies failed to describe the care settings from which data was obtained, and nearly two thirds of studies failed to critically reflect on the potential generalizability of the developed model to new centers or resorted to vague statements that further validation was needed.
Our findings are in line with Kahan’s, who reviewed multicenter randomized trials and found that the reporting of key aspects was poor and only 29% of studies adjusted for center in the analysis [77]. They are also in line with numerous reviews that signaled incomplete reporting and poor design and analysis for prediction models for diabetes [78], kidney disease [79, 80], cancer [81], neurological disorders [82, 83], and fetal and maternal outcomes in obstetrics [84]. Some have criticized vagueness of reported selection criteria, such that it remained unclear whether participants were selected in an unbiased way [85], and specialist fields contributing the majority of models, which are unlikely to be generalizable to general hospital populations [80]. A common grievance is the paucity of external validations [79,80,81,82,83,84,85]. The proportion of multicenter studies was rarely reported in published reviews but seems to vary by specialty [80, 82, 83]. Researchers in neurology have identified the common use of single-center data as one of the main causes for the lack of generalizability of models in their fields [82, 83].
An obvious limitation of our review is that it is limited to 50 risk prediction studies in cardiovascular disease. We expect that in other fields, multicenter studies also differ widely in terms of numbers of included centers and analyses techniques used. A second limitation is that publication bias may have influenced the results. Large-scale studies with many participating centers may have had a higher likelihood of being published, potentially leading to overestimated study sizes and numbers of participating centers in this review.
Recommendations for research practice
The strengths of a multicenter design are the ability to speed up data collection and the coverage of a broader population. However, the successful conduct of prospective multicenter studies requires careful study organization and coordination, motivated study staff at the participating centers, and a dedicated and experienced method center [86]. Based on our findings, we can make some suggestions to improve the analysis and reporting of multicenter studies.
Model development
We recommend using appropriate statistical techniques to analyze clustered data. Studies have shown that standard logistic regression yields suboptimal results when data is clustered [3,4,5]. First, mixed and fixed effects regression provide valuable insights into the differences in event rate between centers, after adjustment for the patient-level predictors [6]. Second, it is well known that ignoring clustering yields incorrect standard errors (often underestimated). This also affects stepwise regression, which is known to induce “testimation bias” even in unclustered data [87], but is still common. Third, standard regression methods yield regression coefficients and predicted probabilities with a marginal interpretation, averaged in the population, ignoring centers [74, 75]. As illustrated here and elsewhere [4, 76], this has a negative impact on model calibration. Miscalibration gets worse as the differences between centers increase. In contrast, fixed (with center dummies) and mixed effects regression methods yield correct standard errors for patient-level predictors and better calibrated predictions to support decision-making at the center level, even if average center effects are used in new centers [3, 4]. Fourth, modeling center effects also improves discrimination between two patients from different centers [4, 88].
During model building, center-level covariates can be used to tailor predictions to centers with specific characteristics. This captures the combined effects of omitted predictors that vary by center and are not fully captured by the patient-specific covariates (e.g., regional differences in population health, different patient spectra due to referral mechanisms). Those may be extremely difficult to capture otherwise. For example, one may incorporate a predictor in the model that specifies the center type or specialty (e.g., tertiary centers versus others) [8, 89, 90]. Note, however, that standard regression techniques that ignore clustering yield standard error estimates for effects of center characteristics that are typically too small.
When using fixed or mixed effects regression, one may check for center-predictor interaction (or random slopes). This type of interaction may be rare and have little influence on predictions [91].
Fixed and mixed effects regression models can easily be fitted with commonly used statistical software. To apply such models in a new center, an average intercept can be used (which is a random intercept of zero in a mixed effects model; in a fixed effects model, this is not straightforward) and simple updating techniques can yield predictions tailored to new centers [13, 92].
Model validation
A geographical split into a single-center development dataset and a validation set consisting of data from the other centers may alleviate the need to address clustering during model building but is very inefficient as only a part of the available data is used for developing the model.
In contrast, leave-center-out cross-validation (also known as internal-external cross-validation) makes efficient use of all available data and offers an excellent opportunity to test the generalizability of the model to new centers [9]. To summarize the predictive performance in each center, a simple average performance [9] or an optimism-corrected performance measure can be computed [93]. At the very least, however, the difference in performance between centers should be inspected [94]. Meta-analytic techniques summarize performance and simultaneously quantify between-center heterogeneity, by distinguishing between sampling variability within centers and variance in predictive performance between centers [18, 95]. Note that leave-center-out cross-validation does not resolve data clustering in the development sets and does not guarantee transportability to distinct care settings or populations not represented in the multicenter dataset.
Reporting
We recommend identifying the patients and care settings for which the model is intended and transparently reporting the threats to generalizability to this target population. This includes careful reporting of center-level characteristics such as types of centers (e.g., secondary versus tertiary care) and the sample sizes per center. Describing the limitations of the study, including non-representativeness of study samples, is already a key element on the TRIPOD checklist [1]. The STROBE checklist includes the requirement to address potential sources of bias and states that the direction and magnitude of any potential biases should be addressed [96]. We fully endorse these recommendations. Selection bias is of special interest in prediction model studies. The study sample may not be representative of the clinical population in which the developed model is intended to be applied, for example, because data was collected in teaching hospitals by physicians with a research interest in the disease under study (referral bias). The performance of a prediction model is influenced by the case-mix and the disease prevalence [15, 97, 98]. Hence, if specialized centers collected the study data, selection bias may lead to overestimated disease probabilities (miscalibration-in-the-large) or lower specificity. Note that in half of the studies reporting information on study settings, all participating centers were tertiary or academic centers. Arguably, prediction models and decision-support systems are most useful at lower levels of care (secondary and primary), where the patient case-mix is broad, and practitioners need to triage and refer patients efficiently.
Design for generalizability
Lastly, multicenter studies have most potential if generalizability is not an afterthought but considered at study design. Frequently, researchers develop prediction models from databases collected for other purposes. However, if prediction is considered in the design phase, centers from diverse settings can participate to ensure the dataset is representative of the intended target population. Moreover, researchers can aim to collect consecutive patients from each participating center, whilst maintaining a good balance in center sample sizes.
Conclusion
Multicenter designs are very common in prediction model development. Although multicenter studies may provide better insight in the generalizability of developed prediction models, this potential often remains untapped due to the use of unsuitable analysis methods and lack of transparent reporting.
Abbreviations
- CPM:
-
Clinical predictive model
- IPD-MA :
-
Individual patient data meta-analysis
- LR :
-
Logistic regression
References
Moons KGM, Altman DG, Reitsma JB, Ioannidis JPA, Macaskill P, Steyerberg EW, Vickers AJ, Ransohoff DF, Collins GS. Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med. 2015;162(1):W1–W73.
Collins GS, Reitsma JB, Altman DG, Moons KG. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): the TRIPOD statement. BMJ. 2015;350:g7594.
Bouwmeester W, Twisk J, Kappen T, Klei W, Moons K, Vergouwe Y. Prediction models for clustered data: comparison of a random intercept and standard regression model. BMC Med Res Methodol. 2013;13(1):19.
Wynants L, Vergouwe Y, Van Huffel S, Timmerman D, Van Calster B: Does ignoring clustering in multicenter data influence the performance of prediction models? A simulation study. Stat Methods Med Res. 2018;27(6):1723-36.
Meisner A, Parikh CR, Kerr KF: Biomarker combinations for diagnosis and prognosis in multicenter studies: principles and methods. Stat Methods Med Res 2017, 0(0):0962280217740392.
Austin PC, Merlo J. Intermediate and advanced topics in multilevel logistic regression analysis. Stat Med. 2017;36(20):3257–77.
Lei L, RM A, Abhik D. Comments on “Intermediate and advanced topics in multilevel logistic regression analysis”. Stat Med. 2018;37(19):2902–6.
Ding X, Gellad ZF, Mather IIIC, Barth P, Poon EG, Newman M, Goldstein BA. Designing risk prediction models for ambulatory no-shows across different specialties and clinics. J Am Med Inform Assoc. 2018;25(8):924–30.
Konig IR, Malley JD, Weimar C, Diener HC, Ziegler A. Practical experiences on the necessity of external validation. Stat Med. 2007;26(30):5499–511.
Steyerberg EW. Validation in prediction research: the waste by data splitting. J Clin Epidemiol. 2018;103:131–3.
Moons KGM, Altman DG, Vergouwe Y, Royston P. Prognosis and prognostic research: application and impact of prognostic models in clinical practice. Br Med J. 2009;338:b606.
Altman DG, Vergouwe Y, Royston P, Moons KG. Prognosis and prognostic research: validating a prognostic model. BMJ. 2009;338:b605.
Steyerberg EW. Clinical prediction models: a practical approach to development, validation, and updating. New York: Springer US; 2009.
Ward H, Toledano MB, Shaddick G, Davies B, Elliott P. Oxford handbook of epidemiology for clinicians. Oxford: OUP; 2012.
Leeflang MM, Rutjes AW, Reitsma JB, Hooft L, Bossuyt PM. Variation of a test's sensitivity and specificity with disease prevalence. CMAJ. 2013;185(11):E537–44.
Willis BH, Hyde CJ. What is the test's accuracy in my practice population? Tailored meta-analysis provides a plausible estimate. J Clin Epidemiol. 2015;68(8):847–54.
Willis BH, Hyde CJ. Estimating a test's accuracy using tailored meta-analysis; how setting-specific data may aid study selection. J Clin Epidemiol. 2014;67(5):538–46.
Wynants L, Riley RD, Timmerman D, Van Calster B. Random-effects meta-analysis of the clinical utility of tests and prediction models. Stat Med. 2018;37(12):2034–52.
Wessler BS, Paulus J, Lundquist CM, Ajlan M, Natto Z, Janes WA, Jethmalani N, Raman G, Lutz JS, Kent DM. Tufts PACE Clinical Predictive Model Registry: update 1990 through 2015. Diagnostic Prognostic Res. 2017;1(1):20.
Wessler BS, Lai Yh L, Kramer W, Cangelosi M, Raman G, Lutz JS, Kent DM. Clinical prediction models for cardiovascular disease: Tufts Predictive Analytics and Comparative Effectiveness Clinical Prediction Model Database. Circ Cardiovasc Qual Outcomes. 2015;8(4):368–75.
Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6(7):e1000097.
Weimar C, Diener HC, Alberts MJ, Steg PG, Bhatt DL, Wilson PW, Mas JL, Rother J. The Essen stroke risk score predicts recurrent cardiovascular events: a validation within the REduction of Atherothrombosis for Continued Health (REACH) registry. Stroke. 2009;40(2):350–4.
Wynants L, Timmerman D, Verbakel JY, Testa A, Savelli L, Fischerova D, Franchi D, Van Holsbeke C, Epstein E, Froyman W, et al. Clinical utility of risk models to refer patients with adnexal masses to specialized oncology care: multicenter external validation using decision curve analysis. Clin Cancer Res. 2017;23(17):5082–90.
Agarwal R, Shah SJ, Foreman AJ, Glassner C, Bartolome SD, Safdar Z, Coslet SL, Anderson AS, Gomberg-Maitland M. Risk assessment in pulmonary hypertension associated with heart failure and preserved ejection fraction. J Heart Lung Transplant. 2012;31(5):467–77.
Alonso A, Krijthe BP, Aspelund T, Stepas KA, Pencina MJ, Moser CB, Sinner MF, Sotoodehnia N, Fontes JD, Janssens AC, et al. Simple risk model predicts incidence of atrial fibrillation in a racially and geographically diverse population: the CHARGE-AF consortium. J Am Heart Assoc. 2013;2(2):e000102.
Amin ST, Morrow DA, Braunwald E, Sloan S, Contant C, Murphy S, Antman EM. Dynamic TIMI risk score for STEMI. J Am Heart Assoc. 2013;2(1):e003269.
Bekelis K, Missios S, Mackenzie TA, Fischer A, Labropoulos N, Eskey C. A predictive model of outcomes during cerebral aneurysm coiling. J Neurointerv Surg. 2014;6(5):342–8.
Carluccio E, Dini FL, Biagioli P, Lauciello R, Simioniuc A, Zuchi C, Alunni G, Reboldi G, Marzilli M, Ambrosio G. The ‘Echo Heart Failure Score’: an echocardiographic risk prediction score of mortality in systolic heart failure. Eur J Heart Fail. 2013;15(8):868–76.
Chung-Esaki H, Rodriguez RM, Alter H, Cisse B. Validation of a prediction rule for endocarditis in febrile injection drug users. Am J Emerg Med. 2014;32(5):412–6.
Cuschieri JR, Drawz P, Falck-Ytter Y, Wong RC. Risk factors for acute gastrointestinal bleeding following myocardial infarction in veteran patients who are prescribed clopidogrel. J Dig Dis. 2014;15(4):195–201.
de la Camara AG, Guerravales JM, Tapia PM, Esteban EA, del Pozo SV, Sandubete EC, Ortega FJ, Puerto AN, Marin-Leon I. Role of biological and non biological factors in congestive heart failure mortality: PREDICE-SCORE: a clinical prediction rule. Cardiol J. 2012;19(6):578–85.
de Man-van Ginkel JM, Hafsteinsdottir TB, Lindeman E, Ettema RG, Grobbee DE, Schuurmans MJ. In-hospital risk prediction for post-stroke depression: development and validation of the Post-stroke Depression Prediction Scale. Stroke. 2013;44(9):2441–5.
den Exter PL, Gomez V, Jimenez D, Trujillo-Santos J, Muriel A, Huisman MV, Monreal M. A clinical prognostic model for the identification of low-risk patients with acute symptomatic pulmonary embolism and active cancer. Chest. 2013;143(1):138–45.
Dodson JA, Reynolds MR, Bao H, Al-Khatib SM, Peterson ED, Kremers MS, Mirro MJ, Curtis JP. Developing a risk model for in-hospital adverse events following implantable cardioverter-defibrillator implantation: a report from the NCDR (National Cardiovascular Data Registry). J Am Coll Cardiol. 2014;63(8):788–96.
Eichinger S, Heinze G, Kyrle PA. D-dimer levels over time and the risk of recurrent venous thromboembolism: an update of the Vienna prediction model. J Am Heart Assoc. 2014;3(1):e000467.
Enajat M, Damen TH, Geenen A, Timman R, van der Hulst RR, Mureau MA. Pulmonary embolism after abdominal flap breast reconstruction: prediction and prevention. Plast Reconstr Surg. 2013;131(6):1213–22.
Farooq V, Vergouwe Y, Raber L, Vranckx P, Garcia-Garcia H, Diletti R, Kappetein AP, Morel MA, de Vries T, Swart M, et al. Combined anatomical and clinical factors for the long-term risk stratification of patients undergoing percutaneous coronary intervention: the Logistic Clinical SYNTAX score. Eur Heart J. 2012;33(24):3098–104.
Felker GM, Leimberger JD, Califf RM, Cuffe MS, Massie BM, Adams KF Jr, Gheorghiade M, O'Connor CM. Risk stratification after hospitalization for decompensated heart failure. J Card Fail. 2004;10(6):460–6.
Goff DC Jr, Lloyd-Jones DM, Bennett G, Coady S, D'Agostino RB Sr, Gibbons R, Greenland P, Lackland DT, Levy D, O'Donnell CJ, et al. 2013 ACC/AHA guideline on the assessment of cardiovascular risk: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol. 2014;63(25 Pt B):2935–59.
Grant SW, Hickey GL, Grayson AD, Mitchell DC, McCollum CN. National risk prediction model for elective abdominal aortic aneurysm repair. Br J Surg. 2013;100(5):645–53.
Gupta PK, Ramanan B, Lynch TG, Sundaram A, MacTaggart JN, Gupta H, Fang X, Pipinos II. Development and validation of a risk calculator for prediction of mortality after infrainguinal bypass surgery. J Vasc Surg. 2012;56(2):372–9.
Hannan EL, Farrell LS, Walford G, Jacobs AK, Berger PB, Holmes DR Jr, Stamato NJ, Sharma S, King SB 3rd. The New York State risk score for predicting in-hospital/30-day mortality following percutaneous coronary intervention. JACC Cardiovasc Interv. 2013;6(6):614–22.
Hubert S, Thuny F, Resseguier N, Giorgi R, Tribouilloy C, Le Dolley Y, Casalta JP, Riberi A, Chevalier F, Rusinaru D, et al. Prediction of symptomatic embolism in infective endocarditis: construction and validation of a risk calculator in a multicenter cohort. J Am Coll Cardiol. 2013;62(15):1384–92.
Iannuzzi JC, Young KC, Kim MJ, Gillespie DL, Monson JR, Fleming FJ. Prediction of postdischarge venous thromboembolism using a risk assessment model. J Vasc Surg. 2013;58(4):1014–20 e1011.
Iida O, Soga Y, Yamauchi Y, Hirano K, Kawasaki D, Tazaki J, Yamaoka T, Suematsu N, Suzuki K, Shintani Y, et al. Anatomical predictors of major adverse limb events after infrapopliteal angioplasty for patients with critical limb ischaemia due to pure isolated infrapopliteal lesions. Eur J Vasc Endovasc Surg. 2012;44(3):318–24.
Iung B, Laouenan C, Himbert D, Eltchaninoff H, Chevreul K, Donzeau-Gouge P, Fajadet J, Leprince P, Leguerrier A, Lievre M, et al. Predictive factors of early mortality after transcatheter aortic valve implantation: individual risk assessment using a simple score. Heart. 2014;100(13):1016–23.
Kay J, Dorbala S, Goyal A, Fazel R, Di Carli MF, Einstein AJ, Beanlands RS, Merhige ME, Williams BA, Veledar E, et al. Influence of sex on risk stratification with stress myocardial perfusion Rb-82 positron emission tomography: results from the PET (positron emission tomography) Prognosis Multicenter Registry. J Am Coll Cardiol. 2013;62(20):1866–76.
Kramer DB, Friedman PA, Kallinen LM, Morrison TB, Crusan DJ, Hodge DO, Reynolds MR, Hauser RG. Development and validation of a risk score to predict early mortality in recipients of implantable cardioverter-defibrillators. Heart Rhythm. 2012;9(1):42–6.
Ky B, French B, Levy WC, Sweitzer NK, Fang JC, Wu AH, Goldberg LR, Jessup M, Cappola TP. Multiple biomarkers for risk prediction in chronic heart failure. Circ Heart Fail. 2012;5(2):183–90.
Liu LP, Zheng HG, Wang DZ, Wang YL, Hussain M, Sun HX, Wang AX, Zhao XQ, Dong KH, Wang CX, et al. Risk assessment of deep-vein thrombosis after acute stroke: a prospective study using clinical factors. CNS Neurosci Ther. 2014;20(5):403–10.
Myint PK, Clark AB, Kwok CS, Davis J, Durairaj R, Dixit AK, Sharma AK, Ford GA, Potter JF. The SOAR (Stroke subtype, Oxford Community Stroke Project classification, Age, prestroke modified Rankin) score strongly predicts early outcomes in acute stroke. Int J Stroke. 2014;9(3):278–83.
Palmerini T, Genereux P, Caixeta A, Cristea E, Lansky A, Mehran R, Della Riva D, Fahy M, Xu K, Stone GW. A new score for risk stratification of patients with acute coronary syndromes undergoing percutaneous coronary intervention: the ACUITY-PCI (Acute Catheterization and Urgent Intervention Triage Strategy-Percutaneous Coronary Intervention) risk score. JACC Cardiovasc Interv. 2012;5(11):1108–16.
Pannucci CJ, Laird S, Dimick JB, Campbell DA, Henke PK. A validated risk model to predict 90-day VTE events in postsurgical patients. Chest. 2014;145(3):567–73.
Park KL, Budaj A, Goldberg RJ, Anderson FA Jr, Agnelli G, Kennelly BM, Gurfinkel EP, Fitzgerald G, Gore JM. Risk-prediction model for ischemic stroke in patients hospitalized with an acute coronary syndrome (from the global registry of acute coronary events [GRACE]). Am J Cardiol. 2012;110(5):628–35.
Perry JJ, Sharma M, Sivilotti ML, Sutherland J, Worster A, Emond M, Stotts G, Jin AY, Oczkowski WJ, Sahlas DJ, et al. A prospective cohort study of patients with transient ischemic attack to identify high-risk clinical characteristics. Stroke. 2014;45(1):92–100.
Piccini JP, Stevens SR, Chang Y, Singer DE, Lokhnygina Y, Go AS, Patel MR, Mahaffey KW, Halperin JL, Breithardt G, et al. Renal dysfunction as a predictor of stroke and systemic embolism in patients with nonvalvular atrial fibrillation: validation of the R(2)CHADS(2) index in the ROCKET AF (Rivaroxaban Once-daily, oral, direct factor Xa inhibition Compared with vitamin K antagonism for prevention of stroke and Embolism Trial in Atrial Fibrillation) and ATRIA (AnTicoagulation and Risk factors In Atrial fibrillation) study cohorts. Circulation. 2013;127(2):224–32.
Pros N, Cambou JP, Aboyans V, Malloizel Delaunay J, Constans J, Lacroix P, Saint Lebes B, Curaudeau E, Bura A. A hospital discharge risk score for 1-year all-cause mortality or non-fatal cardiovascular events in patients with lower-extremity peripheral artery disease, with and without revascularisation. Eur J Vasc Endovasc Surg. 2013;45(5):488–96.
Rao SV, McCoy LA, Spertus JA, Krone RJ, Singh M, Fitzgerald S, Peterson ED. An updated bleeding model to predict the risk of post-procedure bleeding among patients undergoing percutaneous coronary intervention: a report using an expanded bleeding definition from the National Cardiovascular Data Registry CathPCI Registry. JACC Cardiovasc Interv. 2013;6(9):897–904.
Rizzi M, Ravasio V, Carobbio A, Mattucci I, Crapis M, Stellini R, Pasticci MB, Chinello P, Falcone M, Grossi P, et al. Predicting the occurrence of embolic events: an analysis of 1456 episodes of infective endocarditis from the Italian Study on Endocarditis (SEI). BMC Infect Dis. 2014;14:230.
Schellinger PD, Kohrmann M, Liu S, Dillon WP, Nogueira RG, Shuaib A, Liebeskind DS. Favorable vascular profile is an independent predictor of outcome: a post hoc analysis of the Safety and Efficacy of NeuroFlo Technology in Ischemic Stroke trial. Stroke. 2013;44(6):1606–8.
Singer DE, Chang Y, Borowsky LH, Fang MC, Pomernacki NK, Udaltsova N, Reynolds K, Go AS. A new risk scheme to predict ischemic stroke and other thromboembolism in atrial fibrillation: the ATRIA study stroke risk score. J Am Heart Assoc. 2013;2(3):e000250.
Stiell IG, Clement CM, Brison RJ, Rowe BH, Borgundvaag B, Aaron SD, Lang E, Calder LA, Perry JJ, Forster AJ, et al. A risk scoring system to identify emergency department patients with heart failure at high risk for serious adverse events. Acad Emerg Med. 2013;20(1):17–26.
Takagi Y, Takahashi J, Yasuda S, Miyata S, Tsunoda R, Ogata Y, Seki A, Sumiyoshi T, Matsui M, Goto T, et al. Prognostic stratification of patients with vasospastic angina: a comprehensive clinical risk score developed by the Japanese Coronary Spasm Association. J Am Coll Cardiol. 2013;62(13):1144–53.
Than M, Flaws D, Sanders S, Doust J, Glasziou P, Kline J, Aldous S, Troughton R, Reid C, Parsonage WA, et al. Development and validation of the Emergency Department Assessment of Chest pain Score and 2 h accelerated diagnostic protocol. Emerg Med Australas. 2014;26(1):34–44.
Tolenaar JL, Froehlich W, Jonker FH, Upchurch GR Jr, Rampoldi V, Tsai TT, Bossone E, Evangelista A, O'Gara P, Pape L, et al. Predicting in-hospital mortality in acute type B aortic dissection: evidence from International Registry of Acute Aortic Dissection. Circulation. 2014;130(11 Suppl 1):S45–50.
Trujillo-Santos J, Lozano F, Lorente MA, Adarraga D, Hirmerova J, Del Toro J, Mazzolai L, Barillari G, Barron M, Monreal M. A prognostic score to identify low-risk outpatients with acute deep vein thrombosis in the lower limbs. Am J Med. 2015;128(1):90 e99–15.
Van De Bruaene A, Moons P, Belmans A, Post MC, Luermans JG, Delcroix M, Pasquet A, De Backer J, Paelinck B, Morissens M, et al. Predictive model for late atrial arrhythmia after closure of an atrial septal defect. Int J Cardiol. 2013;164(3):318–22.
Van Hattum ES, Tangelder MJ, Lawson JA, Moll FL, Algra A. Long-term risk of vascular events after peripheral bypass surgery. A cohort study. Thromb Haemost. 2012;108(3):543–53.
Wang JW, Chen YD, Wang CH, Yang XC, Zhu XL, Zhou ZQ. Development and validation of a clinical risk score predicting the no-reflow phenomenon in patients treated with primary percutaneous coronary intervention for ST-segment elevation myocardial infarction. Cardiology. 2013;124(3):153–60.
Wilson PW, D'Agostino R Sr, Bhatt DL, Eagle K, Pencina MJ, Smith SC, Alberts MJ, Dallongeville J, Goto S, Hirsch AT, et al. An international model to predict recurrent cardiovascular disease. Am J Med. 2012;125(7):695–703 e691.
Wimmer NJ, Resnic FS, Mauri L, Matheny ME, Piemonte TC, Pomerantsev E, Ho KK, Robbins SL, Waldman HM, Yeh RW. Risk-treatment paradox in the selection of transradial access for percutaneous coronary intervention. J Am Heart Assoc. 2013;2(3):e000174.
Wimmer NJ, Yeh RW, Cutlip DE, Mauri L. Risk prediction for adverse events after carotid artery stenting in higher surgical risk patients. Stroke. 2012;43(12):3218–24.
Zheng Z, Zhang L, Li X, Hu S. SinoSCORE: a logistically derived additive prediction model for post-coronary artery bypass grafting in-hospital mortality in a Chinese population. Front Med. 2013;7(4):477–85.
Neuhaus JM. Statistical methods for longitudinal and clustered designs with binary responses. Stat Methods Med Res. 1992;1(3):249–73.
Zeger SL, Liang K-Y, Albert PS. Models for longitudinal data: a generalized estimating equation approach. Biometrics. 1988;44(4):1049–60.
Pavlou M, Ambler G, Seaman S, Omar RZ. A note on obtaining correct marginal predictions from a random intercepts model for binary outcomes. BMC Med Res Methodol. 2015;15:59.
Kahan BC, Harhay MO. Many multicenter trials had few events per center, requiring analysis via random-effects models or GEEs. J Clin Epidemiol. 2015;68(12):1504–11.
Collins GS, Mallett S, Omar O, Yu LM. Developing risk prediction models for type 2 diabetes: a systematic review of methodology and reporting. BMC Med. 2011;9:103.
Collins GS, Omar O, Shanyinde M, Yu LM. A systematic review finds prediction models for chronic kidney disease were poorly reported and often developed using inappropriate methods. J Clin Epidemiol. 2013;66(3):268–77.
Hodgson LE, Sarnowski A, Roderick PJ, Dimitrov BD, Venn RM, Forni LG: Systematic review of prognostic prediction models for acute kidney injury (AKI) in general hospital populations. BMJ Open. 2017;7(9):e016591.
Mallett S, Royston P, Waters R, Dutton S, Altman DG. Reporting performance of prognostic models in cancer: a review. BMC Med. 2010;8:21.
Mushkudiani NA, Hukkelhoven CW, Hernandez AV, Murray GD, Choi SC, Maas AI, Steyerberg EW. A systematic review finds methodological improvements necessary for prognostic models in determining traumatic brain injury outcomes. J Clin Epidemiol. 2008;61(4):331–43.
Jaja BN, Cusimano MD, Etminan N, Hanggi D, Hasan D, Ilodigwe D, Lantigua H, Le Roux P, Lo B, Louffat-Olivares A, et al. Clinical prediction models for aneurysmal subarachnoid hemorrhage: a systematic review. Neurocrit Care. 2013;18(1):143–53.
Kleinrouweler CE, Cheong-See FM, Collins GS, Kwee A, Thangaratinam S, Khan KS, Mol BW, Pajkrt E, Moons KG, Schuit E. Prognostic models in obstetrics: available, but far from applicable. Am J Obstet Gynecol. 2016;214:79–90 e36.
Bouwmeester W, Zuithoff NP, Mallett S, Geerlings MI, Vergouwe Y, Steyerberg EW, Altman DG, Moons KG. Reporting and methods in clinical prediction research: a systematic review. PLoS Med. 2012;9(5):1–12.
Sprague S, Matta JM, Bhandari M, Dodgin D, Clark CR, Kregor P, Bradley G, Little L. Multicenter collaboration in observational research: improving generalizability and efficiency. J Bone Joint Surg Am. 2009;91(Suppl 3):80–6.
Steyerberg EW, Eijkemans MJ, Habbema JD. Stepwise selection in small data sets: a simulation study of bias in logistic regression analysis. J Clin Epidemiol. 1999;52(10):935–42.
Van Oirbeek R, Lesaffre E. Assessing the predictive ability of a multilevel binary regression model. Comput Stat Data Anal. 2012;56(6):1966–80.
Van Calster B, Van Hoorde K, Valentin L, Testa AC, Fischerova D, Van Holsbeke C, Savelli L, Franchi D, Epstein E, Kaijser J, et al. Evaluating the risk of ovarian cancer before surgery using the ADNEX model to differentiate between benign, borderline, early and advanced stage invasive, and secondary metastatic tumours: prospective multicentre diagnostic study. BMJ. 2014;349:g5920.
Timmerman D, Van Calster B, Testa A, Savelli L, Fischerova D, Froyman W, Wynants L, Van Holsbeke C, Epstein E, Franchi D, et al. Predicting the risk of malignancy in adnexal masses based on the simple rules from the International Ovarian Tumor Analysis (IOTA) group. Am J Obstet Gynecol. 2016;214(4):424–37.
Austin PC, van Klaveren D, Vergouwe Y, Nieboer D, Lee DS, Steyerberg EW. Validation of prediction models: examining temporal and geographic stability of baseline risk and estimated covariate effects. Diagnostic Prognostic Res. 2017;1(1):12.
Debray TP, Vergouwe Y, Koffijberg H, Nieboer D, Steyerberg EW, Moons KG. A new framework to enhance the interpretation of external validation studies of clinical prediction models. J Clin Epidemiol. 2015;68(3):279–89.
Royston P, Parmar MK, Sylvester R. Construction and validation of a prognostic model across several studies, with an application in superficial bladder cancer. Stat Med. 2004;23(6):907–26.
Debray TPA, Moons KGM, Ahmed I, Koffijberg H, Riley RD. A framework for developing, implementing, and evaluating clinical prediction models in an individual participant data meta-analysis. Stat Med. 2013;32(18):3158–80.
Riley RD, Ahmed I, Debray TP, Willis BH, Noordzij JP, Higgins JP, Deeks JJ. Summarising and validating test accuracy results across multiple studies for use in clinical practice. Stat Med. 2015;34(13):2081–103.
Vandenbroucke JP, von Elm E, Altman DG, Gøtzsche PC, Mulrow CD, Pocock SJ, Poole C, Schlesselman JJ, Egger M, Initiative ftS. Strengthening the Reporting of Observational Studies in Epidemiology (STROBE): explanation and elaboration. Epidemiology. 2007;18(6):805–35.
Vergouwe Y, Moons KGM, Steyerberg EW. External validity of risk models: use of benchmark values to disentangle a case-mix effect from incorrect coefficients. Am J Epidemiol. 2010;172(8):971–80.
van Klaveren D, Gonen M, Steyerberg EW, Vergouwe Y. A new concordance measure for risk prediction models in external validation settings. Stat Med. 2016;35(23):4136–52.
Acknowledgements
Not applicable.
Funding
The study was supported by the Research Foundation–Flanders (FWO) projects G049312 N/G0B4716N, and Internal Funds KU Leuven (project C24/15/037). DT is a senior clinical investigator of FWO. LW is a postdoctoral fellow of the FWO. This work was partially supported through a Patient-Centered Outcomes Research Institute (PCORI) Award (ME-1606-35555). All statements in this report, including its findings and conclusions, are solely those of the authors and do not necessarily represent the views of the Patient-Centered Outcomes Research Institute (PCORI), its Board of Governors, or Methodology Committee. The funding bodies had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
The studies analyzed are available in the Tufts PACE Clinical Predictive Model Registry, http://pace.tuftsmedicalcenter.org/cpm/default.aspx/.
Author information
Authors and Affiliations
Contributions
LW, BVC, and DK conceived and designed the study. CL and DK provided access to the full CPM registry. CL selected studies from the registry. LW extracted the data from the publications. LW analyzed the data, with assistance from CL. DT provided the data for the illustrations. LW wrote the first draft, which was then critically reviewed and revised by BVC and DK. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Simulation code. (PDF 204 kb)
Appendix (Fig. 7)
Appendix (Fig. 7)
Simulation details
Illustration 1 Ignoring clustering
We generated a dataset of 500,000 patients. A single standard normal predictor X (which may be thought of as a linear combination of multiple predictors) had a true effect of 0.8. The true intercept was − 1. Random effects (ri) of 500 centers (1000 patients each) were generated with a mean of zero and a standard deviation of 1 or 0.5 (two scenarios). The true disease probability was calculated by applying the inverse logit transformation to − 1 + ri + 0.8 X. By comparing the disease probability to a random draw from a uniform distribution on the interval [0,1], the binary outcome vector Y was created. The event rate is 0.33 in scenario 1 and 0.31 in scenario 2.
In the generated datasets, a standard and mixed effects (random intercept) logistic regression model were fitted. We plotted the predicted probabilities of each model as a function of X. We calculated the C-statistic of both. In addition, we plot the predicted probabilities against the true probabilities in an average-prevalence center, a high-prevalence center, and a low-prevalence center. The R code is provided online.
The results of scenario 1, which has extreme clustering, are shown in the main manuscript for didactical reasons. The plot below shows the results of scenario 2, with small between-center differences, akin to what may be found in a study using data from various similar centers (e.g., all tertiary centers).
Illustration 2 Non-transportability
The estimated random intercept logistic regression models in a clinical database (3439 patients in 12 oncology referral centers and 1664 patients in 8 general hospitals) were taken to be the true models in this example. In tertiary care, this is log(p(Y = 1)/(p(Y = 0)) = − 5.77 + 0.04 × age + 0.85 × log(biomarker). The proportion of residual variance in Y attributable to differences between centers is 0.06. In secondary care, this is log(p(Y = 1)/(p(Y = 0)) = − 7.77 + 0.05 × age + 0.91 × log(biomarker). The proportion of residual variance in Y attributable to differences between centers is < 0.01.
A dataset to develop a clinical prediction model was constructed by sampling age and biomarker values from the tertiary care dataset. This way, we assure a realistic joint distribution of predictor values. The properties of the multicenter model development dataset were informed by findings of the review. We first sampled 10 centers without replacement, and within each center, we sampled patients such that the total sample size was 2263 and the ratio between the largest and the smallest center sample size was 4 to 1. After predictor values were sampled, Y values were generated according to the true model in tertiary care, using center-specific intercepts.
In the created development dataset, a random intercept logistic regression model was fitted to generate the prediction model.
Next, a simple random sample of 10,000 patients was taken with replacement from the tertiary care database, again generating Y values according to the sampled predictor values and the true model coefficients, but this time assuming an average center intercept. By creating such a large validation set (“augmenting” or “upstrapping” the real tertiary care database), we can trust that observed differences in predictive performance in this illustration are not due to random sampling error. Generating a 0/1 outcome based on the model-based true probability for a certain vector of predictor values (e.g., age 49 and log-biomarker value 4) assures that the event is observed with a certain probability each time this predictor value combination is sampled,and avoids exact replicates of observations and deterministic relations in the dataset. The generated dataset is the validation set to assess discrimination and calibration in tertiary care.
Another simple random sample of 10,000 patients was taken with replacement from the secondary care database, while Y values were generated according to the true model coefficients in secondary care. This is the validation set to assess discrimination and calibration in secondary care.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Wynants, L., Kent, D.M., Timmerman, D. et al. Untapped potential of multicenter studies: a review of cardiovascular risk prediction models revealed inappropriate analyses and wide variation in reporting. Diagn Progn Res 3, 6 (2019). https://doi.org/10.1186/s41512-019-0046-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41512-019-0046-9