Abstract
Background
With the continuation of the COVID-19 pandemic, some COVID-19 patients have become reinfected with the virus. Viral gene sequencing has found that some of these patients were reinfected by the different and others by same strains. This has raised concerns about the effectiveness of immunity after infection and the reliability of vaccines. To this end, we conducted a systematic review to assess the characteristics of patients with reinfection and possible causes.
Methods
A systematic search was conducted across eight databases: PubMed, Embase, Web of Science, The Cochrane Library, CNKI, WanFang, VIP and SinoMed from December 1, 2019 to September 1, 2021. The quality of included studies were assessed using JBI critical appraisal tools and Newcastle–Ottawa Scale.
Results
This study included 50 studies from 20 countries. There were 118 cases of reinfection. Twenty-five patients were reported to have at least one complication. The shortest duration between the first infection and reinfection was 19 days and the longest was 293 days. During the first infection and reinfection, cough (51.6% and 43.9%) and fever (50% and 30.3%) were the most common symptoms respectively. Nine patients recovered, seven patients died, and five patients were hospitalized, but 97 patients’ prognosis were unknown. B.1 is the most common variant strain at the first infection. B.1.1.7, B.1.128 and B.1.351 were the most common variant strains at reinfection. Thirty-three patients were infected by different strains and 9 patients were reported as being infected with the same strain.
Conclusions
Our research shows that it is possible for rehabilitated patients to be reinfected by SARS-COV-2. To date, the causes and risk factors of COVID-19 reinfection are not fully understood. For patients with reinfection, the diagnosis and management should be consistent with the treatment of the first infection. The public, including rehabilitated patients, should be fully vaccinated, wear masks in public places, and pay attention to maintaining social distance to avoid reinfection with the virus.
Similar content being viewed by others
Introduction
As COVID-19 epidemic continues to spread worldwide, it has caused 263,563,622 confirmed cases of COVID-19, including 5,232,562 deaths as of 3 December 2021 [1]. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a single-stranded positive-strand RNA virus that belongs to the Coronaviridae family [2, 3]. Coronaviruses (CoVs) were previously known to be present in the environment and to infect humans, for example SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV) have appeared in the past two decades. SARS-CoV-2 is characterized by efficient transmission despite having a lower mortality rate compared with the other two CoVs [4]. A number of animal experiments have shown reinfection with the same or a different strain after initial infection with SARS-CoV-2 for more than or equal to 21 [5, 6] and 28 days [7]. This suggests that humans can also be at risk of being reinfected.
In fact, reinfected people have been reported during the present outbreak. The first case of COVID-19 reinfection was described in Hong Kong in August 2020, a thirty-three years old male was asymptomatic during the second infection and different strains of SARS-CoV-2 were identified in the two infections [8]. Subsequently, many countries, such as the United States [9] and Italy [10], have also reported the emergence of reinfected patients.
The SARS-CoV-2 continues to mutate, and new mutations have appeared in the Netherlands [11], the United States [12], India [13] and elsewhere. World Health Organization (WHO) has announced new easy-to-remember labels for Variants of Interest (VOIs) and Variants of Concern (VOC) to facilitate public communication about SARS-CoV-2 variants, these currently include Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2) and Omicron (B.1.1.529) [14]. The emergence of a variant may affect the retransmission of the disease, its severity and doctors’ ability to diagnose, treat, prevent, and control the infection [15, 16]. However, studies have shown that compared to other variants, the Omicron variants pose an increased risk of reinfection [17]. It has also caused public concern and controversy, which includes questions about the contagious nature of reinfected patients, the effectiveness of vaccines and their usefulness against virus variants. Knowing the frequency and natural course of reinfections is important for developing strategies to control SARS-CoV-2.
Many studies have defined re-positive RT-PCR as reinfection which may not always be the case, or have not reported viral gene sequencing results or have omitted clear epidemiological data of patients with reinfections, which will greatly distort the description of the number and characteristics of reinfected patients. Knowledge about reinfected patients is still inadequate and limited. Therefore, because of the need to target confirmed reinfections in patients we have done this review in order to provide clear information for this paper. The present study provides an independent definition of reinfected persons: laboratory confirmation of two infections with the same or different virus strains by lineage, clades, phylogenetic analysis (proof of two distinct virus variants with any sequence variation between the two episodes) for the first and second infections. If there are no laboratory data on the first infection, clear epidemiological data are needed (eg. there are clear epidemiological data to indicate that the virus reinfecting the patient was not spreading locally at the time of the patient's initial infection, so as to prove that the virus strains of the two infections are unrelated).
The purpose of this systematic review is to summarize the characteristics of patients with proven reinfection, including details of clinical symptoms, viral load, and viral gene sequencing of primary infections and subsequent reinfections, and whether or not these patients are contagious. In addition, we will discuss the potential reasons for reinfection to provide advice on management of reinfected patients.
Methods
The study protocol was registered at PROSPERO, which is an ongoing systematic review registry (ID: CRD42021265333) [18]. This review was performed and reported in accordance with Preferred Reporting Items for Systematic Reviews and Meta-analyses 2020 (PRISMA 2020) [19].
Data sources and search strategy
We searched the following eight databases: PubMed, Embase, Web of Science, The Cochrane Library, CNKI, WanFang, VIP and SinoMed from December 1, 2019 to September 1, 2021. At the same time, we checked the previous relevant systematic reviews on the topic to ensure that no eligible articles were missed [20,21,22,23,24,25,26,27]. We constructed a detailed search strategy to fully capture the reinfected patients, and Additional file 1: Table S1 provides the search strategy for databases. We applied no restrictions for language of publications. Studies were selected for further consideration through screening of titles, abstracts, and methods for relevance based on the eligibility criteria after excluding duplications. Two independent researchers (XY Ren and J Zhou) screened retrieved articles and both of them reviewed each article. These investigators then independently assessed full texts of records deemed eligible for inclusion. Any discrepancies were resolved by discussion with other co-authors.
Eligibility criteria
Studies were selected based on the following inclusion criteria: (1) papers recruited patients that met our definition of reinfection; (2) reported outcomes of interest included description of clinical symptoms of both infections, viral gene sequencing, virus load, or infectivity; (3) original research with any type of observational study (cohort study, cross-sectional study, case–control study, case report and case series).
Exclusion criteria are: (1) articles focusing on animal experiments; (2) Full texts of studies were not available.
Data extraction
Two independent reviewers (XY Ren and J Zhou) extracted data from each eligible study and then cross-checked the results. Disagreements between reviewers regarding extracted data were resolved through discussion and consensus with the third reviewer (J Guo). We extracted data about the constructed indices from all papers that met the inclusion criteria, which included first author name, date of publication, country, type of study, age, sex and co-morbidities of the reinfected patients, the proportion of reinfected patients among discharged patients, the time interval between the first and second clinical symptoms, results of virus gene sequencing and the cycle threshold (Ct) value of both infections, vaccination status, and the patient outcomes.
Quality assessment
Included articles were independently assessed for quality by two reviewers (CM Hao and MX Zheng) using criteria based on the standard principles of quality assessment. The methodological quality of the included case reports, case series, cross-sectional and case–control studies were assessed based on JBI critical appraisal tools [28]. The quality of each checklist item was graded as Yes, No, Unclear or Not applicable. The methodological quality for the cohort studies was assessed based on Newcastle–Ottawa Scale [29]. The quality was ranked as: unsatisfactory (0–4 points), satisfactory (5–6 points), and good (7–8 points), or very good (9–10 points) [30]. The three reviewers then shared the quality assessment checklist results and reached consensus through discussion.
Results
Search results
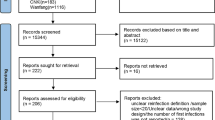
A total of 2788 records were identified in the initial literature search. After removing 1708 duplicates, 1080 articles were screened by titles and abstracts, and 837 articles were excluded. 243 studies were reviewed using the full texts and finally 50 articles met the inclusion criteria and were analyzed in the systematic review (Fig. 1). In these studies, there were 46 case reports [8,9,10, 31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73], 2 cross-sectional studies [74, 75], 1 cohort study [76] and 1 case–control study [77]. Ten papers were from Brazil, 7 from the United States, 5 from India, 4 from Italy, 3 from the United Kingdom, 12 studies, 2 each from Spain, Belgium, Ecuador, Netherlands, Iran and France. The remaining 9 studies came from Panama, Qatar, Luxembourg, South Korea, Saudi Arabia, Switzerland, Colombia, Germany and China.
Study quality assessment
Overall, the methodological quality of 46 case reports (Additional file 1: Table S2) and 1 cohort study (Additional file 1: Table S5) were moderate to high, 1 case–control study (Additional file 1: Table S4) was moderate because it did not identify and deal with the confounding factors. The methodological quality of 2 cross-sectional studies were moderate (Additional file 1: Table S3) because neither of them had clear exposure factors.
Characteristics of reinfected patients
A total of 118 reinfected patients were included in 50 studies. These reinfected patients have a wide age distribution (a range of 16–92 years), with a gender distribution of 62 (52.5%) male and 54 (45.8%) female (two case reports did not report gender), including 24 healthcare staff (9 male and 15 female). 25 patients were reported as having at least one comorbidity (such as hypertension, end-stage renal disease, asthma.). Patients often presented with overt symptoms upon reinfection. Characteristics of reinfected patients are presented in Table 1. Figure 2 shows the duration of symptoms between the two infections and outcomes in reinfected patients. The corresponding patient information in Fig. 2 is shown in the Additional file 1: Table S6.
Symptoms of reinfected patients
Most reinfected patients show clinical symptoms, and only a few studies have reported patients being asymptomatic at both the first and secondary infections.
In the 36 studies (n = 51) [8,9,10, 32,33,34,35,36,37, 39, 43,44,45, 47,48,49,50,51,52,53,54, 56, 57, 59,60,61,62, 64,65,66,67,68,69, 71,72,73], which reported details of patients’ symptoms during the first infection, these commonly included cough (30, 62.3%), fever (31, 58.5%), headache (20, 37.7%), diarrhea (13, 24.5%), sore throat (12, 22.6%), myalgia (12, 22.6%), dyspnea (11, 20.8%), rhinitis (9, 17%), fatigue (7, 13.2%), chills (6, 11.3%), anosmia (5, 9.4%), ageusia (5, 9.4%), malaise (4, 7.5%), chest pain (4, 7.5%), nasal congestion (4, 7.5%), odynophagia (4, 7.5%), nausea (3 5.7%), vomiting (2, 3.8%), anxiety (2, 3.8%), lethargy (2, 3.8%), panic attacks (1, 1.9%), sneezing (1, 1.9%), confusion (1, 1.9%), body pain (1, 1.9%), arthralgia (1, 1.9%), exertional tachycardia (1, 1.9%), dizziness (1, 1.9%), and arthromyalgia (1, 1.9%), and 10 (18.9%) patients [31, 33, 36, 41, 46, 48, 55, 70] were asymptomatic.
At reinfection, 36 studies reported 54 patients [9, 32,33,34,35,36,37,38,39, 41, 43, 45,46,47,48,49,50,51,52,53,54,55,56,57, 59, 60, 62,63,64,65, 67,68,69,70,71] with common symptoms including cough (29, 51.8%), fever (26, 46.4%), headache (19, 33.9%), dyspnea (18, 32.1%), fatigue (17, 30.4%), myalgia (14, 25%), anosmia (10, 17.9%), diarrhea (8, 14.3%), sore throat (8, 14.3%), rhinitis(7, 12.5%), body pain(6, 10.7%), ageusia(6, 10.7%), odynophagia(6, 10.7%), malaise(4, 7.1%), nasal congestion (4, 7.1%), chill (3, 5.4%), dizziness (3, 5.4%), arthralgia (3, 5.4%), nausea (2, 3.6%), abdominal pain (2, 3.6%), anorexia (1, 1.8%), back pain (1, 1.8%), muscle fatigue (1, 1.8%), insomnia (1, 1.8%), hypoxia (1, 1.8%), gastrointestinal symptoms (1, 1.8%), leg pain (1, 1.8%), swelling (1, 1.8%), sneezing (1, 1.8%), lethargy (1, 1.8%), chest pain (1, 1.8%), shivering (1, 1.8%), respiratory failure (1, 1.8%),, and 9 (15.4%) patents [8, 31, 43, 66, 76] were asymptomatic.
Time from first to second clinical symptom
The shortest time from first infection to reinfection was 19 days [59] and the longest was 293 days [71].
Co-morbidity of reinfected patients
Thirty-four studies reported comorbidities in 64 patients [8,9,10, 34,35,36, 38, 41,42,43,44, 48,49,50,51,52,53,54, 56,57,58,59, 61, 63,64,65,66,67,68,69, 71, 73, 75, 77]. Among patients with co-morbidity, 10 had a combination of two or more chronic conditions [38, 41,42,43,44, 51, 59, 61, 66, 77]. Of these patients having comorbidities the youngest was 16 years old [58] and the oldest was 92 [43]. Hypertension and obesity were the most common comorbidities, followed by end-stage renal disease, asthma, chronic obstructive pulmonary disease (COPD), dementia, dyslipidemia and type 2 diabetes.
Vaccination
Two case reports reported on patients who had been vaccinated before reinfection. One patient developed reinfection 10 days after the first dose bur did not report the vaccine type [68]. Another patient developed reinfection 13 days after the first dose of Pfizer vaccination was administered [41].
Patient outcomes
Among the 21 studies that reported patient outcomes [9, 10, 34, 35, 38, 40,41,42,43,44, 52,53,54, 58, 61, 64, 66, 67, 73, 75, 77], nine patients (an age range from 16 to 54) recovered after reinfection [34, 40, 53, 58, 67, 73]. Seven patients died (aged 44–92): one died of septic shock and respiratory failure [10], another one died of respiratory failure [77], and the cause of death was not reported for the remaining five patients [43, 44, 54, 75]. Five patients were reported as still being hospitalized [38, 41, 61], and five patients had been discharged from hospital [35, 42, 52, 66].
Infectivity of reinfected patients
One case report showed that two days after diagnosis, one of the patient’s co-workers was also diagnosed with COVID-19 [63].
Treatment of first and second infections
At the first infection for the patients with reinfection, nine studies reported that 12 patients with COVID-19 were not treated [10, 38, 40, 47, 51, 52, 56, 60, 65]. Among the 9 studies reporting on 9 patients who had treatment [35, 41, 42, 48, 50, 53, 58, 61, 71], most patients received corticosteroids [61], including methylprednisolone [58], dexamethasone [41], and prednisone [50, 58]. Treatment with atazanavir and other antiviral drugs [35, 48], and tocilizumab [35, 41], and hydroxychloroquine was also common [35, 42]. Some patients also received levofloxacin [61], paracetamol [71], acetaminophen [53], and low molecular weight heparin [61]. And 4 patients were using a combination of drugs [35, 41, 58, 61].
For reinfected patients, 11 patients in 8 studies were untreated [8, 10, 35, 38, 40, 46, 51, 60]. Among the treated patients, most received prednisone [42, 61]and dexamethasone [42, 56, 69]. Treatment with remdesivir [42, 56], tocilizumab [42, 69], enoxaparin [42, 61], and azithromycin was also common [42, 61]. A few patients received inhaled salmeterol [42], amoxicillin-clavulanate [42] and convalescent plasma [66]. All of them were using combination drugs [42, 56, 61, 69].
Sequence analysis of reinfection cases
The B.1 variant strain was the most common one in the first infection. Variants B.1.1.7, B.1.128 and B.1.351 were the most common strains in reinfection. In the studies reporting the gene sequencing results in detail, 33 cases were infected by different strains [8, 10, 35, 36, 38, 39, 41, 47,48,49, 51, 52, 55,56,57, 59,60,61, 63,64,65, 67, 68, 71,72,73, 77]. Among them, the virus gene sequence of the first infection could not be detected in 2 cases, but epidemiological reports showed that the virus lineage of reinfection did not spread locally at the time of first infection [53, 58]. Eight patients were reported as being infected with the same strain (see Table 1) [9, 32, 37, 45, 46, 48, 62, 70].
Viral mutations of reinfected cases
In the included studies, viral gene sequencing revealed mutations among some patients. Of the 29 studies that reported mutations in details, D614G was the most common mutation [10, 34,35,36, 38, 39, 42, 47,48,49, 52, 60, 62, 64, 65, 67, 68, 70, 71], and other mutations such as N440K [70] and E484K [50, 68, 69] were also detected. See Additional file 1: Table S7.
Discussion
We have systematically summarized and analyzed the characteristics of COVID-19 reinfected patients and the infecting viral gene sequences. In the current included studies, we found that reinfected patients usually have clinical symptoms. Reinfection events can occur within a short time, and there is a wide age distribution among reinfected patients. The B.1 variant strain was the most common one in the first infection, B.1.1.7, B.1.128 and B.1.351 variant strain were the most common strains in reinfection. And D614G was the most common mutation. Thirty-nine patients had no comorbidities and 10 had a combination of two or more chronic conditions. Nine patients (an age range from 16 to 54 years) recovered and 7 patients died after reinfection.
One cohort study reported that the incidence rate of reinfection was estimated at 0.66 per 10,000 person-weeks (95% CI: 0.56–0.78) [76]. Most reinfections constitute infection by different virus strains, but the virus gene sequencing of some patients showed that they were reinfected with the same strain as the first infection. Relevant animal experiments showed that after the second inoculation of the virus, no viral shedding from nasal, oropharyngeal, and rectal cavities was observed in these animals, and the virus was not transmitted to other animals [5, 6]. In our systematic review, there is only one study report of a patient infecting others. Thus, whether reinfected patients are infectious remains to be determined.
We think that reinfection is one of the reasons for re-detectable positive RNA test. Beyond that, the reason of patients with re-detectable positive RNA test including the results of Reverse Transcription-polymerase Chain Reaction (RT-PCR) may be a false negative at discharge or incomplete elimination of the virus [78]. The chief reasons for patients becoming reinfected are potentially as follows:
-
(1).
Insufficient immune capacity after the first infection. Individuals who recovered from COVID-19 have generally been thought to generate a robust immune response to clear the virus. Some studies have shown that the presence of SARS-CoV-2 antibodies confers subsequent immunity in most people for at least six to eight months [79, 80]. However, due to SARS-COV-2’s high variability, different genotypes and some human’s weak or non-lasting immune response, it remains to be determined whether the first infection confers protective immunity to subsequent infections.
-
(2).
Mutant viral strains. New virus variants such as B.1.1.7, P.1, and B.1.351 have emerged and become the main virus variants prevalent in many countries [12, 81, 82]. Some studies have indicated that P.1 has a 25–61% capacity to evade the immunity elicited by a previous infection caused by non-P.1 viruses [83]. The E484k mutation in these virus variants can, to a certain extent, escape recognition by people’s rehabilitation serum antibodies and make the virus variants have higher transmissibility [84, 85]. And the D614G mutation might help to increase the viral fitness in all emerging variants where this mutation is present. With the help of this mutation (D614G), the SARS-CoV-2 variants have gained viral fitness to enhance viral replication and increase transmission [86]. These S protein variants recently reported pose new potential challenges for the efficacy of vaccination, antibody-based therapies and viral diffusion control [87, 88].
With the continued emergence of variants of SARS-CoV-2, and the increased rate of disease transmission due to new variants, concerns have been raised about the practical effectiveness of vaccines [89]. Most COVID-19 vaccines elicit high levels of antibodies that target diverse regions of the spike protein, so some of the molecules are likely to be able to block variants of the virus [90]. One study found that the spike protein of the UK variant B.1.1.7 had little effect on sera from 16 subjects who received Pfizer vaccine injections [91]. By increasing the levels of cross-neutralizing antibodies, SARS-CoV-2 vaccination may strengthen protection, especially against variants harboring antibody escape mutations like B1.351 [92]. Protective immunity conferred by the mRNA vaccines is most likely to be retained against the B.1.617.1 and B.1.617.2 variants [93]. However, with the continuous mutation of the virus, the effectiveness of the vaccine for different variants remains to be studied.
Based on this study, we suggest that the management of reinfected patients should be consistent with the treatment of the first infection. These cases should be divided into mild, moderate and severe infection and given antiviral treatment. As a highly infectious virus, the modes of transmission include airborne, droplet, contact with contaminated surfaces, oral and fecal secretions [94]. With the emergence of new varieties, the transmission ability of new variants is increasing [95]. Thus, the public, including rehabilitated patients, should be fully vaccinated, wear masks in public places, and maintain social distance to avoid reinfection with the virus.
At the same time, our results found that the cause of death among patients who died was septic shock and respiratory failure. According to existing studies, lung disease is the most common long-term complication in patients with COVID-19 [96, 97], and the virus may also affect the cardiovascular system and nervous system [98]. Therefore, it is still necessary to conduct long-term follow-up studies to determine the various complications and prognosis of COVID-19 patients.
The current concept of reinfection is still not consistent. According to the European Centre for Disease Prevention and Control, reinfection is defined as “laboratory confirmation of two infections by two different strains (minimum distance to be determined or supported by phylogenetic and epidemiological data) with timely separated illness/infection episodes” [99]. The Centers for Disease Control and Prevention (CDC) uses the following criteria to define reinfection with SARS-CoV-2: detection of SARS-CoV-2 RNA (with Ct values < 33 if detected by RT-PCR) > 90 days after the first detection of viral RNA whether or not symptoms were present and paired respiratory specimens from each episode that belong to different clades of virus or have genomes with > 2 nucleotide differences per month [100]. The reinfection rate may vary greatly according to the different definitions of reinfection used. In screening the literature, we found that many studies, use RT-PCR positive as the standard for reinfection, but it has been stated that RT-PCR is meaningless when detecting reinfection as a positive RT-PCR test can only reflect the detection of RNA fragments that could be related to either a new viral infection, viral persistence with the reappearance of virus in mucosae, or viable viral debris [101]. Therefore, a positive RT-PCR test cannot be assumed to represent new viral infections in all situations.
Eight systematic reviews have already been published [20,21,22,23,24,25,26,27], but they have many limitations, such as not reporting the results of viral gene sequencing [20,21,22], or defining reinfection based on RT-PCR results [20, 27]. Thus, we decided to conduct this current review to address these limitations.
However, this current review also has some limitations. First, we only included data reported in the studies, and did not contact the authors for unreported data. Thus, we could not report the outcome measures concerned, such as the reinfection rate. In addition, the available evidence is still insufficient, and some relevant results, such as the infectivity of reinfected patients, the results of gene sequencing and vaccination, have not been reported. Second, In the cohort and cross-sectional studies, the possible factors for reinfection were not discussed. This also limits our discussion of factors posing a risk for reinfection. Third, for reports in which a patient was reinfected with the same strain, we relied on the report by the authors of the original study. But they did not report in detail how to distinguish between prolonged shedding of the virus and reinfection with the same strain. In addition, as patients with asymptomatic reinfections are usually found through the community testing for COVID-19 cases or Entry-exit screening of people at airport examinations, the number of reinfected persons may be seriously underestimated.
Conclusions
In conclusion, our study shows that for some patients, the immune response to the first infection was not adequate to protect against reinfection. And reinfection is not specific to any specific strain. Therefore, individuals, regardless of history of prior infection, should continue to participate in mitigating the spread of infection by practicing social distancing and mask-wearing. More high-quality cohort studies based on viral gene sequencing are needed in the future to help us better understand the causes of reinfection and formulate vaccination strategies.
Availability of data and materials
The data used in this study were gathered from publicly available studies.
Abbreviations
- COVID-19:
-
Coronavirus Disease 2019
- SARS-CoV-2:
-
Severe acute respiratory syndrome coronavirus 2
- MERS-CoV:
-
Middle East respiratory syndrome coronavirus
- WHO:
-
World Health Organization
- VOIs:
-
Variants of Interest
- VOC:
-
Variants of Concern
- Ct:
-
Cycle threshold
- RT-PCR:
-
Reverse Transcription-polymerase Chain Reaction
- CDC:
-
The Centers for Disease Control and Prevention
References
WHO. WHO Coronavirus Disease (COVID-19) Situation Dashboard. https://covid19.who.int/. Accessed 21 Nov 2021.
Boni MF, Lemey P, Jiang X, et al. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat Microbiol. 2020;5(11):1408–17.
Wang MY, Zhao R, Gao LJ, Gao XF, Wang DP, Cao JM. SARS-CoV-2: structure, biology, and structure-based therapeutics development. Front Cell Infect Microbiol. 2020;10:587269.
Hu T, Liu Y, Zhao M, Zhuang Q, Xu L, He Q. A comparison of COVID-19, SARS and MERS. PeerJ. 2020;8:e9725.
Gaudreault NN, Carossino M, Morozov I, et al. Experimental re-infected cats do not transmit SARS-CoV-2. Emerg Microbes Infect. 2021;10(1):638–50.
Brustolin M, Rodon J, Rodriguez de la Concepcion ML, et al. Protection against reinfection with D614-or G614-SARS-CoV-2 isolates in golden Syrian hamster. Emerg Microbes Infect. 2021;10(1):797–809.
Deng W, Bao L, Liu J, et al. Primary exposure to SARS-CoV-2 protects against reinfection in rhesus macaques. Science. 2020;369(6505):818.
To KK, Hung IF, Ip JD, et al. COVID-19 re-infection by a phylogenetically distinct SARS-coronavirus-2 strain confirmed by whole genome sequencing. Clin Infect Dis. 2020. https://doi.org/10.1093/cid/ciaa1275.
Tillett RL, Sevinsky JR, Hartley PD, et al. Genomic evidence for reinfection with SARS-CoV-2: a case study. Lancet Infect Dis. 2021;21(1):52–8.
Borgogna C, De Andrea M, Griffante G, et al. SARS-CoV-2 reinfection in a cancer patient with a defective neutralizing humoral response. J Med Virol. 2021. https://doi.org/10.1002/jmv.27200.
Urhan A, Abeel T. Emergence of novel SARS-CoV-2 variants in the Netherlands. Sci Rep. 2021;11(1):6625.
Paul P, France AM, Aoki Y, et al. Genomic surveillance for SARS-CoV-2 variants circulating in the United States, December 2020-May 2021. MMWR Morb Mortal Wkly Rep. 2021;70(23):846–50.
Hoffmann M, Hofmann-Winkler H, Krüger N, et al. SARS-CoV-2 variant B.1.617 is resistant to bamlanivimab and evades antibodies induced by infection and vaccination. Cell Rep. 2021;36(3):109415.
WHO. Tracking SARS-CoV-2 variants. WHO. https://www.who.int/zh/activities/tracking-SARS-CoV-2-variants. Accessed 7 Sept 2021.
Davies NG, Abbott S, Barnard RC, et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372(6538):eabg3055.
Graham MS, Sudre CH, May A, et al. Changes in symptomatology, reinfection, and transmissibility associated with the SARS-CoV-2 variant B.1.1.7: an ecological study. Lancet Public Health. 2021;6(5):e335–45.
Pulliam JRC, van Schalkwyk C, Govender N, et al. Increased risk of SARS-CoV-2 reinfection associated with emergence of the Omicron variant in South Africa. medRxiv. 2021:2021.2011.2011.21266068.
Yinghui Jin XR, Jie Zhou, Jing Guo, Chunmei Hao, Mengxue Zheng, Rong Zhang, Qiao Huang, Ruiling Li, Xiaomei Yao. Patients showing reinfection with COVID-19: a systematic review and meta-analysis protocol. PROSPERO. 2021. CRD42021265333. https://www.crd.york.ac.uk/prospero/display_record.php?ID=CRD42021265333. Accessed 15 Mar 2022.
PRISMA 2020 statement. https://www.bmj.com/content/372/bmj.n71. Accessed 9 Mar 2022.
Murchu EO, Byrne P, Carty PG, et al. Quantifying the risk of SARS-CoV-2 reinfection over time. Rev Med Virol. 2021;32:2260.
Pinto LM, Nanda V, Sunavala A, Rodriques C. Reinfection in COVID-19: a scoping review. Med J Armed Forces India. 2021;77(Suppl 2):S257-s263.
SeyedAlinaghi S, Oliaei S, Kianzad S, et al. Reinfection risk of novel coronavirus (COVID-19): a systematic review of current evidence. World J Virol. 2020;9(5):79–90.
Roberts AT, Piani F, Longo B, Andreini R, Meini S. Reinfection of SARS-CoV-2—analysis of 23 cases from the literature. Infect Dis (Lond). 2021;53(7):479–85.
Wang J, Kaperak C, Sato T, Sakuraba A. COVID-19 reinfection: a rapid systematic review of case reports and case series. J Investig Med. 2021;69(6):1253–5.
Massachi J, Donohue KC, Kelly JD. Severe acute respiratory syndrome coronavirus 2 reinfection cases corroborated by sequencing. Am J Trop Med Hyg. 2021. https://doi.org/10.4269/ajtmh.21-0365.
Choudhary MC, Crain CR, Qiu X, Hanage W, Li JZ. SARS-CoV-2 sequence characteristics of COVID-19 persistence and reinfection. Clin Infect Dis. 2021. https://doi.org/10.1093/cid/ciab380.
Dhillon RA, Qamar MA, Gilani JA, et al. The mystery of COVID-19 reinfections: a global systematic review and meta-analysis. Ann Med Surg (Lond). 2021;72:103130.
Institute JB. JBI critical appraisal tools. https://jbi.global/. Accessed 23 Sept 2021.
Ma LL, Wang YY, Yang ZH, Huang D, Weng H, Zeng XT. Methodological quality (risk of bias) assessment tools for primary and secondary medical studies: what are they and which is better? Mil Med Res. 2020;7(1):7.
Wells GA, Shea B, O'Connell D, Peterson J, Welch V, Losos M, Tugwell P. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp. Accessed 23 Sept 2021.
Gupta V, Bhoyar RC, Jain A, et al. Asymptomatic reinfection in two healthcare workers from India with genetically distinct SARS-CoV-2. Clin Infect Dis. 2020. https://doi.org/10.1093/cid/ciaa1451.
Larson D, Brodniak SL, Voegtly LJ, et al. A case of early re-infection with SARS-CoV-2. Clin Infect Dis. 2020. https://doi.org/10.1093/cid/ciaa1436.
Staub T, Arendt V, Lasso de la Vega EC, et al. Case series of four re-infections with a SARS-CoV-2 B1351 variant, Luxembourg, February 2021. Euro Surveill. 2021;26(18):2100423.
Salehi-Vaziri M, Jalali T, Farahmand B, et al. Clinical characteristics of SARS-CoV-2 by re-infection vs. reactivation: a case series from Iran. Eur J Clin Microbiol Infect Dis. 2021;40(8):1713–9.
Klein J, Brito AF, Trubin P, et al. Longitudinal immune profiling of a severe acute respiratory syndrome coronavirus 2 reinfection in a solid organ transplant recipient. J Infect Dis. 2022;225(3):374–84.
Shastri J, Parikh S, Agrawal S, et al. Clinical, serological, whole genome sequence analyses to confirm SARS-CoV-2 reinfection in patients from Mumbai, India. Front Med (Lausanne). 2021;8:631769.
Vetter P, Cordey S, Schibler M, et al. Clinical, virologic and immunologic features of a mild case of SARS-CoV-2 reinfection. Clin Microbiol Infect. 2021;27(5):791.e791-794.
Harrington D, Kele B, Pereira S, et al. Confirmed reinfection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant VOC-202012/01. Clin Infect Dis. 2021. https://doi.org/10.1093/cid/ciab014.
Prado-Vivar B, Becerra-Wong M, Guadalupe JJ, et al. COVID-19 re-infection by a phylogenetically distinct SARS-CoV-2 variant, first confirmed event in South America. In: SSRN; 2020. https://doi.org/10.2139/ssrn.3686174.
Moschetta MO, Hadi RA, Franco RF, et al. COVID-19 Reinfection by the gamma variant in kidney transplant recipients. Transplantation. 2021. https://doi.org/10.1097/tp.0000000000003924.
Scarpati G, Piazza O, Pagliano P, Rizzo F. COVID-19: a confirmed case of reinfection in a nurse. BMJ Case Rep. 2021;14(7):e244507.
Massanella M, Martin-Urda A, Mateu L, et al. Critical presentation of a severe acute respiratory syndrome coronavirus 2 reinfection: a case report. Open Forum Infect Dis. 2021;8(7):ofab329.
Garvey MI, Casey AL, Wilkinson MAC, et al. Details of SARS-CoV-2 reinfections at a major UK tertiary centre. J Infect. 2021;82(6):e29–30.
Siqueira JD, Goes LR, Alves BM, et al. Distinguishing SARS-CoV-2 bonafide re-infection from pre-existing minor variant reactivation. Infect Genet Evol. 2021;90:104772.
Sevillano G, Ortega-Paredes D, Loaiza K, Zurita-Salinas C, Zurita J. Evidence of SARS-CoV-2 reinfection within the same clade in Ecuador: a case study. Int J Infect Dis. 2021;108:53–6.
Kulkarni O, Narreddy S, Zaveri L, Kalal IG, Tallapaka KB, Sowpati DT. Evidence of SARS-CoV-2 reinfection without mutations in Spike protein. Clin Infect Dis. 2021. https://doi.org/10.1093/cid/ciab136.
Lee JS, Kim SY, Kim TS, et al. Evidence of severe acute respiratory syndrome coronavirus 2 reinfection after recovery from mild coronavirus disease 2019. Clin Infect Dis. 2020. https://doi.org/10.1093/cid/ciaa1421.
Fintelman-Rodrigues N, da Silva APD, Dos Santos MC, et al. Genetic evidence and host immune response in persons reinfected with SARS-CoV-2, Brazil. Emerg Infect Dis. 2021;27(5):1446–53.
Fonseca V, de Jesus R, Adelino T, et al. Genomic evidence of SARS-CoV-2 reinfection case with the emerging B.1.2 variant in Brazil. J Infect. 2021;83(2):237–79.
Nonaka CKV, Franco MM, Gräf T, et al. Genomic evidence of SARS-CoV-2 reinfection involving E484K spike mutation, Brazil. Emerg Infect Dis. 2021;27(5):1522–4.
Ramírez JD, Muñoz M, Ballesteros N, et al. Phylogenomic evidence of reinfection and persistence of SARS-CoV-2: first report from Colombia. Vaccines (Basel). 2021;9(3):282.
Alshukairi AN, El-Kafrawy SA, Dada A, et al. Re-infection with a different SARS-CoV-2 clade and prolonged viral shedding in a hematopoietic stem cell transplantation patient. Int J Infect Dis. 2021;110:267–71.
Aguilar-Shea AL, Gutiérrez-Martín-Arroyo J, Vacas-Córdoba M, Gallardo-Mayo C. Reinfection by SARS-CoV-2: the first one in a family reported in Spain. Med Clin (Barc). 2021. https://doi.org/10.1016/j.medcli.2021.04.009.
Mulder M, Vegt D, Oude Munnink B, et al. Reinfection of severe acute respiratory syndrome coronavirus 2 in an immunocompromised patient: a case report. Clin Infect Dis. 2020. https://doi.org/10.1093/cid/ciaa1538.
Dhar MS, Asokachandran V, Uppili B, et al. Reinfection or reactivation: genome-based two distinct SNP profile of SARS-CoV2 repositivity in an Indian case. J Med Virol. 2021;93(7):4152–5.
Goldman JD, Wang K, Roltgen K, et al. Reinfection with SARS-CoV-2 and failure of humoral immunity: a case report. medRxiv. 2020. https://doi.org/10.1101/2020.09.22.20192443.
Marquez L, Koy T, Spinler JK, et al. Reinfection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) B.1.1.7 variant in an immunocompromised adolescent. Infect Control Hosp Epidemiol. 2021:1–2. https://doi.org/10.1017/ice.2021.195
Buddingh EP, Vossen A, Lamb HJ, van der Palen RLF, Brinkman DMC. Reinfection with severe acute respiratory syndrome coronavirus 2 without recurrence of multisystem inflammatory syndrome in children. Pediatr Infect Dis J. 2021. https://doi.org/10.1097/inf.0000000000003280.
Tang CY, Wang Y, McElroy JA, et al. Reinfection with two genetically distinct SARS-CoV-2 viruses within 19 days. J Med Virol. 2021;93(10):5700–3.
Amorim MR, Souza WM, Barros ACG Jr, et al. Respiratory viral shedding in healthcare workers reinfected with SARS-CoV-2, Brazil, 2020. Emerg Infect Dis. 2021;27(6):1737–40.
Novazzi F, Baj A, Genoni A, et al. SARS-CoV-2 B.1.1.7 reinfection after previous COVID-19 in two immunocompetent Italian patients. J Med Virol. 2021;93(9):5648–9.
Salehi-Vaziri M, Omrani MD, Pouriayevali MH, et al. SARS-CoV-2 presented moderately during two episodes of the infection with lack of antibody responses. Virus Res. 2021;299:198421.
Romano CM, Felix AC, Paula AV, et al. SARS-CoV-2 reinfection caused by the P.1 lineage in Araraquara city, Sao Paulo State, Brazil. Rev Inst Med Trop Sao Paulo. 2021;63:e36.
Camargo CH, Gonçalves CR, Pagnoca E, et al. SARS-CoV-2 reinfection in a healthcare professional in inner Sao Paulo during the first wave of COVID-19 in Brazil. Diagn Microbiol Infect Dis. 2021;101(4):115516.
Brehm TT, Pfefferle S, von Possel R, et al. SARS-CoV-2 Reinfection in a Healthcare Worker Despite the Presence of Detectable Neutralizing Antibodies. Viruses. 2021;13(4):661.
Tomkins-Tinch CH, Daly JS, Gladden-Young A, et al. SARS-CoV-2 reinfection in a liver transplant recipient. Ann Intern Med. 2021;174(8):1178–80.
Díaz Y, Ortiz A, Weeden A, et al. SARS-CoV-2 reinfection with a virus harboring mutation in the Spike and the Nucleocapsid proteins in Panama. Int J Infect Dis. 2021;108:588–91.
Yu ALF, Liphaus BL, Ferreira PM, et al. SARS-CoV-2 reinfection: report of two cases in Southeast Brazil. Rev Inst Med Trop Sao Paulo. 2021;63:e50.
Zucman N, Uhel F, Descamps D, Roux D, Ricard JD. Severe reinfection with South African SARS-CoV-2 variant 501Y.V2: A case report. Clin Infect Dis. 2021. https://doi.org/10.1093/cid/ciab129.
Rani PR, Imran M, Lakshmi JV, et al. Symptomatic reinfection of SARS-CoV-2 with spike protein variant N440K associated with immune escape. J Med Virol. 2021;93(7):4163–5.
Loconsole D, Sallustio A, Accogli M, et al. Symptomatic SARS-CoV-2 reinfection in a healthy healthcare worker in Italy confirmed by whole-genome sequencing. Viruses-Basel. 2021;13(5):899.
Selhorst P, Van Ierssel S, Michiels J, et al. Symptomatic SARS-CoV-2 reinfection of a health care worker in a Belgian nosocomial outbreak despite primary neutralizing antibody response. Clin Infect Dis. 2020. https://doi.org/10.1093/cid/ciaa1850.
Van Elslande J, Vermeersch P, Vandervoort K, et al. Symptomatic severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) reinfection by a phylogenetically distinct strain. Clin Infect Dis. 2021;73(2):354–6.
Jeffery-Smith A, Iyanger N, Williams SV, et al. Antibodies to SARS-CoV-2 protect against re-infection during outbreaks in care homes, September and October 2020. Euro Surveill. 2021;26(5):2100092.
Brouqui P, Colson P, Melenotte C, et al. COVID-19 re-infection. Eur J Clin Investig. 2021;51(5):e13537.
Abu-Raddad LJ, Chemaitelly H, Coyle P, et al. SARS-CoV-2 antibody-positivity protects against reinfection for at least seven months with 95% efficacy. EClinicalMedicine. 2021;35:100861.
Adrielle Dos Santos L, Filho PGG, Silva AMF, et al. Recurrent COVID-19 including evidence of reinfection and enhanced severity in thirty Brazilian healthcare workers. J Infect. 2021;82(3):399–406.
Ren X, Ren X, Lou J, et al. A systematic review and meta-analysis of discharged COVID-19 patients retesting positive for RT-PCR. EClinicalMedicine. 2021;34:100839.
Breathnach AS, Riley PA, Cotter MP, Houston AC, Habibi MS, Planche TD. Prior COVID-19 significantly reduces the risk of subsequent infection, but reinfections are seen after eight months. J Infect. 2021;82(4):e11–2.
Griffin S. Covid-19: antibodies protect against reinfection for at least six months, study finds. BMJ. 2020;371:m4961.
Baric RS. Emergence of a highly fit SARS-CoV-2 variant. N Engl J Med. 2020;383(27):2684–6.
Cerutti G, Rapp M, Guo Y, et al. Structural basis for accommodation of emerging B.1.351 and B.1.1.7 variants by two potent SARS-CoV-2 neutralizing antibodies. Structure. 2021;29(7):655-663.e654.
Faria NR, Mellan TA, Whittaker C, et al. Genomics and epidemiology of the P1 SARS-CoV-2 lineage in Manaus, Brazil. Science. 2021;372(6544):815–21.
Jangra S, Ye C, Rathnasinghe R, et al. SARS-CoV-2 spike E484K mutation reduces antibody neutralisation. Lancet Microbe. 2021;2(7):e283–4.
Augusto G, Mohsen MO, Zinkhan S, Liu X, Vogel M, Bachmann MF. In vitro data suggest that Indian variant B.1.617 of SARS-CoV-2 escapes neutralization by both receptor affinity and immune evasion. Allergy. 2021. https://doi.org/10.1111/all.15065.
Chakraborty C, Saha A, Sharma AR, Bhattacharya M, Lee SS, Agoramoorthy G. D614G mutation eventuates in all VOI and VOC in SARS-CoV-2: Is it part of the positive selection pioneered by Darwin? Mol Ther Nucleic Acids. 2021;26:237–41.
Triveri A, Serapian SA, Marchetti F, et al. SARS-CoV-2 spike protein mutations and escape from antibodies: a computational model of epitope loss in variants of concern. J Chem Inf Model. 2021. https://doi.org/10.1021/acs.jcim.1c00857.
Ding C, He J, Zhang X, et al. Crucial mutations of spike protein on SARS-CoV-2 evolved to variant strains escaping neutralization of convalescent plasmas and RBD-specific monoclonal antibodies. Front Immunol. 2021;12:693775.
Callaway E. Could new COVID variants undermine vaccines? Labs scramble to find out. Nature. 2021;589(7841):177–8.
Callaway E. Fast-spreading COVID variant can elude immune responses. Nature. 2021;589(7843):500–1.
Muik A, Wallisch AK, Sänger B, et al. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine-elicited human sera. Science. 2021;371(6534):1152–3.
Gallais F, Gantner P, Bruel T, et al. Evolution of antibody responses up to 13 months after SARS-CoV-2 infection and risk of reinfection. EBioMedicine. 2021;71:103561.
Edara VV, Pinsky BA, Suthar MS, et al. Infection and vaccine-induced neutralizing-antibody responses to the SARS-CoV-2 B.1.617 variants. N Engl J Med. 2021;385(7):664–6.
Mehraeen E, Salehi MA, Behnezhad F, Moghaddam HR, SeyedAlinaghi S. Transmission modes of COVID-19: a systematic review. Infect Disord Drug Targets. 2021;21(6):e170721187995.
Dadras O, Alinaghi SAS, Karimi A, et al. Effects of COVID-19 prevention procedures on other common infections: a systematic review. Eur J Med Res. 2021;26(1):67.
SeyedAlinaghi S, Afsahi AM, MohsseniPour M, et al. Late complications of COVID-19; a systematic review of current evidence. Arch Acad Emerg Med. 2021;9(1):e14.
Al-Jahdhami I, Al-Mawali A, Bennji SM. Respiratory complications after COVID-19. Oman Med J. 2022;37(1):e343.
Wang J, Zhu K, Xue Y, Wen G, Tao L. Research progress in the treatment of complications and sequelae of COVID-19. Front Med (Lausanne). 2021;8:757605.
European Centre for Disease Prevention and Control (2020) Reinfection with SARS-CoV-2: Considerations for Public Health Response. European Centre for Disease Prevention and Control. https://www.ecdc.europa.eu/en/publications-data/threat-assessment-brief-reinfection-sars-cov-2. Accessed 22 Oct 2021.
Investigative Criteria for Suspected Cases of SARS-CoV-2 Reinfection (ICR) Centers for Disease Control and Prevention. https://www.cdc.gov/coronavirus/2019-ncov/php/invest-criteria.html. Accessed 22 Oct 2021.
Atkinson B, Petersen E. SARS-CoV-2 shedding and infectivity. Lancet. 2020;395(10233):1339–40.
Acknowledgements
We thank Jean Glover from Tianjin Golden Framework Consulting Company for English editing. This work was supported (in part) by the Emergency Special Project for COVID-19 of Wuhan Municipal Health Commission (EG20A02).
Differences between protocol and review
The definition of reinfected persons has been modified in current review. The reason is that eight patients reported by eight articles were re-infected by the same strain and they were verified as reinfected cases by viral gene sequencing, so after deep discussion among the research group we add “same strain” in the definition.
Funding
Emergency Special Project for COVID-19 of Wuhan Municipal Health Commission (EG20A02). The funder of the study had no role in data collection, data analysis, or data interpretation. The corresponding authors have had full access to all the data in the study and have final responsibility for the decision to submit for publication.
Author information
Authors and Affiliations
Contributions
YH Jin, XM Yao and RL Li conceived and designed the study. XY Ren, J Zhou and J Guo were involved in the search process, study selection and data extraction, and wrote the manuscript. Q Huang and R Zhang were involved in data analysis, data handling, and commented on drafts of the manuscript. CM Hao and MX Zheng were involved in the quality assessment and commented on the manuscript. YH Jin, XM Yao and RL Li revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
YH Jin, XM Yao, XY Ren, and Q Huang conducted clinical practice guidelines on COVID-19. YH Jin reported research projects involving infection of healthcare workers during this epidemic, which was supported by Special Project for Emergency of Hubei Province (2020FCA008). All other authors declare they have nothing to disclose and have no conflicts of interest.
Supplementary Information
Additional file 1.
Table S1. Search strategy. Table S2. JBI assessment results of case reports. Table S3. JBI assessment results of cross-sectional studies. Table S4. JBI assessment results of case-control studies. Table S5. NOS assessment results of cohort studies. Table S6. Patients’ information. Table S7. Viral mutations of reinfection cases.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ren, X., Zhou, J., Guo, J. et al. Reinfection in patients with COVID-19: a systematic review. glob health res policy 7, 12 (2022). https://doi.org/10.1186/s41256-022-00245-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41256-022-00245-3