Abstract
Alternative splicing is a regulated process whereby one gene can generate multiple mRNA isoforms susceptible to be translated into protein isoforms of various functions. Several publications report the aberrant expression of splicing isoforms in cancer cells and tissues. However, in most cases, their function remains to be established. In this review article, I will discuss the molecular tool available to perform isoform-specific functional genomics, the methodologies to quantify their effectiveness and the resulting isoform-specific phenotype in human cancer cell lines.
Similar content being viewed by others
Introduction to the splicing molecular toolbox
Generalities about alternative splicing
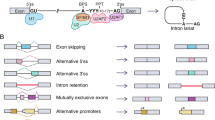
Alternative splicing is a regulated process whereby one gene can generate multiple mRNA isoforms susceptible to be translated into protein isoforms of various functions. In human, most multi-exon genes undergo alternative splicing [1]. As illustrated in Fig. 1, two 5’ss (5′ splice site) could compete for a single 3’ss (3′ splice site) (Fig. 1a) or inversely two 3’ss for a single 5’ss (Fig. 1b). These examples are alternative 5′ (alt5’) or 3′ (alt3’) splicing events (ASEs), respectively. Complete exon exclusion (cassette exon) (Fig. 1c) is the most frequent ASE type in human. There are also cases where two or more consecutive exons are excluded as a whole (multiple exon skipping, Fig. 1d) or mutually exclusive (Fig. 1e). Alternative splicing can also be coupled to other RNA regulatory mechanism such as polyadenylation [2, 3] (Fig. 1f) or non-sense mediated decay (NMD) [4] (Fig. 1g). The later happen when a stop codon is introduced too early in the mRNA sequence relative to the last exon. It is predicted that up to one third of splicing isoforms is linked to the NMD mechanism [5, 6]. Of note, even if the ASE is located outside the coding region, the untranslated region still contains regulatory sequences [7]. Alternative initiation of transcription does produce mRNA isoforms but is not due to alternative splicing (Fig. 1h). Thus, the average gene is transcribed into several mRNA isoforms and the majority of them are susceptible to be translated into protein isoforms with widely different functions. Although there is no consensus nomenclature on splicing isoforms, this review will use the convention where, for a particular ASE, the resulting isoform without the alternative sequence is named “short isoform” and the isoform with the alternative sequence is named the “long isoform” [8,9,10].Whereas the consensus sequence defining the 5’ss, 3’ss and branch point define constitutive splicing, less well defined exonic and intronic regulatory sequence in cis, as well as trans-acting splicing factors, dictate the final outcome of an ASE [11].
Type of alternative splicing event. a Alternative 5′ splice site. Two 5’ss are in competition for a single 3’ss. b Alternative 3′ splice site. Two 3’ss are in competition for a single 5’ss. c Cassette exon. An exon may be skipped. d Multiple exon skipping. A stretch of exons may be skipped together. e Mutually exclusive exon skipping. A choice between two alternative exons. f Alternative splicing event coupled to alternative polyadenylation g Alternative splicing event coupled to non-sense mediated decay. h Alternative transcription initiation
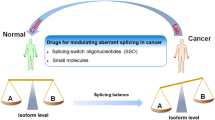
Several publications report the aberrant expression of splicing isoforms in cancer cells and tissues [12, 13]. However, in most cases, their function remains to be established [14]. At least 15% of DNA mutations disrupt splice site and hence interfere with splicing regulation and gene expression [15]. It is likely that this percentage is higher but evaluating the functional consequence of gene mutation on RNA isoforms and ultimately protein isoforms remain a significant challenge [16]. Aberrant splicing isoforms level may also be the indirect results of unbalanced splicing factors expression or mutated splicing factors rather than the direct cause of DNA mutation in cis [17,18,19]. Importantly, splicing modulators (SMs) and isoform-specific inhibitors (ISIs) can be used to either correct splicing defect due to DNA mutation or to shift splicing due to expression change in trans acting factor.
Generalities about antisense oligonucleotides
Theoretically, interfering with any gene function (enzymatic activity, protein-protein interaction) is amenable to rational drug design by providing a molecule that ultimately decreases its protein level or activity. In practice, designing a small molecule inhibitor is a daunting task. In contrast, designing an antisense oligonucleotide is straightforward thanks to the base pairing rule. One simply need to design an oligonucleotide having a complimentary sequence to the mRNA target. The mechanism of action of antisense depend mainly on its binding location on the mRNA (or pre-mRNA) and the oligonucleotide backbone chemistry used. The RNA strand of a DNA/RNA duplex can be recognized and cleaved by endogenous and ubiquitously expressed RNase H [20, 21]. Therefore, targeting specific mRNA with a DNA molecule (or a DNA-like oligonucleotide retaining the capacity to form a B type helix such as phosphorothioate [22] leads to decreased level of this mRNA and ultimately a lower protein level (for protein-coding genes). Since the minimal requirement for the RNase H is a stretch of 4 base pair [23], a novel class of antisense with as few as 4 DNA bases flanked by modified bases (to increase nuclease resistance) was developed and named “gapmer” [24]. Most if not all modifications of the oligonucleotide backbone known to date [2’O-alkyl, morpholinos, peptide nucleic acid (PNA), locked nucleic acid (LNA)] confer resistance to various RNase (including RNase H) as well as increase the binding affinity to RNA [25,26,27] and use therefore RNase H-independent mechanism. Oligonucleotide targeting the start codon interfere with ribosome initiation and hence reduce the protein translation of specific mRNA [28]. Antisense targeting alternative splice site or splicing regulatory sequence may affect splicing regulation. To reach its target, the antisense have to cross the cellular membrane and localized to the cytoplasm (mRNA) or nucleus (pre-mRNA). The challenge is even greater in vivo where other physiological barriers (vasculature, blood-brain barrier, epithelial layer) and pharmacokinetic issues restrict the efficient delivery of oligonucleotides. Several strategies are currently investigated [29, 30].Thus, the binding location on the mRNA (or pre-mRNA) and the oligonucleotide backbone chemistry used dictate the mechanism of action of antisense oligonucleotides.
Splicing modulators (SM)
Splice-switching oligonucleotides (SSO) are antisense oligonucleotides that reprogram splicing of endogenous pre-mRNA target. Initially, SSOs were designed to correct aberant splicing defect due to DNA mutation. For example, b-thalasemia is caused by a mutation in the second intron of b-globin, which result in defective hemoglobin and hence anemia. Most b-globin mutations expose a cryptic 3’ss and a new 5’ss favoring the inclusion of a novel exon. Targeting the cryptic splice site redirect splicing toward the original splice site and restore protein function [31,32,33]. In the case of the neuromuscular disease such as Duchenne muscular dystrophy (DMD), nonsense mutation drastically decreases the level of dystrophin; whereas in the mild version, the Becker muscular dystrophy, mutation generates a truncated form of dystrophin that retains partial activity [34, 35]. This suggests that DMD patients would benefit from a strategy that would skip the premature stop codon-containing exon to restore the reading frame. This idea implies the establishment of antisense design rule. Intron-exon (Fig. 2a) or exon-intron (Fig. 2b) boundary-spanning antisense over the alternative cassette exon promote exon exclusion. Targeting an exclusively exonic sequence generally promote exon exclusion (Fig. 2c) although it also promotes exon inclusion in some cases, most likely due to masking of an exonic splicing silencer (Fig. 2e) [8, 36]. Targeting an exclusively intronic sequence is less common but may be approached rationally by targeting putative intronic splicing regulatory sequence (Fig. 2d, f) [37,38,39]. Importantly, bioinformatic prediction of splicing regulatory sequences are used as guidelines for finding a robust SSO usually require testing several antisense sequences [40, 41]. Thus, in most cases, SSOs favor the production of one isoform over another without significant impact on the overall gene expression [8].
Mechanism of action of Splicing modulators and Isoform-specific inhibitors. a A splicing modulator typically favor exon exclusion and can be designed as a a boundary-spanning (intron-exon) antisense, b covering an exon-intron junction, c on an exonic splicing enhancer, or d on an intronic splicing enhancer. A splicing modulator can also promote exon inclusion when masking e an exonic splicing silencer or f an intronic splicing silencer. g The TOSS is a fully exonic bifunctional splicing modulator that redirect splicing to favor exon exclusion. h The TOES is a fully exonic bifunctional splicing modulator that redirect splicing to favor exon inclusion. i The isoform-specific inhibitor (gapmer, siRNA) is design on the alternative sequence to decrease the long isoform expression. j The isoform-specific inhibitor (gapmer, siRNA) is design on the unique exon-exon junction of the short isoform to decrease its expression. Blue = Splicing enhancer sequence. Green = Splicing silencer sequence
The “Targeted Oligonucleotide Silencer of Splicing” (TOSS) is a bifunctional SSO (Fig. 2g). An antisense targeting the alternative sequence is used to artificially display a non-hybridizing exonic splicing silencer sequence recruiting HNRNPA1 [42]. Initially used to reprogram the alt5’ of Bcl-x [42], the TOSS design was later generalized to cassette exon, alternative 3’ ASE, and complex ASEs [8, 43, 44]. In all cases, the TOSS is designed to promote the short isoform. Mechanistically, it probably acts by blocking the upstream proximal 3’ss and the downstream proximal 5’ss [45]. The main advantage is the high success rate of TOSS as most of the time there is no need to design and test multiple oligonucleotides [8], most probably because the strong silencer of the TOSS overrides the context-dependent regulatory sequence mask by the antisense platform. Theoretically, one can conceptually imagine the complementary tool: the “Targeted Oligonucleotide Enhancer of Splicing” (TOES) (Fig. 2h). Indeed, this concept was reported in the literature [46,47,48]. However, the generalization of the TOES concept as remain elusive so far [8, 36, 44].
Isoform-specific inhibitors (ISI)
As opposed to SMs, RNase H-dependent antisense (e.g. gapmer) trigger the degradation of their target mRNA. If designed to target a splicing isoform, it becomes an isoform-specific inhibitor (ISI) (Fig. 2h, i). Similarly, siRNAs targeting specific splicing isoform fall in the same category. These tools are powerful and complementary to the use of SM. Importantly, they are intended to decrease the expression of specific isoform and indirectly, to alter the ratio of one isoform over another in addition to the overall gene expression. One of the challenges is to design ISIs against the short isoform as the only unique sequence is an exon-exon junction (Fig. 2i). Studies on siRNA distinguishing single nucleotide polymorphism revealed the critical role of an RNA helix type A integrity. In brief, one mismatch is generally not sufficient to discriminate between highly similar sequences [49] unless it is positioned at the cleavage site [50, 51].
Methodologies to measure the effectiveness of isoform-specific tool
End-point PCR
The gold standard method to quantify splicing isoform modulation is end-point PCR (Fig. 3a). Briefly, two primers are designed on flanking constitutive exons in such a way that it will generate two PCR products. Fractionation on an agarose gel will allow the quantification of each band. Capillary electrophoresis allows to reduce hazards and increase the throughput significantly [52]. The ratio or the “percent splicing index” (Psi) is calculated as the ratio of the long isoform over the sum of the long and short isoforms [52].
Methodologies to validate isoform-specific tool. a End-point PCR: A single PCR reaction amplifying simultaneously the long and short isoform is ran for every treatment. Quantification by the Psi formula is output as bar graph. b Real-time PCR. Two PCR reaction (one for the short isoform, one for the long isoform) is ran for every treatment. No gel is needed. Quantification by the QPsi formula is output as bar graph. In all cases, the splicing shift represent the difference between control (ctrl) and Splicing modulator (SM) or isoform-specific Inhibitor (ISI)
To determine the splicing shift of a SM or an ISI, Psi values are compared over a control oligonucleotide of the same length and same chemistry (Psimodulator – Psicontrol = splicing shift). Importantly, the end-point PCR assay is a competitive PCR reaction meaning that the individual PCR products (Long PCR productmodulator vs Long PCR productcontrol) can’t be directly compared. Thus, it is impossible with this technique to assess the impact of a SM on the overall gene expression.
Real-time PCR
Real-time PCR is routinely used to monitor gene expression but much less frequently to quantify splicing isoforms (Fig. 3b). This is partly due to the challenge of designing isoform-specific PCR primers [53]. In this case, each isoform needs to be amplified in a separate tube. For the long isoform, at least one primer needs to be located in the alternative sequence. For the short isoform, the only unique region is the exon-exon sequence and hence, boundary-spanning primer.
The extent of modulation can be quantified in two ways. The first method is to run one PCR reaction per isoform plus one PCR reaction for a normalizing gene (long, short and normalizing gene) for each sample (control and SM) and apply the deltadeltaCt relative quantification method [54, 55]. This analysis will give the extent of modulation as a fold change for each isoform. However, for the purpose of comparison to the gold standard, a real-time PCR version of the Psi was derived (QPsi) as a second method [43, 44]. To determine the splicing shift, QPsi values are compared over a control (QPsimodulator – QPsicontrol = splicing shift). Of note, the QPsi formula doesn’t include any normalizing gene. Although antisense oligonucleotides should have a minimal off-target effect, confirmation by unbiased and genome-wide methodologies such as mass spectrometry and RNA sequencing have not been reported [56, 57].
Western-blot
In most cases, alternatively spliced isoforms are predicted to be translated into protein. Therefore, it is theoretically possible to detect the splicing shift at the protein level by western-blot. Indeed, the effectiveness of SMs to modulate the splicing ratio can be performed using antibodies that recognize both isoforms [43, 44, 58]. In practice, the resulting protein size of the isoforms is often approaching the resolution limit on a polyacrylamide gel. Alternatively, this issue can be circumvented using isoform-specific antibodies [59]. Conceptually similar to the Real-time PCR technique, the ratio or absolute level of each isoform can be estimated by quantification of individual bands.
When designing an experiment using oligonucleotide, proper control is needed to assess the on-target effect. The ideal control is an oligonucleotide of the same length and same chemistry. This could be a scrambled version of the oligonucleotide or a version with few mismatches. This is particularly useful when a phenotype is evaluated. To control for the off-target effect, splicing shift to unrelated ASEs can be examined.
Initially, antisense was developed to block the elongation of reverse transcriptase [60]. If remaining antisense post-treatment can survive the RNA extraction, then it is possible that some antisense may bloc the cDNA synthesis step of the PCR assay required for splicing shift evaluation. Since the antisense is hybridized to the long isoform, it may lead to an underestimation of the long isoform and hence, artefactual splicing shift. Indeed, a dose-response curve of SM spiked on total RNA extract (without cell transfection) is sufficient to deduce artefactual splicing shift [44]. However, it was demonstrated that RNA extraction using Trizol® reagent, as opposed to column-based strategy, remove any residual oligonucleotide. A column-based approach is attractive to perform high throughput SM validation but typically overestimate the splicing shift. Confirmation by further Trizol extraction is recommended, particularly for moderate SM (deltaPsi < 25) [44].
Prior to any phenotypic analysis, the induced-splicing shift of a SM or an ISI have to be validated (Fig. 4). The first step is to develop an assay to monitor the level of splicing isoform. In brief, RNA is extracted from a cell line of interest and the ASE target is monitored. Then, SMs and ISIs are designed and transiently transfected. Finally, the splicing shift is evaluated based on the detection method previously established in step 1. Validated SM and ISIs can then be used for functional genomics.
Workflow to validate isoform-specific tool. STEP 1: The splicing isoform detection pipeline is used to select cancer cell lines with adequate splicing isoform ratio and expression. STEP 2: Splicing Modulator (SM) and Isoform-Specific Inhibitor (ISI) are designed and transfected in the desired cancer cell lines. STEP 3: The splicing isoform detection pipeline is used to quantify the splicing shift and validate isoform-specific tools. If needed, SM and ISI are re-designed
Splicing isoforms specific phenotype in human cancer cell lines
The functional relationship between splicing isoforms of a gene
Bioinformatic analyses and experimental evidence suggest that the majority of ASEs in coding region will ultimately produce protein isoforms [61]. To uncover the functions of endogenous splicing isoforms in vitro using transient transfection of an oligonucleotide, three complementary class of tools can be used. ISIs, SMs and global targeting of all isoforms (Global siRNAs or global Gapmers) [62]. Directly comparing the phenotype of an ISI (induce a splicing shift and a decrease in global expression) vs a SM (induce a splicing shift) or vs a strategy targeting all isoforms (decrease global expression) may shed light on the functional relationship of splicing isoforms of a gene [43]. In theory, there are at least three possible scenarios to illustrate the relationship between two splicing isoforms (Table 1). In scenario 1, alternative splicing produces an isoform without a functional domain or triggering NMD. In this case, only one of the two isoforms have a function and its abundance will drive the phenotype. Therefore, SMs, ISIs and global targeting of all isoforms should all yield similar phenotypic results. Of note, similar results are expected if the two isoforms have a widely different function because only one of them will respond in a defined functional assay. In scenario 2, alternative splicing produces an isoform with antagonistic functions or one isoform act as a dominant negative. In this case, the relative abundance of one isoform over the other (i.e. the isoform ratio) will dictate the phenotype. Therefore, SMs and ISIs should yield similar results to scenario 1 except that they can’t be discriminated by an inhibitor targeting all isoforms. In scenario 3, alternative splicing produces an isoform with compensatory function. In this case, the overall expression level will drive the phenotype. Therefore, SM should not produce a phenotype as they do not impact on the overall gene expression level. This framework may help to decipher the role of splicing isoforms with unknown function.
Functional genomic of splicing isoforms
Although there is example of splicing isoforms promoting each hallmark of cancer [13, 63, 64], the phenotype of a limited number of cancer-related genes targeted by SMs (Tables 2 and 4) or ISIs (Table 3) were reported. Most of these splicing isoforms are functionally linked to the apoptotic pathway. Apoptosis is a regulated programmed cell death to clear out undesired cells. The Bcl-2 family of proteins contains about 20 members; some having anti-apoptotic and other pro-apoptotic properties. Interestingly, the pro- vs anti-apoptotic function of some Bcl-2 members such as Bcl-2, Bcl-x and Mcl-1 is finely regulated through alternative splicing (MCL-1S/MCL-1 L; BCL-2α/BCL-2β and BCl-xS/BCl-xL [65, 66].
RAP1GDS1 is a guanine nucleotide exchange factor that activates RhoA and RhoC in vitro. Skipping of exon C generate isoforms (smgGDS-558 and smgGDS-607) with different biochemical properties for small GTPases. To uncover their role in breast cancer, Hauser et al. systematically target the short, long and all isoforms using siRNAs in multiple breast cancer cell lines [67]. Very consistently, they observed a decrease in proliferation using siRNAs against the short isoform or all isoform but not siRNAs against the long isoform. These results indicate that the phenotype is driven by the quantity of the short isoform (scenario 1 in Table 1).
Protein kinase C (PKC) is a family of a protein involved in cellular proliferation and apoptosis. During all-trans retinoic acid-induced differentiation of NT2 teratocarcinoma cells, Jiang et al. observed that generation of a new PKCdelta isoform [68]. Usage of an alternative 5’ss downstream of the constitutive 5’ss of exon 10 produces PKCdeltaVIII. Jiang et al. extensively optimized multiple oligonucleotides to finally obtain antisense strategies that promote almost exclusively either the distal 5’ss (ASO 71) or the proximal 5’ss (ASO 75) on exon 10 [68]. Consistently, favoring the production of PKCdeltaVIII through ASO71 decrease apoptosis whereas targeting PKCdeltaVIII increase apoptosis of NT2 cells [68]. Thus, a splicing shift is sufficient to induce an apoptosis phenotype. Altogether, the PKCdelta exon 10 ASE generate isoforms with antagonistic function (scenario 2 in Table 1).
There is experimental evidence suggesting that the exon 69 of SYK encode a nuclear localization signal [69]. Consequently, reprogramming SYK splicing to favor the short form (without exon 69) redistribute the protein to the cytoplasm and hence affect cell survival [43]. Indeed, a SM or ISI is sufficient to induce apoptosis in multiple cancer cell lines but not a strategy targeting all isoforms [43]. Therefore, alternative splicing of SYK leads to isoform with antagonistic functions (Table 1, scenario 2). Interestingly, the SYK isoforms do not appear to act antagonistically in the context of adherent independent growth in soft agar. Indeed, both ISIs targeting the long isoform as well as siRNAs targeting all isoforms reduce the growth of cancer cells. The theory predicts that targeting SYK with siRNAs against the short isoform or with SMs would allow to delineate between scenario 1 (monofunctional) or scenario 3 (redundancy). Unfortunately, the particular exon-exon junction was refractory to siRNA design and the duration of the assay prevent the use of SM, respectively [43]. Thus, isoform functional relationship is not an intrinsic property and may vary depending on the context.
One of the well studied model gene in alternative splicing is the gene BCL2L1 (best known as Bcl-x). Two 5’ss are in competition in the second exon to produce a shorter and pro-apoptotic version (Bcl-xS) or a longer and anti-apoptotic molecule (Bcl-xL). Several groups have successfully targeted the Bcl-xL isoform and witnessed a concomitant increase in apoptosis in multiple cancer cell lines (Table 4). Since both isoforms possesses opposite function; the isoform ratio should dictate the phenotypic outcome and hence either a SM or ISI targeting Bcl-xL should be effective at inducing apoptosis. However, SMs stimulating the use of the distal 5’ss was not as robust at inducing apoptosis despite an obvious splicing shift (Table 4). This can be reconciled if one takes account of the much more transient effect of a SMs compared to ISIs. Indirect support to this theory comes from sustain Bcl-xS overexpression that sensitizes cancer cells to apoptosis [70, 71]. In addition, there is a strong correlation between the overall Bcl-x expression level in a defined cell line and the efficiency of apoptosis induction, suggesting that generation of Bcl-xS and hence the Bcl-xL / Bcl-xS ratio drives the apoptotic phenotype [72]. Also, worth mentioning is that phosphorothioate-modified oligonucleotides can lead to false positive splicing and phenotypic effects [44, 73]. Thus, Bcl-xS and BclxL possess antagonistic apoptotic functions (Table 1, scenario 2).
Perspectives
This review article focusses on studies that investigate the phenotype of cancer-related splicing isoform through the use of SMs and/or ISIs. Doing so, it excludes studies where the functional role of splicing isoform was uncovered through other approaches such as overexpression of specific isoforms through expression vectors in vitro and isoform-specific shRNA expressing vector [74]. It also excludes reports where SMs were used but the consequence on the phenotype was not examined in cancer cells [37, 48, 75, 76].
Initially, antisense oligonucleotides were used to correct splicing defect due to a mutation in the splicing regulatory sequence (e.g. b-thalassemia) [32]. It was later applied to reprogram alternative splicing of cancer-related genes independently of the mutational status of the gene (Table 2, Table 4). As high-throughput genomic studies continue to reveal mutations directly causing splicing defect [16, 19, 77], it would be beneficial to restore the expression and splicing pattern of some of those genes through antisense oligonucleotides to uncover their functional role in cancer.
Recently, re-analysis of TCGA datasets indicates that intron retention is abundant in cancer [78,79,80]. Their enrichment in tumor suppressor gene suggests that intron retention and hence the inclusion of premature stop codon is an effective way of shutting off gene expression. Therefore, efforts aiming at restoring tumor suppressor genes expression level through antisense-based strategies, including enhancing intron splicing are worth pursuing.
Conclusions
In summary, SMs and ISIs are powerful molecular tool to dissect the functional contribution of cancer-related splicing isoform in cancer cell lines. End-point PCR and isoform-specific real-time PCR are complementary and scalable assays to evaluate the performance of SMs and ISIs. Direct comparison of SM, ISI and siRNA targeting all isoforms help to deduce the functional contribution of individual splicing isoforms. Importantly, there is a need to develop antisense-based strategies promoting splicing (TOES-like strategies) as there is no design rule established so far.
Abbreviations
- 2’-MOE:
-
2’ OMethoxyethyl
- 2’OMe:
-
2’OMethyl
- alt3’:
-
Alternative 3’
- alt5’:
-
Alternative 5’
- ASE:
-
Alternative splicing event
- DMD:
-
Duchenne muscular dystrophy
- ISI:
-
Isoform-specific inhibitor
- LNA:
-
Locked nucleic acid
- NMD:
-
Non-sense mediated decay
- PNA:
-
Peptide nucleic acid
- PS:
-
Phosphorothioate
- Psi:
-
Percent splicing index
- QPsi:
-
Quantitative Psi
- Ref:
-
Reference
- siRNA:
-
Small interfering RNA
- SM:
-
Splicing modulator
- SSO:
-
Splice-swithing oligonucleotide
- TOES:
-
Targeted oligonucleotide enhancer of splicing
- TOSS:
-
Targeted oligonucleotide silencer of splicing
References
Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456(7221):470–6.
Lutz CS. Alternative polyadenylation: a twist on mRNA 3′ end formation. ACS Chem Biol. 2008;3(10):609–17.
Masamha CP, Wagner EJ. The contribution of alternative polyadenylation to the cancer phenotype. Carcinogenesis. 2018;39(1):2–10.
Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8(2):113–26.
Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci U S A. 2003;100(1):189–92.
da Costa PJ, Menezes J, Romao L. The role of alternative splicing coupled to nonsense-mediated mRNA decay in human disease. Int J Biochem Cell Biol. 2017;91(Pt B):168–75.
Hughes TA. Regulation of gene expression by alternative untranslated regions. Trends Genet. 2006;22(3):119–22.
Brosseau JP, et al. Redirecting splicing with bifunctional oligonucleotides. Nucleic Acids Res. 2014;42(6):e40.
Madgwick A, et al. Neural differentiation modulates the vertebrate brain specific splicing program. PLoS One. 2015;10(5):e0125998.
Vitting-Seerup K, Sandelin A. The landscape of isoform switches in human cancers. Mol Cancer Res. 2017;15(9):1206–20.
Wang Y, et al. Intronic splicing enhancers, cognate splicing factors and context-dependent regulation rules. Nat Struct Mol Biol. 2012;19(10):1044–52.
Brosseau JP, et al. Tumor microenvironment-associated modifications of alternative splicing. RNA. 2014;20(2):189–201.
Urbanski LM, Leclair N, Anczukow O. Alternative-splicing defects in cancer: splicing regulators and their downstream targets, guiding the way to novel cancer therapeutics. Wiley Interdiscip Rev RNA. 2018;9(4):e1476.
Climente-Gonzalez H, et al. The functional impact of alternative splicing in Cancer. Cell Rep. 2017;20(9):2215–26.
Krawczak M, Reiss J, Cooper DN. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum Genet. 1992;90(1–2):41–54.
Jayasinghe RG, et al. Systematic analysis of splice-site-creating mutations in Cancer. Cell Rep. 2018;23(1):270–81 e3.
Bonnal S, Vigevani L, Valcarcel J. The spliceosome as a target of novel antitumour drugs. Nat Rev Drug Discov. 2012;11(11):847–59.
Dvinge H, et al. RNA splicing factors as oncoproteins and tumour suppressors. Nat Rev Cancer. 2016;16(7):413–30.
Seiler M, et al. Somatic mutational landscape of splicing factor genes and their functional consequences across 33 Cancer types. Cell Rep. 2018;23(1):282–96 e4.
Walder RY, Walder JA. Role of RNase H in hybrid-arrested translation by antisense oligonucleotides. Proc Natl Acad Sci U S A. 1988;85(14):5011–5.
Schultz SJ, Champoux JJ. RNase H activity: structure, specificity, and function in reverse transcription. Virus Res. 2008;134(1–2):86–103.
Nakamura H, et al. How does RNase H recognize a DNA.RNA hybrid? Proc Natl Acad Sci U S A. 1991;88(24):11535–9.
Donis-Keller H. Site specific enzymatic cleavage of RNA. Nucleic Acids Res. 1979;7(1):179–92.
Leech SH, et al. Induction of apoptosis in lung-cancer cells following bcl-xL anti-sense treatment. Int J Cancer. 2000;86(4):570–6.
Manoharan M. 2′-carbohydrate modifications in antisense oligonucleotide therapy: importance of conformation, configuration and conjugation. Biochim Biophys Acta. 1999;1489(1):117–30.
Kurreck J. Antisense technologies. Improvement through novel chemical modifications. Eur J Biochem. 2003;270(8):1628–44.
Crooke ST. Molecular mechanisms of antisense oligonucleotides. Nucleic Acid Ther. 2017;27(2):70–7.
Summerton J. Morpholino antisense oligomers: the case for an RNase H-independent structural type. Biochim Biophys Acta. 1999;1489(1):141–58.
Juliano RL. The delivery of therapeutic oligonucleotides. Nucleic Acids Res. 2016;44(14):6518–48.
van der Bent ML, et al. Assisted delivery of antisense therapeutics in animal models of heritable neurodegenerative and neuromuscular disorders: a systematic review and meta-analysis. Sci Rep. 2018;8(1):4181.
Lacerra G, et al. Restoration of hemoglobin a synthesis in erythroid cells from peripheral blood of thalassemic patients. Proc Natl Acad Sci U S A. 2000;97(17):9591–6.
Dominski Z, Kole R. Restoration of correct splicing in thalassemic pre-mRNA by antisense oligonucleotides. Proc Natl Acad Sci U S A. 1993;90(18):8673–7.
Oikonomidou PR, Casu C, Rivella S. New strategies to target iron metabolism for the treatment of beta thalassemia. Ann N Y Acad Sci. 2016;1368(1):162–8.
Muntoni F, Torelli S, Ferlini A. Dystrophin and mutations: one gene, several proteins, multiple phenotypes. Lancet Neurol. 2003;2(12):731–40.
Gao QQ, McNally EM. The dystrophin complex: structure, function, and implications for therapy. Compr Physiol. 2015;5(3):1223–39.
Owen N, et al. Design principles for bifunctional targeted oligonucleotide enhancers of splicing. Nucleic Acids Res. 2011;39(16):7194–208.
Bruno IG, Jin W, Cote GJ. Correction of aberrant FGFR1 alternative RNA splicing through targeting of intronic regulatory elements. Hum Mol Genet. 2004;13(20):2409–20.
Bruun GH, et al. Blocking of an intronic splicing silencer completely rescues IKBKAP exon 20 splicing in familial dysautonomia patient cells. Nucleic Acids Res. 2018;46(15):7938–52.
Hua Y, et al. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am J Hum Genet. 2008;82(4):834–48.
Aartsma-Rus A, et al. Functional analysis of 114 exon-internal AONs for targeted DMD exon skipping: indication for steric hindrance of SR protein binding sites. Oligonucleotides. 2005;15(4):284–97.
Hua Y, et al. Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. PLoS Biol. 2007;5(4):e73.
Villemaire J, et al. Reprogramming alternative pre-messenger RNA splicing through the use of protein-binding antisense oligonucleotides. J Biol Chem. 2003;278(50):50031–9.
Prinos P, et al. Alternative splicing of SYK regulates mitosis and cell survival. Nat Struct Mol Biol. 2011;18(6):673–9.
Brosseau, J.P., Detection, functional annotation and regulation of splicing isoforms associated to ovarian cancer, in Department of Biochemistry. 2012, Universite de Sherbrooke: Sherbrooke p 293.
Wang Z, et al. General and specific functions of exonic splicing silencers in splicing control. Mol Cell. 2006;23(1):61–70.
Cartegni L, Krainer AR. Correction of disease-associated exon skipping by synthetic exon-specific activators. Nat Struct Biol. 2003;10(2):120–5.
Skordis LA, et al. Bifunctional antisense oligonucleotides provide a trans-acting splicing enhancer that stimulates SMN2 gene expression in patient fibroblasts. Proc Natl Acad Sci U S A. 2003;100(7):4114–9.
Ghigna C, et al. Pro-metastatic splicing of Ron proto-oncogene mRNA can be reversed: therapeutic potential of bifunctional oligonucleotides and indole derivatives. RNA Biol. 2010;7(4):495–503.
Dahlgren C, et al. Analysis of siRNA specificity on targets with double-nucleotide mismatches. Nucleic Acids Res. 2008;36(9):e53.
Schwarz DS, et al. Designing siRNA that distinguish between genes that differ by a single nucleotide. PLoS Genet. 2006;2(9):e140.
Huang H, et al. Profiling of mismatch discrimination in RNAi enabled rational design of allele-specific siRNAs. Nucleic Acids Res. 2009;37(22):7560–9.
Klinck R, et al. Multiple alternative splicing markers for ovarian cancer. Cancer Res. 2008;68(3):657–63.
Brosseau JP, et al. High-throughput quantification of splicing isoforms. RNA. 2010;16(2):442–9.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25(4):402–8.
Hellemans J, et al. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8(2):R19.
Malcolm DW, et al. Evaluating side effects of nanoparticle-mediated siRNA delivery to mesenchymal stem cells using next generation sequencing and enrichment analysis. Bioeng Transl Med. 2016;1(2):193–206.
Malioutov D, et al. Quantifying homologous proteins and proteoforms. Mol Cell Proteomics. 2018.
Koh CM, et al. MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature. 2015;523(7558):96–100.
Weidle UH, et al. Differential splicing generates new transmembrane receptor and extracellular matrix-related targets for antibody-based therapy of cancer. Cancer Genomics Proteomics. 2011;8(5):211–26.
Boiziau C, et al. Antisense oligonucleotides inhibit in vitro cDNA synthesis by HIV-1 reverse transcriptase. Antisense Nucleic Acid Drug Dev. 1996;6(2):103–9.
Blencowe BJ. The relationship between alternative splicing and proteomic complexity. Trends Biochem Sci. 2017;42(6):407–8.
Kole R, Krainer AR, Altman S. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat Rev Drug Discov. 2012;11(2):125–40.
Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74.
Akgul C, Moulding DA, Edwards SW. Alternative splicing of Bcl-2-related genes: functional consequences and potential therapeutic applications. Cell Mol Life Sci. 2004;61(17):2189–99.
Lin JC, Tsao MF, Lin YJ. Differential impacts of alternative splicing networks on apoptosis. Int J Mol Sci. 2016;17(12)E2097.
Hauser AD, et al. The SmgGDS splice variant SmgGDS-558 is a key promoter of tumor growth and RhoA signaling in breast cancer. Mol Cancer Res. 2014;12(1):130–42.
Jiang K, et al. Identification of a novel antiapoptotic human protein kinase C delta isoform, PKCdeltaVIII in NT2 cells. Biochemistry. 2008;47(2):787–97.
Wang L, et al. Alternative splicing disrupts a nuclear localization signal in spleen tyrosine kinase that is required for invasion suppression in breast cancer. Cancer Res. 2003;63(15):4724–30.
Sumantran VN, et al. Overexpression of Bcl-XS sensitizes MCF-7 cells to chemotherapy-induced apoptosis. Cancer Res. 1995;55(12):2507–10.
Clarke MF, et al. A recombinant bcl-x s adenovirus selectively induces apoptosis in cancer cells but not in normal bone marrow cells. Proc Natl Acad Sci U S A. 1995;92(24):11024–8.
Mercatante DR, Mohler JL, Kole R. Cellular response to an antisense-mediated shift of Bcl-x pre-mRNA splicing and antineoplastic agents. J Biol Chem. 2002;277(51):49374–82.
Winkler J, et al. Off-target effects related to the phosphorothioate modification of nucleic acids. ChemMedChem. 2010;5(8):1344–52.
Paschalis A, et al. Alternative splicing in prostate cancer. Nat Rev Clin Oncol. 2018;15(11):663–75.
Anczukow O, et al. BRCA2 deep intronic mutation causing activation of a cryptic exon: opening toward a new preventive therapeutic strategy. Clin Cancer Res. 2012;18(18):4903–9.
Karras JG, et al. Deletion of individual exons and induction of soluble murine interleukin-5 receptor-alpha chain expression through antisense oligonucleotide-mediated redirection of pre-mRNA splicing. Mol Pharmacol. 2000;58(2):380–7.
Shiraishi Y, et al. A comprehensive characterization of cis-acting splicing-associated variants in human cancer. Genome Res. 2018;28(8):1111–25.
Dvinge H, Bradley RK. Widespread intron retention diversifies most cancer transcriptomes. Genome Med. 2015;7(1):45.
Jung H, et al. Intron retention is a widespread mechanism of tumor-suppressor inactivation. Nat Genet. 2015;47(11):1242–8.
Supek F, et al. Synonymous mutations frequently act as driver mutations in human cancers. Cell. 2014;156(6):1324–35.
Shiraishi T, Eysturskarth J, Nielsen PE. Modulation of mdm2 pre-mRNA splicing by 9-aminoacridine-PNA (peptide nucleic acid) conjugates targeting intron-exon junctions. BMC Cancer. 2010;10:342.
Owen LA, et al. Morpholino-mediated increase in soluble Flt-1 expression results in decreased ocular and tumor neovascularization. PLoS One. 2012;7(3):e33576.
Uehara H, et al. Dual suppression of hemangiogenesis and lymphangiogenesis by splice-shifting morpholinos targeting vascular endothelial growth factor receptor 2 (KDR). FASEB J. 2013;27(1):76–85.
Shieh JJ, et al. Modification of alternative splicing of mcl-1 pre-mRNA using antisense morpholino oligonucleotides induces apoptosis in basal cell carcinoma cells. J Invest Dermatol. 2009;129(10):2497–506.
Catena R, et al. Increased expression of VEGF121/VEGF165-189 ratio results in a significant enhancement of human prostate tumor angiogenesis. Int J Cancer. 2007;120(10):2096–109.
Wilhelm E, et al. TAF6delta controls apoptosis and gene expression in the absence of p53. PLoS One. 2008;3(7):e2721.
Nielsen TO, et al. Directing HER4 mRNA expression towards the CYT2 isoform by antisense oligonucleotide decreases growth of breast cancer cells in vitro and in vivo. Br J Cancer. 2013;108(11):2291–8.
Wang Z, et al. Manipulation of PK-M mutually exclusive alternative splicing by antisense oligonucleotides. Open Biol. 2012;2(10):120133.
Zammarchi F, et al. Antitumorigenic potential of STAT3 alternative splicing modulation. Proc Natl Acad Sci U S A. 2011;108(43):17779–84.
Dewaele M, et al. Antisense oligonucleotide-mediated MDM4 exon 6 skipping impairs tumor growth. J Clin Invest. 2016;126(1):68–84.
Lin J, et al. Induced-decay of Glycine decarboxylase transcripts as an anticancer therapeutic strategy for non-small-cell lung carcinoma. Mol Ther Nucleic Acids. 2017;9:263–73.
Izaguirre DI, et al. PTBP1-dependent regulation of USP5 alternative RNA splicing plays a role in glioblastoma tumorigenesis. Mol Carcinog. 2012;51(11):895–906.
Smith LD, et al. Novel splice-switching oligonucleotide promotes BRCA1 aberrant splicing and susceptibility to PARP inhibitor action. Int J Cancer. 2017;140(7):1564–70.
Mogilevsky M, et al. Modulation of MKNK2 alternative splicing by splice-switching oligonucleotides as a novel approach for glioblastoma treatment. Nucleic Acids Res. 2018.
Liu J, et al. Overcoming imatinib resistance conferred by the BIM deletion polymorphism in chronic myeloid leukemia with splice-switching antisense oligonucleotides. Oncotarget. 2017;8(44):77567–85.
Fletcher L, et al. Live cell imaging reveals distinct roles in cell cycle regulation for Nek2A and Nek2B. Biochim Biophys Acta. 2005;1744(2):89–92.
Kim CJ, et al. Anti-oncogenic activities of cyclin D1b siRNA on human bladder cancer cells via induction of apoptosis and suppression of cancer cell stemness and invasiveness. Int J Oncol. 2018;52(1):231–40.
Camats M, et al. P19 H-ras induces G1/S phase delay maintaining cells in a reversible quiescence state. PLoS One. 2009;4(12):e8513.
Johnson RM, et al. The alternative splicing of cytoplasmic polyadenylation element binding protein 2 drives Anoikis resistance and the metastasis of triple negative breast Cancer. J Biol Chem. 2015;290(42):25717–27.
Crnkovic-Mertens I, et al. Isoform-specific silencing of the Livin gene by RNA interference defines Livin beta as key mediator of apoptosis inhibition in HeLa cells. J Mol Med (Berl). 2006;84(3):232–40.
Goldberg MS, Sharp PA. Pyruvate kinase M2-specific siRNA induces apoptosis and tumor regression. J Exp Med. 2012;209(2):217–24.
Kim CJ, et al. Cyclin D1b variant promotes cell invasiveness independent of binding to CDK4 in human bladder cancer cells. Mol Carcinog. 2009;48(10):953–64.
Nitz MD, et al. RREB1 transcription factor splice variants in urologic cancer. Am J Pathol. 2011;179(1):477–86.
Wu D, et al. An alternative splicing isoform of eukaryotic initiation factor 4H promotes tumorigenesis in vivo and is a potential therapeutic target for human cancer. Int J Cancer. 2011;128(5):1018–30.
Tatsumi N, et al. Identification of a novel C-terminal truncated WT1 isoform with antagonistic effects against major WT1 isoforms. PLoS One. 2015;10(6):e0130578.
Yoshida M, et al. RPAP3 splicing variant isoform 1 interacts with PIH1D1 to compose R2TP complex for cell survival. Biochem Biophys Res Commun. 2013;430(1):320–4.
Mei H, et al. Alternative splicing of S6K1 promotes non-small cell lung cancer survival. Tumour Biol. 2016;37(10):13369–76.
Hosoya T, et al. A novel splice variant of the nuclear coactivator p120 functions strongly for androgen receptor: characteristic expression in prostate disease. Endocr J. 2008;55(4):657–65.
Ibanez-Costa A, et al. In1-ghrelin splicing variant is overexpressed in pituitary adenomas and increases their aggressive features. Sci Rep. 2015;5:8714.
DiFeo A, et al. A functional role for KLF6-SV1 in lung adenocarcinoma prognosis and chemotherapy response. Cancer Res. 2008;68(4):965–70.
Marzese DM, et al. Brain metastasis is predetermined in early stages of cutaneous melanoma by CD44v6 expression through epigenetic regulation of the spliceosome. Pigment Cell Melanoma Res. 2015;28(1):82–93.
Marcel V, et al. Modulation of p53beta and p53gamma expression by regulating the alternative splicing of TP53 gene modifies cellular response. Cell Death Differ. 2014;21(9):1377–87.
Datta D, et al. Ras-induced modulation of CXCL10 and its receptor splice variant CXCR3-B in MDA-MB-435 and MCF-7 cells: relevance for the development of human breast cancer. Cancer Res. 2006;66(19):9509–18.
Byun HJ, et al. A splice variant of CD99 increases motility and MMP-9 expression of human breast cancer cells through the AKT-, ERK-, and JNK-dependent AP-1 activation signaling pathways. J Biol Chem. 2006;281(46):34833–47.
Rouleau M, et al. Dual roles for splice variants of the glucuronidation pathway as regulators of cellular metabolism. Mol Pharmacol. 2014;85(1):29–36.
Giglio S, et al. Identification of an aberrantly spliced form of HDMX in human tumors: a new mechanism for HDM2 stabilization. Cancer Res. 2005;65(21):9687–94.
Clayburgh DR, et al. A differentiation-dependent splice variant of myosin light chain kinase, MLCK1, regulates epithelial tight junction permeability. J Biol Chem. 2004;279(53):55506–13.
Taylor JK, et al. Induction of endogenous Bcl-xS through the control of Bcl-x pre-mRNA splicing by antisense oligonucleotides. Nat Biotechnol. 1999;17(11):1097–100.
Mercatante DR, et al. Modification of alternative splicing of Bcl-x pre-mRNA in prostate and breast cancer cells. Analysis of apoptosis and cell death. J Biol Chem. 2001;276(19):16411–7.
Simoes-Wust AP, et al. Bcl-xl antisense treatment induces apoptosis in breast carcinoma cells. Int J Cancer. 2000;87(4):582–90.
Wilusz JE, Devanney SC, Caputi M. Chimeric peptide nucleic acid compounds modulate splicing of the bcl-x gene in vitro and in vivo. Nucleic Acids Res. 2005;33(20):6547–54.
Dodier P, Piche A. Bcl-X(L) is functionally non-equivalent for the regulation of growth and survival in human ovarian cancer cells. Gynecol Oncol. 2006;100(2):254–63.
Li Z, et al. Pro-apoptotic effects of splice-switching oligonucleotides targeting Bcl-x pre-mRNA in human glioma cell lines. Oncol Rep. 2016;35(2):1013–9.
Acknowledgements
The author would like to thank Jean-Pierre Perreault, Benoit Chabot and Sherif Abou Elela.
Funding
Not applicable.
Availability of data and materials
Not applicable.
Author information
Authors and Affiliations
Contributions
The author read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The author declares that he has no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Brosseau, JP. Splicing isoform-specific functional genomic in cancer cells. Appl Cancer Res 38, 18 (2018). https://doi.org/10.1186/s41241-018-0068-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41241-018-0068-6