Abstract
Glucose-6-phosphate dehydrogenase (G6PD) deficiency is a group of X-linked, hereditary genetic disorders caused by mutations in the G6PD gene and results in functional variants of about 400 biochemical and clinical phenotypes. Among them, more than 215 genotypes have been identified so far. In this review, specific features of the genotype distribution in different communities and countries are discussed based on multiple reports and our molecular epidemiological studies of Southeast Asian countries. Particularly, in Indonesia, the frequency distribution of G6PD deficiency variants was distinct between western and eastern Indonesian populations, suggesting two different gene flows during Indonesian expansions.
Similar content being viewed by others
Background
Glucose-6-phosphate dehydrogenase (G6PD) deficiency is a group of X-linked, hereditary genetic disorders caused by mutations in the G6PD gene. These mutations result in protein variants with decreased levels of enzyme activities and are correlated with a wide range of biochemical and clinical phenotypes [1]. The G6PD gene spans 18 kb on the X chromosome (Xq28), contains an open reading frame of 1545 base pairs and encodes 13 exons and 12 introns. Most instances of G6PD deficiency are caused by a single nucleotide mutation resulting in one amino acid change among 515 amino acids. To date, more than 400 biochemical variants have been described, and more than 215 mutations among them have been discovered at the molecular level [2,3,4].
In Southeast Asia, there is significant heterogeneity of the dominant variants in different areas, which appears to be closely related to the geographic locations and associated with different racial and ethnic groups [5, 6]. Since 1997, we have been surveying malaria and G6PD deficiency in malaria endemic areas in Southeast Asian countries [6,7,8,9,10,11,12,13] using two rapid diagnostic methods [14, 15]. With these methods, malaria patients can be informed of the diagnostic results on-site, usually within 30 min of a blood examination, and they are prescribed antimalarial drugs, including primaquine if their G6PD activity is normal. Blood samples collected from G6PD-deficient volunteers were analyzed at the molecular level, and distributing variants of G6PD deficiency were identified in each population. All G6PD-deficient blood samples obtained in our field surveys were confirmed as having deficient genotypes without exception. In this review, we show the precise distribution and distinct features of the distribution of G6PD deficiency genotypes among several populations in Southeast Asia by referring to published reports.
Distribution of G6PD deficiency genotypes in Vietnam, Laos and Cambodia
Four studies on Vietnam [6, 12, 16, 17] reported G6PD Viangchan (871G > A, 1311C > T, IVS11 nt93T > C; WHO Class II) as the most dominant variant in the Kinh population (largest ethnic group; 164/275, 59.6%) and in two minority populations, K’Ho and Stieng (19/41, 46.3%) (Table 1). This variant is also very common in the Lao (245/298, 82.2%) and Khmer (637/675, 94.4%) populations of Laos and Cambodia, respectively. In the Kinh population, variants of Chinese origins, such as G6PD Kaiping (1388G > A; WHO Class II), G6PD Canton (1376G > T; WHO Class II), G6PD Chinese-5 (1024C > T; WHO Class III), G6PD Quing Yuan (or Chinese-4; 392G > T; WHO Class III), and G6PD Gaohe (95A > G; WHO Class III), were also detected (45/275, 16.4%). In addition, G6PD Union (1360C > T; WHO Class II), which originated from Oceania, was also observed, suggesting that the Kinh population has many different ancestral sources from continental Southeast Asia, China and Oceania. Indeed, it is well known that Vietnam had a strong historical connection with China for more than 2000 years. Until recently, marriages between the Kinh and Chinese were common, which would allow G6PD variants of Chinese origins, such as G6PD Viangchan, being introduced into the Kinh population, reflecting another ancestral source for Southeast Asian populations. However, these Chinese variants are very rare or absent in the K’Ho and the Stieng populations, suggesting that these minorities remained in remote areas, as seen in the present day, and/or that they rejected relations or marriages with other tribes or foreigners. This tendency is also seen in Lao and Khmer ethnic groups, which would indicate a more simple and homogeneous ancestry than the Kinh ethnic group.
G6PD 1311C > T with IVS11 nt93T > C (designated here as G6PD 1311T/93C; WHO Class III) was found as another common genotype in Vietnam (71/316, 22.5%). This mutation was also detected among the Kinh and the Stieng populations. G6PD 1311T/93C is a silent mutation that does not change any amino acids in exon 11, but it causes lower G6PD activity by an unknown mechanism. It was reported first in Filipinos [44], and later, many cases were discovered in populations of the Javanese [41], Chinese [46], southern Thai [26], Malaysian Malay [38], Orang Asli (Malaysian aborigine [38,39,40]) and Kachin (a minority in Myanmar [5]). The Orang Asli is one of the “Proto”-Malay groups and well known as an ancient ethnic group in Southeast Asia, suggesting this variant has existed since antiquity along with G6PD Viangchan. Quite interestingly, however, G6PD 1311T/93C is not distributed in Laos and Cambodia despite their geographical continuity.
G6PD Viangchan is always accompanied with mutations at 1311C > T in exon 11 and IVS 11 nt93T > C in intron 11, whereas G6PD Jammu (871G > A; WHO Class II), which is dominantly distributed in India, has the wild type sequence at 1311C with IVS11 nt93T. These two variants are thought to have arisen independently [47]. It is worth noting that G6PD Mediterranean (563C > T; WHO Class II) also has two subtypes at polymorphisms 1311 and IVS 11 nt93: The Mediterranean subtype accompanying “1311C > T with IVS11 nt93T > C” and the Indo-Pakistan subtype with the wild type sequence “1311C with IVS11 nt93T (see below).
Among the Lao population in Laos, the frequency distribution of G6PD Union is fairly high (25/298, 8.4%) in comparison with the Kinh (9/275, 3.3%) or the Khmer (4/675, 0.6%) populations for unknown reasons. G6PD Union is dominantly detected from countries in the Pacific Ocean such as the Philippines [21, 44], Papua New Guinea [48], Solomon Islands [49] and Vanuatu archipelago [50]. It is probable that a high prevalence of this variant in the Lao population may be related to foreign migrant workers from the Philippines.
Recently, an analysis of G6PD genotypes in the Lao Theung (or the Lao Mon-Khmer) population was reported from Laos [51]. This tribe is the second largest ethnic group, and their language (the Mon-Khmer language) is different from the Lao ethnic group, who speaks languages belonging to the Thai-Kra Dai family. Surprisingly, the most dominant variant in the Lao Theung tribe is G6PD Aures (143T > C; WHO Class III; 17/29, 58.6%), which is commonly detected in Mediterranean and North African populations. Next common are G6PD Union (3/29, 10.3%) and G6PD Jammu (2/29, 6.9%), but G6PD Vianghan was rare (1/29, 3.4%) among this tribe. All these results taken together strongly suggest that the Lao Theung tribe has different ancestral origins from the Lao ethnic group [51].
G6PD deficiency genotypes in Thailand, Myanmar, Malaysia and the Philippines
Among the Thai population, G6PD Viangchan is also the most common variant (169/397, 42.6%), followed by various genotypes of Chinese origins (101/397, 25.4%), G6PD Mahidol (487G > A; WHO Class III; 97/397, 24.4%) and G6PD Union (19/397, 4.8%). Therefore, the frequency distribution of G6PD variants in the Thai population is quite similar to that in the Kinh population, except for the high prevalence of G6PD Mahidol. As in the Kinh group, the Thai population is derived from complicated ancestries that involve intermarriages with other tribes or foreigners.
In Myanmar, G6PD Viangchan is very rare (12/1140, 1.1%). G6PD Mahidol is the most dominant in the Burman population (701/762, 92.0%) and in many minority populations, such as the Mon, Karen, Rakhine, Shan, etc. [5, 7, 17, 31,32,33]. Because these minorities have their own languages, the presence of new genotypes or distinct distribution patterns of variants was anticipated, as seen in the Lao Theung population in Laos. However, almost variants detected from these minorities were G6PD Mahidol only [7]. This observation indicates that, in spite of their different cultural backgrounds, these populations probably share the same ancestry origin with the Burman group before migration from southern China to the present territories. It is interesting to note that this variant was entirely absent in Vietnam and rare in Laos (7/298, 2.3%) and Cambodia (10/675, 1.5%). This observation suggests that all these ethnic groups have different origins from each other. In Myanmar, a high prevalence of G6PD 1311T/93C (95/182; 52.2%) was reported among the Kachin ethnic group and only this group for reasons unknown [5].
In Malaysia, Malaysian Malay, Chinese and Indian people make up the three major ethnic groups. In addition to these groups, minor indigenous communities called Orang Asli inhabit the Malay Peninsula. In the Malay population, G6PD Viangchan (36/101, 35.6%), G6PD Mahidol (18/101, 17.8%) and G6PD Mediterranean (25/101, 24.8%) are most common, followed by G6PD Coimbra (592C > T; WHO Class II) and G6PD Chatham (1003G > A;WHO Class II). A few cases of G6PD Vanua Lava (383 T > C; WHO Class II; a variant reported first from Melanesia [50]) and G6PD Union have also been identified. Regarding G6PD Mediterranean, the Malay population possess both subtypes [38]: The Mediterranean subtype (563 C > T with 1311T/93C), which may have originated from Mediterranean countries, and the Indo-Pakistan subtype (563 C > T with the wild sequence of 1311C/93T), which may have originated from India independently.
G6PD Coimbra is one of the oldest variants originated from Europe and is found in several Asian countries, including Vietnam, Cambodia, Myanmar, Malaysia, and the Orang Asli population. In contrast to the wide distribution of G6PD Coimbra, however, another old variant of Europe origin, G6PD Chatham, is found only in the Malaysian Malay among Southeast Asia populations. This variant as well as the two subtypes of G6PD Mediterranean may have been introduced to Malaysia by European people and/or Indian immigrants who settled through British colonization. On the other hand, variants of Chinese origins in the Malay population are rare (6/101, 5.9%) when compared with the Kinh (45/275, 16.4%) and Thai (101/397, 25.4%) populations. This observation may reflect Malay and Chinese people not intermarrying, possibly for religious reasons.
The frequency distribution of G6PD genotypes in the Philippines is remarkably different from continental Southeast Asian populations and Indonesian populations in that it has the G6PD Union as the most common variant (38/58, 65.5%). One case of G6PD Vanua Lava was reported from a Filipino immigrant boy in Spain [45].
G6PD deficiency genotypes in Indonesia
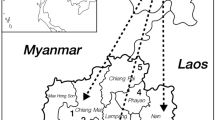
According to wildlife belonging to Asia and to Australia, the Indonesian archipelago is divided mainly into two parts, western and eastern Indonesia, by the Wallace line [52] at the Lombok Strait between the Bali and Lombok islands (Fig. 1). During the last Ice Age, all the islands of western Indonesia up to Bali were linked to Southeast Asian continent and created a unique peninsula named “Sundaland”, where the “Deutro”-Malay people were able to walk between the Asian continent and the islands of Sumatra, Java, Borneo (Kalimantan) and Bali. Therefore, genetic interactions may have arisen among the Deutro-Malay population, and similar genes flows, including G6PD genes, may have been shared among the Malaysian Malay and western Indonesian populations such as the Sumatra Malay, Sundanese, Javanese, Balinese, etc.
With respect to the molecular analyses of G6PD genotypes among western Indonesian populations, 24 cases of Java islanders (17 Javanese, 6 Betawi and 1 Sundanese) have been reported so far (Tables 1 and 2; [7, 41, 42]). The presence of G6PD Viangchan, G6PD Mahidol, G6PD Mediterranean and G6PD 1311T/93C in these Java islanders may imply a resemblance to the frequency distribution of the Malaysian Malay population (Table 2).
In eastern Indonesia population, however, the most common variant is G6PD Vanua Lava (95/202, 47.0%), which is unlike the Java islanders (Table 2). This genotype was first reported in Vanuatu [50] and found later to spread from Papua New Guinea [48] to eastern Indonesia [7], Malaysia [38] and the Philippines [45] (Table 1). This variant was detected as the most dominant genotype in all eastern Indonesian islands, including the Halmahera, Buru, Flores, Sumba, West Timor, Banka and Sulawesi islands (Table 2), and it accounts for nearly half of all detected variants. However, G6PD Vanua Lava, as well as G6PD Chatham, was never detected in continental Southeast Asia populations except the Malaysian Malay population (Table 1). On the contrary, G6PD Mahidol, which is widely distributed from Myanmar, Thailand, Malaysia to Java, did not reach the eastern Indonesia islands or the Philippines. Furthermore, G6PD Union and G6PD 1311T/93C were also not detected in eastern Indonesian populations, and G6PD Mediterranean and Chinese genotypes are very rare or absent (except for G6PD Kaiping in the Sikka population in the Flores island). It is somewhat strange that the G6PD Union mutation was never observed in eastern Indonesian populations, despite the possible connection between this population and Filipino and Melanesian groups. A probable reason for the lack of G6PD Union in eastern Indonesian populations is the genetic drift caused by their long-term isolation [7].
All these results strongly suggest that the frequency distribution of G6PD variants in Indonesia may differ greatly between western and eastern populations. The separation of distributed G6PD variants between western and eastern Indonesia was also proposed by a bioinformatics analysis [53] using the data cited in this review. To support this hypothesis further, we conducted field surveys for G6PD deficiency (unpublished) in a western Indonesian tribe, the Balinese group of Bali Island, and an eastern Indonesian tribe, the Sasak group of Lombok Island, and compared the distributions of G6PD variants. The Bali and Lombok islands face across the Wallace line from the east side of the western Indonesian islands and the west side of the eastern Indonesian islands, respectively (Fig. 1).
As expected, G6PD Vanua Lava was found to be the most dominant variant in the Sasak population (n = 18; 9 G6PD Vanua Lava + 7 G6PD Viangchan + 2 G6PD Chatham). In contrast, this variant was absent in the Balinese population (n = 9; 4 G6PD Mediterranean + 3 G6PD Viangchan + 2 G6PD Chatham). However, the sample size for the Balinese population was too small, meaning more field surveys of this island are needed. Nevertheless, these preliminary results support our hypothesis that G6PD Vanua Lava is distributed in eastern Indonesian populations but not in western populations, implying two different gene flows during the expansions of the Indonesian peoples: In the western Indonesian islands, the Deutro-Malay populations had genetic interactions without involvement of the G6PD Vanua Lava mutation, while eastern Indonesian islanders, who carried this mutated gene, spread around eastern Indonesian islands and Melanesia.
According to a recent new theory, the migration and expansion of Melanesian (Austronesian language speakers) into the Pacific Ocean [54, 55] are thought to have begun with Taiwan aborigines, a type of Austronesian group, who expanded from Taiwan to the Pacific through the Philippines, Sulawesi and other Indonesian islands. Therefore, there is a possibility that Taiwan aborigines are also the origin of eastern Indonesian islanders. G6PD deficiency genotypes in the Taiwan aboriginal tribes were investigated by Tang et al. [56], who identified 5 cases of G6PD Coimbra and 6 cases of G6PD Chinese-3 and G6PD Canton. Further molecular investigations on the distribution of G6PD genotypes in the Taiwan aborigines, particularly on whether the G6PD Vanua Lava mutation exists or not, and on how deep this variant is spreading into Filipino populations, are awaited for understanding gene flows during Indonesian expansions.
Conclusions
Based on published reports and our molecular epidemiological studies of Southeast Asian countries, specific features of distribution of G6PD deficiency genotypes in each community and country are discussed. Particularly, in Indonesia, the frequency distribution of genotypes was distinct between western and eastern Indonesian populations, suggesting two different gene flows during expansions of Indonesian peoples.
Availability of data and materials
Not applicable.
Abbreviations
- G6PD:
-
Glucose 6 phosphate dehydrogenase
References
Cappellini MD, Fiorelli G. Glucose-6-phosphate dehydrogenase deficiency. Lancet. 2008;371:64–74.
Beutler E, Vulliamy TJ. Hematologically important mutations, glucose-6-phosphate dehydrogenase. Blood Cells Mol Dis. 2002;28:93–103.
Minucci A, Moradkhani K, Hwang MJ, Zuppi C, Giardina B, Capoluongo E. Glucose-6- phosphate dehydrogenase (G6PD) mutations database: review of the “old” and update of the new mutations. Blood Cells Mol Dis. 2012;48:154–65.
Gomez-Manzo S, Marcial-Quino J, Vanoye-Carlo A, Serrano-Posada H, Ortega-Cuellar D, Gonzalez-Valdez A, et al. Glucose-6-phosphate dehydrogenase: update and analysis of new mutations around the world. Int J Mol Sci. 2016;17:2069.
Li Q, Yang F, Liu R, Luo L, Yang Y, Zhang L, et al. Prevalence and molecular characterization of glucose-6-dehydrogenase deficiency at the China-Myanmar border. PLoS ONE. 2015;10:e0134593.
Kawamoto F, Matsuoka H, Pham NM, Hayashi T, Kasahara Y, Dung NT, et al. Further molecular analysis on G6PD deficiency variants in southern Vietnam and a novel variant designated as G6PD Ho Chi Minh (173 A>G; 58 Asp>Gly). Acta Med Okayama. 2017;71:325–32.
Iwai K, Hirono A, Matsuoka H, Kawamoto F, Horie T, Lin K, et al. Distribution of glucose-6-phosphate dehydrogenase mutations in Southeast Asia. Hum Gen. 2001;108:445–9.
Matsuoka H, Arai M, Yoshida S, Tantular IS, Pusarawati S, Kerong H, et al. Five different glucose-6-phophate dehydrogenase (G6PD) variants found among 11 G6PD deficient persons in Flores Island, Indonesia. J Hum Genet. 2003;48:541–4.
Matsuoka H, Wang J, Hirai M, Arai M, Yoshida S, Kobayashi T, et al. Glucose-6- phosphate dehydrogenase (G6PD) mutations in Myanmar: G6PD Mahidol (487G>A) is the most common variant in the Myanmar population. J Hum Genet. 2004;49:544–7.
Matsuoka H, Nguon C, Kanbe T, Jalloh A, Sato H, Yoshida S, et al. Glucose-6-phosphate dehydrogenase (G6PD) mutations in Cambodia: G6PD Viangchan (871G>A) is the most common variant in the Cambodian population. J Hum Genet. 2005;50:468–72.
Kawamoto F, Matsuoka H, Kanbe T, Tantular IS, Pusarawati S, Kerong HI, et al. Further investigations of glucose-6-phosphate dehydrogenase variants in Flores Island, eastern Indonesia. J Hum Genet. 2006;51:952–7.
Matsuoka H, Thuan DTV, van Thien H, Kanbe T, Jalloh A, Hirai M, et al. Seven different glucose-6-phosphate dehydrogenase variants including a new variant distributed in Lam Dong Province in southern Vietnam. Acta Med Okayama. 2007;61:181–5.
Tantular IS, Matsuoka H, Kasahara Y, Pusarawati S, Kanbe T, Tuda JSB, et al. Incidence and mutation analysis of glucose-6-dehydrogenase deficiency in eastern Indonesian populations. Acta Med Okayama. 2010;64:367–73.
Tantular IS, Kawamoto F. An improved, simple screening method for detection of glucose-6-phosphate dehydrogenase deficiency. Trop Med Int Health. 2003;8:569–74.
Tantular IS, Wulansari W, Kido Y, Inaoka DK, Matsuoka H, Lusida MI, et al. MTT formazan replaced WST-8 as a better simple screening method for detection of glucose-6-phosphate dehydrogenase deficiency, Indonesian. J Trop Inf Dis. 2019;7:161–7.
Hue NT, Charlieu JP, Chau TTH, Day N, Farrar JJ, Hien TT, et al. Glucose-6-phosphate dehydrogenase (G6PD) mutations and haemoglobinuria syndrome in the Vietnamese population. Malaria J. 2009;8:152.
Bancone G, Menard D, Khim N, Kim S, Canier L, Nguong C, et al. Molecular characterization and mapping of glucose-6-phosphate dehydrogenase (G6PD) mutations in the Greater Mekong Subregion. Malaria J. 2019;18:20.
Kanchanavithayakul A, Prasittisa K, Kiat-Amornak P, Chanda M, Kittiwatanasarn P, Nuchprayoon I, et al. Prevalence of glucose-6-phosphate dehydrogenase deficiency and genetic mutations among Karen and Lao populations in Thailand. Southeast Asian J Trop Med Public Health. 2017;48:1308–17.
Lover AA, Dantzer E, Hangvanthong B, Chindavongsa K, Welty S, Reza T, et al. Prevalence and risk factors for asymptomatic malaria and genotyping of glucose-6- phosphate (G6PD) deficiencies in a vivax-predominant setting, Lao PDR: implications for sub-national elimination goals. Malaria J. 2018;17:218.
Ong KIC, Iwagami M, Araki H, Khattignavong P, Soundala P, Keomalaphet S, et al. Prevalence of G6PD Viangchan variant in malaria endemic areas in Lao PDR: an implication for malaria elimination by 2030. Malaria J. 2019;18:75.
Hsia YE, Miyakawa F, Baltazar J, Ching NS, Yuen J, Westwood B, et al. Frequency of glucose-6-phosphate dehydrogenase (G6PD) mutations in Chinese, Filipinos, and Laotians from Hawaii. Hum Genet. 1993;92:470–6.
Louicharoen C, Nuchprayoon I. G6PD Viangchan (871G>A) is the most common G6PD-deficient variant in the Cambodian population. J Human Genet. 2005;50:448–52.
Kim S, Nguon C, Guillard B, Duong S, Chy S, Sum S, et al. Performance of the CareStartTM G6PD deficiency screening test, a Point-of-Care Diagnostic for primaquine therapy screening. PLoS ONE. 2011;6:e28357.
Roca-Feltrer A, Khim N, Kim S, Chy S, Canier L, Kerleguer A, et al. Field trial evaluation of the performances of Point-of-Care tests for screening G6PD deficiency in Cambodia. PLoS ONE. 2014;9:e116143.
Nuchprayoon I, Sanpavat S, Nuchprayoon S. Glucose-6-phosphate dehydrogenase (G6PD) mutations in Thailand: G6PD Viangchan (871G>A) is the most common deficiency variant in the Thai population. Hum Mutat. 2002;19:185.
Laosombat V, Sattayasevana B, Janejindamai W, Viprakasit V, Shirakawa T, Nishiyama K, et al. Molecular heterogeneity of glucose-6-phosphatedehydrogenase (G6PD) variants in the south of Thailand and identification of a novel variant (G6PD Songklanagarind). Blood cells Mol Dis. 2005;34:191–6.
Ninokata A, Kimura R, Samakkarn U, Settheetham-Ishida W, Ishida T. Coexistence of five G6PD variants indicates ethnic complexity of Phuket islanders, Southern Thailand. J Hum Genet. 2006;51:424–8.
Phompradit P, Kuesap J, Chaijaroenkul W, Rueangweerayut R, Hongkaew Y, Yamnuan R, et al. Prevalence and distribution of glucose-6-phosphate dehydrogenase (G6PD) variants in Thai and Burmese populations in malaria endemic areas of Thailand. Malaria J. 2011;10:368.
Nantakomol D, Paul R, Palasuwan A, Day NP, White NJ, Imwong M. Evaluation of the phenotypic test and genetic analysis in the detection of glucose-6-phosphate dehydrogenase deficiency. Malar J. 2013;12:289.
Charoenkwan P, Tantiprabha W, Sirichotiyakul S, Phusua A, Sanguansermsri T. Prevalence and molecular characterization of glucose-6-phosphate dehydrogenase deficiency in northern Thailand, Southeast Asian. J Trop Med Public Health. 2014;45:187–93.
Bancone G, Gilder ME, Chowwiwat N, Gornsawun G, Win E, Cho WW, et al. Prevalences of inherited red blood cell disorders in pregnant women of different ethnicities living along the Thailand-Myanmar border. Wellcome Open Res. 2017;2:72.
Nuchprayoon I, Louicharoen C, Charoenvej W. Glucose-6-phosphate dehydrogenase mutations in Mon and Burmese of southern Myanmar. J Hum Genet. 2008;53:48–54.
Bancone G, Chu CS, Somsakchaicharon R, Chowwiwat N, Parker DM, Charunwatthana P, et al. Characterization of G6PD genotypes and phenotypes on the Northwestern Thailand-Myanmar Border. PLoS ONE. 2014;9:e116063.
Deng Z, Yang F, Bai Y, He L, Li Q, Wu Y, et al. Co-inheritance of glucose-6-phosphate dehydrogenase deficiency mutations and heamoglobin E in a Kachin population in a malaria-endemic region of Southeast Asia. PLoS ONE. 2017;12:e0177917.
Lee J, Kim TI, Kang JM, Jun H, Lê HG, Thái TL, et al. Prevalence of glucose-6-phosphate dehydrogenase (G6PD) deficiency among malaria patients in Upper Myanmar. BMC Infect Dis. 2018;18:131.
May WL, Kyaw MP, Blacksell SD, Pukrittayakamee S, Chotivanich K, Hanboonkunupakarn B, et al. Impact of glucose-6-phosphate dehydrogenase deficiency on dengue infection in Myanmar children. PLoS ONE. 2019;14:e0209204.
Ainoon O, Yu YH, Amir Muhriz AL, Boo NY, Cheong SK, Hamidah NH. Glucose-6-phosphate dehydrogenase (G6PD) variants in Malaysian Malays. Hum Mutat. 2003;21:101.
Wang J, Luo E, Hirai M, Arai M, Abdul-Manan E, Mohamed-Isa Z, et al. Nine different glucose-6-phosphate dehydrogenase (G6PD) variants in a Malaysian population with Malay, Chinese, Indian and Orang Asli (aboriginal Malaysian) backgrounds. Acta Med Okayama. 2008;62:327–32.
Amini F, Ismail E, Zilfalil BA. Prevalence and molecular study of G6PD deficiency in Malaysian Orang Asli. Internal Med J. 2011;41:351–3.
Amini F, Ismail E. 3’-UTR variations and G6PD deficiency. J Hum Genet. 2013;58:189–94.
Soemantri AG, Saha S, Saha N, Tay JS. Molecular variants of red cell glucose-6-phosphate dehydrogenase deficiency in Central Java, Indonesia. Hum Hered. 1995;45:346–50.
Wisnumurti D, Sribudiani Y, Porsch RM, Maskcoen AM, Rahayuningsih SE, Asni EK, et al. G6PD genetic variations in neonatal hyperbilirubinemia in Indonesian deutromalay population. BMC Pediatr. 2019;19:506.
Satyagraha AW, Sadhewa A, Baramuli V, Elvira R, Ridenour C, Elyazar I, et al. G6PD deficiency at Sumba in eastern Indonesia is prevalent, diverse and severe: implications for primaquine therapy against vivax malaria. PLoS Negl Trop Dis. 2015;9:e0003602.
Silao CL, Shirakawa T, Nishiyama K, Padilla C, Matsuo M. Molecular basis of glucose-6-phosphate dehydrogenase deficiency among Filipinos. Pediatrics Int. 1999;41:138–41.
Minucci A, De Luca D, Torti E, Concolino P, Maurizi P, Giardina B, et al. Acute hemolytic crisis to concomitant presence of infection and possible altered acetaminophen catabolism in a Philipino child carrying the G6PD Vanua Lava mutation. Ann Clin Biochem. 2011;48:282–5.
Jiang W, Yu G, Liu P, Geng Q, Chen L, Lin Q, et al. Structure and function of glucose-6-phophate dehydrogenase-deficient variants in Chinese population. Human Genet. 2006;119:463–78.
Beutler E, Westwood B, Kuhl W. Definitions of the mutations of G6PD Wayn, G6PD Viangchan, G6PD Jammu, and G6PD “LeJeune.” Acta Haematol. 1991;86:179–82.
Wagner G, Bhatia K, Board P. Glucose-6-phosphate dehydrogenase deficiency mutations in Papua New Guinea. Human Biol. 1996;68:383–94.
Hirono A, Ishii A, Kere N, Fujii H, Hirono K, Miwa S. Molecular analysis of glucose-6-phosphate dehydrogenase variants in the Solomon Islands. Am J Hum Genet. 1995;56:1243–5.
Ganczakowski M, Town M, Bowden DK, Vulliamy TJ, Kaneko A, Clegg JB, et al. Multiple glucose 6-phosphate dehydrogenase-deficient variants correlate with malaria endemicity in the Vanuatu archipelago (southwestern Pacific). Am J Hum Genet. 1995;56:294–301.
Sanephonasa A, Cheepsunthorn CL, Khaminnsou N, Savonqsy O, Nuchprayoon I, Leecharoenkiat K. Molecular characterization of G6PD mutations reveals the high frequency of G6PD Aures in the Lao Theung population. Malaria J. 2021;20:30.
Wallace AR. The Malay Archipelago: The land of the Orang Utan, and the Bird of Paradise. A narrative of travel, with studies of man and nature, vol 1, Oxford: Oxford University Press; 1986, p. 478.
Omega M, Barnard RT. Phylogeny and origin of glucose-6-phosphate dehydrogenase (G6PD) deficiency mutations in Indonesia. Indonesian J Biotechnol. 2013;18:14–25.
Moodley Y, Linz B, Yamaoka Y, Winsor HM, Breurec S, Wu JY, et al. The peopling of the Pacific from a bacterial perspective. Science. 2009;323:527–30.
Gray RD, Jordan FM. Language trees support the express train sequence of Austronesian expansion. Nature. 2000;405:1052–5.
Tang TK, Huang WY, Tang CJ, Hsu M, Cheng TA, Cheng KH. Molecular basis of glucose-6-phosphate dehydrogenase (G6PD) deficiency in three Taiwan aboriginal tribes. Hum Genet. 1995;95:630–2.
Acknowledgements
We thank Ni L.P.E. Diarthini, Meliance Bria, Urip Yustin and Erlin Yustin for their kind help during this study.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
IST, FK conceived and designed this review. Both authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical approval and consent to participate
This study was approved by the Ethical Committee of Universitas Airlangga, Surabaya, Indonesia (No. 25-995/UN3.14/PPd/2013).
Consent of publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tantular, I.S., Kawamoto, F. Distribution of G6PD deficiency genotypes among Southeast Asian populations. Trop Med Health 49, 97 (2021). https://doi.org/10.1186/s41182-021-00387-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41182-021-00387-z