Abstract
Background
Japanese encephalitis (JE) is a mosquito-borne infectious disease caused by Japanese encephalitis virus (JEV). About 1–10 cases with severe central nervous system symptoms have been constantly reported every year in Japan. To clarify the mechanism of maintenance of JEV, the present study surveyed pigs for serological evidence of JEV infection and isolated JEV strains from pigs and mosquitoes in Isahaya City (Isahaya) and Goto City (Goto) in the islets of Goto in Nagasaki Prefecture from 2008 to 2014.
Results
The serological survey of pigs showed the increase of IgM sero-positivity against JEV in July or August, and it was maintained until October or November in both Isahaya and Goto every year. There were 47 JEV strains isolated in Nagasaki from 2001 to 2014 including the isolates in this study, and they belonged to genotype 1. Thirty four of the isolated strains were from pigs in Isahaya and were classified under six subclusters (1-A-1, 1-A-2, 1-A-3, 1-A-4, 1-A-5, and 1-A-9). Thirteen strains were isolated from pigs and mosquitoes in Goto and were classified into three subclusters (1-A-5 (2008); 1-A-1 (2009); and 1-A-2). In the subcluster 1-A-2, three different monophyletic subgroups, 1-A-2-2 (2010), 1-A-2-3 (2011), and 1-A-2-1 (2013, 2014), appeared in Goto.
Conclusions
These data strongly suggested that JEV appearance in Goto seems to depend on the frequent introduction of JEV from outside of the island and this pattern is different from what has been observed in subtropical islands in the East China Sea such as Okinawa and Taiwan, where the same populations of JEV (1-A-7 (1998–2008) in Okinawa; genotype 3 (until 2012) in Taiwan) have been maintained for a long period.
Similar content being viewed by others

Background
Japanese encephalitis virus (JEV) belongs to the family Flaviviridae, genus Flavivirus. It has a positive-sense single-stranded RNA genome which consists of approximately 11,000 nucleotides. The genome encodes three structural proteins and seven non-structural proteins [1]. JEV has five genotypes (1, 2, 3, 4, and 5) [2, 3] with genotype 3 being previously dominant in Japan up to 1990; however, genotype 1 took over and has become dominant until now [3–7].
Japanese encephalitis (JE) is caused by JEV infection through a mosquito bite. Most cases are asymptomatic; however, one out of 100 to 1000 case patients develops clinical symptoms such as high fever, headache, and vomiting, and some are severe central nervous system (CNS) symptoms such as convulsion, paralysis, coma, and encephalitis, which eventually lead to a fatal outcome. The mortality rate of patients with CNS symptoms is 20–40 %, and severe sequelae such as paralysis of motor function and cognitive abnormalities may remain in the patient after recovery [8, 9]. In Japan, JE has decreased drastically due to the introduction of JEV vaccination since the 1970s; however, still around 1–10 JE patients with CNS symptoms are constantly reported every year in the western part of Japan including Nagasaki Prefecture.
JEV is maintained among pigs, birds, and mosquitoes and occasionally in humans who are dead end host. Pigs are recognized as amplifier animal and the sources of JEV during their viremic phase. Culex tritaeniorhynchus and other Culex spp. are major vector mosquitoes for transmission of JEV [3, 9]. In Japan, seroconversion of pigs against JEV has been being monitored since 1960s and has been observed to occur during summer season (June to September) [10, 11]; however, the maintenance mechanism of JEV circulation during winter is not fully elucidated. JEV appears every summer season in Japan, and there are some reports that JEV circulates within Japan throughout the year [12, 13]. Nabeshima et al. [14] suggested that the sources of JEV are not only within Japan but also from continental Asia where many JE patients and fatal cases have been reported [15].
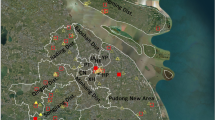
The present study aimed to clarify the ecology of JEV in Japan by two approaches: serological survey of swine sera and molecular epidemiological analysis of JEV isolates from pigs and mosquitoes collected in two observation points in Nagasaki Prefecture from 2008 to 2014. Goto City (Goto) in the islets of Goto was selected as an observation point because of its location—a small remote island in western Japan—and the presence of pig farms and wide rice paddy fields which are important factors for JEV maintenance cycle [16]. Another observation point was Isahaya City (Isahaya) located in Kyushu main island (Fig. 1). Many pig farms, wide rice paddy fields, and many wild boars exist in that area. The influence of the geographical uniqueness and environment of these two observation points for the maintenance of JEV was the major interest of this survey.
Methods
Swine serum samples
A total of 1740 swine serum samples were collected from approximately 6-month-old pigs at the slaughterhouse in Isahaya City (centered at 32° 50’ 36” N, 130° 3’ 11” E) and several pig farms in Goto City (centered at 32° 41’ 43” N, 128° 49’ 45” E, 100 km west from Nagasaki City, 420 km2 land, mean temperature is 17 °C), Nagasaki Prefecture, Japan, from 2009 to 2014 (Fig. 1). A total of 980 and 760 samples were collected, respectively, from Isahaya and Goto as a result of the collection of about 10 to 30 samples from each site every month. Collection of serum samples from pigs to be slaughtered for food consumption was done in accordance with the Guideline on the Conduct of Slaughtering Animals (Ministry of Health, Labour and Welfare) and the Guideline of Animal Welfare (Ministry of the Environment).
Mosquito samples
From 2008 to 2014, a total of 10,989 mosquitoes were collected near the pig farms in Goto by using CDC light traps. Mosquitoes were identified as to their species, sex, and whether they were engorged with blood or not. There were 8661 C. tritaeniorhynchus and Culex spp. that were collected and were put together as 474 mosquito pools (about 20 heads per pool). These mosquito pools were mixed with 2.0 mL of Eagle’s minimum essential medium (MEM), homogenized by using electric grinder, and centrifuged at 6142g for 10 min at 4 °C. The supernatants were filtered through 0.22-μm pore size membrane filters.
Virus and cell lines
A total of 47 JEV strains (Table 1) were used in the present study for phylogenetic analysis. Twenty-two strains were obtained from the JEV repository at the Nagasaki Prefectural Institute for Environmental Science and Public Health (NIEP). These were isolated from swine sera that were collected from 2001 to 2007 in Isahaya. Twenty-five strains were isolated in the present study from swine sera and mosquito samples in Isahaya and Goto. These viruses were propagated in the C6/36 mosquito cell line [17] and Vero African Green monkey epithelial kidney cell line (No. JCRB9013; Health Science Research Resources Bank). Both C6/36 cell line and Vero cell line were used for JEV isolation.
Detection of JEV-IgM
To check JEV infection among pigs, in-house JEV IgM-capture ELISA was carried out by following the protocol described by Bundo and Igarashi [18] but with modification. In the present study, anti-pig-IgM antibody (μ-chain specific; BETHYL, Montgomery, TX) was used as catching antibody, JaGAr strain of JEV as the assay antigen, and the mouse monoclonal antibody 6B6C-1 [19] as the horseradish peroxidase-conjugated anti-flavivirus antibody. The optical density (OD) was read at 492 nm by using Multiscan JX (model No.353, Thermolabsystem, Tokyo, Japan). A P/N (positive control or sample serum OD492 value/negative control serum OD492 value) ratio ≥2.0 was considered positive.
Virus isolation
Swine sera and filtrated mosquito-homogenate samples were inoculated to the monolayer of Vero cells and C6/36 cells which were kept in maintenance medium (Eagle’s MEM containing 2.0 % heat inactivate-fetal bovine serum) and incubated at 37 and 28 °C, respectively, for 7 days. The infected culture fluid (ICF) was harvested when cytopathic effects (CPE) were observed during two passages and was centrifuged at 3700 g for 10 min at 4 °C. The supernatant was then stored at −80 °C until it was used. The presence of virus was verified by reverse transcription-polymerase chain reaction (RT-PCR).
RT-PCR
RNA was extracted from ICF with QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. RT-PCR was carried out using JEV-specific sense primer (JE-NS3-1S; 5′-AGAGCGGGGAAAAAGGTCAT-3′) and anti-sense primer (JE-NS3-4R; 5′-TTTCACGCTCTTTCTACAGT-3′) [20] with SuperScript III One-Step RT-PCR System with Platinum Taq (Thermo Fisher Scientific Inc, Waltham, MA). RT-PCR was performed 1 cycle at 53 °C for 30 min and 94 °C for 2 min; 40 cycles at 94 °C for 15 s, 53 °C for 30 s, and 68 °C for 90 s; and 1 cycle at 68 °C for 5 min.
Sequencing
To complete the nucleotide sequence of the envelope (E) gene of each of the 47 JEV strains isolated in Nagasaki (Table 1), amplified PCR products were purified using QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The primer extension dideoxy chain termination method was used for direct sequencing of PCR product. For each sequencing reaction, 30–90 ng of purified PCR product was combined with 3.2 pmol of specific primer and BigDye Terminator Cycle Sequencing Ready Reaction Mixture v3.1 containing the four dye-labeled deoxynucleotide terminators (Thermo Fisher Scientific Inc, Waltham, MA). The terminal cycle sequencing parameters were used, as described by the manufacturer. The reaction mixture was column-purified (Performa DTR Gel Filtration Cartridges, Edge BioSystems, Gaithersburg, MD) and kept at 4 °C until it was loaded into the sequencer ABI Prism™ 3130 Genetic Analyzer (Thermo Fisher Scientific Inc, Waltham, MA). The nucleotide sequences of the primers used for the E gene were JE821-840F (5′-GAAAGCCACACGGTATCTCA-3′) and JE2851-2817R (5′-GCAAAGAGAATGCTTTTTCCCCATGCTTTCCAGCC-3′) [14] or JE955f (5′-TGYTGGTCGCTCCGGCTTA-3′) and JE2536r (5′-AAGATGCCACTTCCACAYCTC-3′) [21].
Phylogenetic analysis
To compare JEV population dynamics in remote islands of East Asia, we obtained 994 strains of genotype 1 JEV E coding region sequences that were available in Genbank. From the data set, we selected 48 strains from Taiwan, 7 strains from Okinawa, and 10 strains from Nagasaki. From the remaining data set, 151 strains were selected by cd-hit version 4.6.1 [22]. The reference sequence of JEV (NC_001437, Strain JaOArS982, genotype 3) and four genotype 3 strains from Nagasaki were used as out-group. We used a total of 268 strains including 47 Nagasaki strains that were sequenced in this work for phylogenetic analysis. Sequence data were aligned using the MAFFT program version 7.245 [23]. The substitution model was selected by jModelTest2 [24]. The phylogenetic tree was constructed by the maximum-likelihood method using the PhyML, version 3.0 [25]. The tree was drawn by FigTree software, version 1.4 (http://tree.bio.ed.ac.uk/software/figtree/).
DDBJ accession numbers
The DDBJ accession numbers of the JEV Nagasaki strains used in the present study for the construction of the phylogenetic tree as well as date of sample collection, sample collection area in Nagasaki, type of sample, name of strain, and subcluster of strain are listed in Table 1.
Results
Serological survey
During the 6 years (2009 to 2014) of consecutive monitoring of IgM sero-positivity against JEV among 10 to 30 pigs in Isahaya and Goto, a similar pattern was observed almost every year. IgM-positive pigs were detected either in July or August with most of the pigs becoming IgM positive between August and September, and positive detection was sustained up to October or November (Fig. 2). No IgM-positive pig could be detected during winter and spring (December to June) every year during this study. Only a slight difference was observed between Isahaya and Goto, and this was the timing of pig seroconversion which occurred 1 month earlier in Isahaya than in Goto. The seroconversion rates in both Isahaya and Goto in 2014 only reached 40–60 % which was unlike the previous years where the rates were in the range of 80–100 %.
Virus isolation
During the span of 7 years, 16 strains of JEV were successfully isolated from pigs. Twelve strains were isolated from out of 980 swine serum samples collected in Isahaya, and four strains were from out of 760 swine serum samples in Goto (Table 1). Nine strains of JEV were isolated from out of 474 mosquito pools (C. tritaeniorhynchus and other Culex spp.) collected in Goto from 2008 to 2014 (Table 1).
Phylogenetic analysis
Twenty-five strains of JEV isolated in the present study and 22 strains obtained from the JEV repository at the NIEP were analyzed for their E gene sequences (1500 nucleotides). All the 47 strains of JEV Nagasaki isolates belonged to genotype 1. JEV genotype 1 isolates analyzed in the present study were classified into nine subclusters (1-A-1 to 1-A-9, Fig. 3) by following the terminology of Nabeshima et al. [26]. Thirty-four strains isolated from Isahaya were classified into six subclusters which were 1-A-1, 1-A-2, 1-A-3, 1-A-4, 1-A-5, and 1-A-9 (Figs. 3 and 4a, b); and 13 strains isolated from Goto were classified into three subclusters which were 1-A-1, 1-A-2, and 1-A-5. In Goto, subclusters 1-A-1 and 1-A-5 were seen in 2009 and 2008 respectively, and subcluster 1-A-2 has been detected since 2010. The subcluster 1-A-2 contains some monophyletic subgroups. The subgroup 1-A-2-2 appeared in 2010, 1-A-2-3 in 2011, and 1-A-2-1 in 2013 and 2014 (Fig. 3).
Phylogenetic analysis of the E gene of Japanese encephalitis virus. The phylogenetic tree was constructed by the maximum-likelihood method using the PhyML ver. 3.0 Software. Bootstrap analysis was replicated 1000 times, and bootstrap value (%) greater than 50 % are shown above branches. Genotype 1 JEVs (268 strains) are shown in this figure. Other genotype, genotype 3 (five strains) was used as out-group. Labels of strains conform to the following format: (GenBank or DDBJ accession no.)_(strain name)_(country-region)_(year of isolation). Thirty-four Isahaya strains, 13 Goto strains, 48 Taiwan strains, and 7 Okinawa strains are indicated as red, blue, green, and purple color, respectively. JEV strains from China and other areas strains are indicated in black
a Locations and isolation years of JEVs. Geographical locations of genotype 1 JEVs isolated in Isahaya, Goto, Okinawa, Taiwan, and China are shown in red, blue, green, purple, and black color, respectively, with subcluster name and year of isolation. b Geographical locations of the seven subclusters of JEV genotype 1. Seven subclusters of JEV genotype 1 isolated in Isahaya, Goto, Okinawa, Taiwan, and China are shown in red, blue, green, purple, and black color, respectively, with year of isolation
Discussion
In the present study, the results of monthly monitoring of antibody positivity against JEV for six consecutive years from 2009 to 2014 in Isahaya and Goto in Nagasaki Prefecture supported the previous report by the NIEP on pigs sero-surveillance about the pig becoming seropositive during summer season in these areas between July to September (particularly, in the end of July to the beginning of August) [27]. However, their report lacked data on the other three seasons: autumn, winter, and spring (October to June). Thus, the present study clarified pig sero-positivity trend against JEV in all four seasons. During these 6 years, no JEV seropositive pigs were found in both Isahaya and Goto during winter and spring. It suggests that either no transmission of JEV between vector mosquitoes and amplifier animals occurred in these two observation points or it occurred as a very minor incidence which could not be detected by our sampling scale (10~30 heads of pigs per month). The reason of low seroconversion rate in 2014 could be due to unusual climate condition that caused the reduction of sunshine duration to half and the increase in the volume of precipitation twice in August in comparison to the same month in the other years [28].
The genotype shift of JEV from genotype 3 to genotype 1 in Japan and surrounding area was reported before [4, 5, 29, 30], and the current dominant genotype is still genotype 1 [7]. We classified the genotype 1 into nine subclusters (1-A-1 to 1-A-9, Fig. 3). The 47 strains isolated in this study were distributed in six subclusters (1-A-1, 1-A-2, 1-A-3, 1-A-4, 1-A-5, and 1-A-9).
In case of Isahaya, the JEV isolates showed variety of subclusters (1-A-1, 1-A-2, 1-A-3, 1-A-4, 1-A-5, and 1-A-9); furthermore, multiple subclusters of JEV appeared in 2001, 2003, 2004, and 2010. Obara et al. [31] also reported the occasional introduction of minor subcluster "C" (1-A-5 in the present study) in 2007 and the maintenance of major subcluster "A" (1-A-1 in the present study) of JEV in Toyama Prefecture (centered at 36° 41’ 45” N, 137° 12’ 49” E, central north of Japan) from 2005 to 2009 and this pattern was similar to the pattern in Isahaya in the present study.
In Goto, three subclusters were isolated during 7 years (2008 to 2014) in the present study. The subcluster 1-A-5 appeared in 2008, and 1-A-1 appeared in 2009. Since 2010, JEV strains belonging to the subcluster 1-A-2 have been isolated from Goto. In the subcluster 1-A-2, three subgroups, 1-A-2-1, 1-A-2-2, and 1-A-2-3, were distributed not only in Goto but also in China, South Korea, and a main island (Kyushu) of Japan, and each subgroup appeared in Goto in different years (Fig. 3). Replacement of subclusters and subgroups occurred both in Goto and Isahaya, but the subclusters and subgroups that appeared in Isahaya were different from Goto based on the same year of observation. It implies that these subclusters and subgroups were introduced independently to each area, Goto and Isahaya, in 2009 (Isahaya; 1-A-5, Goto; 1-A-1), in 2013 (Isahaya; 1-A-5, Goto; 1-A-2), and in 2011 (Isahaya; 1-A-2-1, Goto; 1-A-2-3).
Goto, Okinawa, and Taiwan are islands in East China Sea. However, in Taiwan, genotype shift from genotype 3 to 1 occurred between 2008 and 2012 [29]. In Okinawa, until 2008, only 1-A-7 JEV strains had been isolated, but in 2012, another subcluster (1-A-5) appeared in Okinawa. These data implied that, in Taiwan and Okinawa, subcluster seemed stable compared with Goto. This could be explained by the mechanism of maintenance of JEV within each area due to the long distance from the mainland China and by the large population of pigs, wide area of rice paddy fields, and subtropical climate in Taiwan and Okinawa as compared to Goto.
It is worthy to note that more frequent replacement of subclusters seems to be occurring in Goto, compared to Taiwan and Okinawa. Nabeshima et al. [14, 26] has proposed the hypothesis for the mechanism of introduction of new subclusters of JEV into Japan from continental Asia by periodical mosquito flying along with the westerlies and overwintering of JEV somewhere in Japan [12, 13]. Otuka [32] reported that planthoppers, Laodelphax striatellus, migrate by westerly winds from east China to Japan along the frontal zone, called Bai-u in the Far East. If JEV is also introduced by migration of mosquitoes, it is possible that more frequent introduction occurs in the area around Goto than in Taiwan or Okinawa.
Remote islands provide better setting to investigate viral ecology because migration of terrestrial organisms is restricted and the size of population is small. We expect that chronological surveillance in Goto which had frequent replacement of viral population in contrast to that from subtropical islands such as Okinawa and Taiwan will provide important data to reveal how new arbovirus population migrate, invade, and settle in newly arrived area.
Conclusions
In conclusion, JEV was being introduced in Goto from outside of the island. The transition patterns of JEV populations in Goto suggested that there were frequent introductions of JEV in Goto island in comparison to Taiwan and Okinawa (subtropical islands in the East China Sea) where the same populations of JEV were being kept for a long time.
References
Sumiyoshi H, Mori C, Fuke I, Morita K, Kuhara S, Kondou J, Kikuchi Y, Nagamatu H and Igarashi A. Complete nucleotide sequence of the Japanese encephalitis virus genome RNA. Virology. 1987;161:497–510.
Uchil PD, Satchidanandam V. Phylogenetic analysis of Japanese encephalitis virus: envelope gene based analysis reveals a fifth genotype, geographic clustering, and multiple introductions of the virus into the Indian subcontinent. Am J Trop Med Hyg. 2001;65:242–51.
Morita K, Nabeshima T, Buerano CC. Japanese encephalitis. Rev Sci Tech. 2015;34:441–52.
Ma SP, Yoshida Y, Makino Y, Tadano M, Ono T, Ogawa M. Short report: a major genotype of Japanese encephalitis virus currently circulating in Japan. Am J Trop Med Hyg. 2003;69:151–4.
Nga PT, del Carmen PM, Cuong VD, Ma SP, Hasebe F, Inoue S, Makino Y, Takagi M, Nam VS and Morita K. Shift in Japanese encephalitis virus (JEV) genotype circulating in northern Vietnam: implications for frequent introductions of JEV from southeast Asia to east Asia. J Gen Virol. 2004;85:1625–31.
Takegami T, Ishak H, Miyamoto C, Shirai Y, Kamimura K. Isolation and molecular comparison of Japanese encephalitis virus in Ishikawa, Japan. Jpn J Infect Dis. 2000;53:178–9.
Pan XL, Liu H, Wang HY, Fu SH, Liu HZ, Zhang HL, Li MH, Gao XY, Wang JL, Sun XH, et al. Emergence of genotype I of Japanese encephalitis virus as the dominant genotype in Asia. J Virol. 2011;85:9847–53.
Solomon T, Dung NM, Kneen R, Thao LTT, Gainsborough M, Nisalak A, Day NPJ, Kirkham FJ, Vaughn DW, Smith S and White NJ. Seizures and raised intracranial pressure in Vietnamese with Japanese encephalitis. Brain. 2002;125:1084–93.
Gubler DJ, Kuno G, Markoff L. Flaviviruses. In: Knipe DM, Howley P, eds. Fields Virology. 5th ed. Philadelphia, PA: Lippincott Williams and Wilkins; 2007. p. 1153–252.
National Institute of Infectious Diseases: National Epidemiological Surveillance of Vaccine-Preventable Diseases (NESVPD). http://www.nih.go.jp/niid/ja/y-graphs/1600-yosoku-index-e.html (2015). Accessed 30 Nov 2015.
Infectious Disease Surveillance Center, National Institute of Infectious Diseases. Annual Report 2012 National Epidemiological Surveillance of Vaccine-Preventable Diseases [in Japanese]. Tokyo: National Institute of Infectious Diseases; 2015. p. 123–49.
Takashima I, Watanabe T, Ouchi N, Hashimoto N. Ecological studies of Japanese encephalitis virus in Hokkaido: interepidemic outbreaks of swine abortion and evidence for the virus to overwinter locally. Am J Trop Med Hyg. 1988;38:420–7.
Takasaki T, Kotaki T, Kurane I, Sawabe K, Hayashi T, Kobayashi M. Monthly report on findings of infection agents in Japan [in Japanese]. IASR. 2009;30(6):156–7.
Nabeshima T, Loan HT, Inoue S, Sumiyoshi M, Haruta Y, Nga PT, Huoung VT, del Carmen Parquet M, Hasebe F and Morita K. Evidence of frequent introductions of Japanese encephalitis virus from south-east Asia and continental east Asia to Japan. J Gen Virol. 2009;90:827–32.
World Health Organization (WHO). Fact sheet No. 386. In: Japanese encephalitis. World Health Organization (WHO). 2015. http://www.who.int/mediacentre/factsheets/fs386/en/. Accessed 10 Jan 2016.
Goto City Office. Goto City Profile Book: 2015 siseiyouran [in Japanese]. Goto: Goto City Office; 2015.
Igarashi A. Isolation of a Singh’s Aedes albopictus cell clone sensitive to Dengue and Chikungunya viruses. J Gen Virol. 1978;40:531–44.
Bundo K, Igarashi A. Antibody-capture ELISA for detection of immunoglobulin M antibodies in sera from Japanese encephalitis and dengue hemorrhagic fever patients. J Virol Methods. 1985;11(1):15–22.
Roehrig JT, Mathews JG, Trent DW. Identification of epitopes on the E glycoprotein of Saint Louis encephalitis virus using monoclonal antibodies. Virology. 1983;128(1):118–26.
Tanaka M. Rapid identification of flavivirus using the polymerase chain reaction. J Virol Methods. 1993;41(3):311–22.
Nerome R, Tajima S, Takasaki T, Yoshida T, Kotaki A, Lim CK, et al. Molecular epidemiological analyses of Japanese encephalitis virus isolates from swine in Japan from 2002 to 2004. J Gen Virol. 2007;88:2762–8.
Li W, Godzik A. Cd-hit: a fast program for clustering and computing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22(13):1658–9.
Katoh K, Standley DM. MAFFT Multiple Sequence Alignment Software Version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–80.
Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more model, new heuristics and parallel computing. Nat Methods. 2012;9(8):772–5.
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel W. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–21.
Nabeshima T, Morita K. Phylogeographic analysis of migration of Japanese encephalitis virus in Asia. Future Virol. 2010;5(3):343–54.
Nagasaki Prefectural Institute for Environmental Research and Public Health: Nagasaki Prefectural Government [in Japanese]. http://www.pref.nagasaki.jp/bunrui/kenseijoho/koho/info/shoho/(2015). Accessed 30 Nov 2015.
Japan Meteorological Agency: Kako no kisyo data kensaku [in Japanese]. http://www.data.jma.go.jp/obd/stats/etrn/view/monthly_s3.php?prec_no=84&block_no=47843&year=&month=&day=&view=p5. Accessed 10 Jan 2016.
Su CH, Yang CF, Teng HJ, Lu LC, Lin C, Tsai KH, Chen YY, Chen LY, Chang SF and Shu PY. Molecular epidemiology of Japanese encephalitis virus in mosquitoes in Taiwan during 2005–2012. Plos Negl Trop Dis. 2014;8(10), e3122.
Saito M, Taira K, Itokazu K, Mori N. Recent change of the antigenicity and genotype of Japanese encephalitis virus distributed on Okinawa Island. Am J Trop Med Hyg. 2007;77:737–46.
Obara M, Yamaguchi T, Watanabe M, Hasegawa S, Ueda Y, Matsuno K, Iwai M, Horimoto E, Kurata T, Takizawa H and Takashima I. Continuity and change of Japanese encephalitis virus in Toyama Prefecture, Japan. Am J Trop Med Hyg. 2011;84:695–708.
Otuka A. Migration of rice planthoppers and their vectored re-emerging and novel rice virus in East Asia. Front Microbiol. 2013;4:309.
Acknowledgements
This work was supported financially by KAKENHI [Grant-in-Aid for Scientific Research (B) (25305015) and Grant-in-Aid for Young Scientists (B) (25870524)] from the Japan Society for the Promotion of Science, a Health and Labor Sciences Research Grant on Emerging and Re-emerging Infectious Diseases from the Japanese Ministry of Health, Labor and Welfare; Health and Labor Sciences Research Grants from J-GRID, AMED, JST, and the U.G.-Japan Cooperative Medical Sciences Program; and the research fund from the NIEP. We thank Yukiko Higa, Toshihiko Sunahara, Kyoko Futami, and Nozomi Imanishi (Department of Vector Ecology and Environment, Institute of Tropical Medicine, Nagasaki University); Yoshihide Maekawa (Department of Medical Entomology, National Institute of Infectious Diseases); and Kenta Okamoto, Yuki Takamatsu, and Leo Uchida (Department of Virology, Institute of Tropical Medicine, Nagasaki University) for the collection of mosquito samples used in this study and Yoshihiro Kato, Kunihiro Mizota, Shinya Nakamura, and Katsuhiko Yamaguchi (Nagasaki Prefectural Isahaya Meat Inspection Center, Japan) and Fujio Ueyama and Hisashi Fuchigami (Sasebo City Meat Inspection Center, Japan) for the collection of swine serum samples used in this study. We also thank Yoshiko Saito for her technical assistance. We also thank the contribution of Dr. Corazon C. Buerano for the improvement of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
AY, MA, and KM designed the study. AY, TN, SI, and MA collected the samples. AY and TN performed the experiments. AY, TN, SI, and MA analyzed the data. AY wrote the manuscript. All authors read and approved the final manuscript.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Yoshikawa, A., Nabeshima, T., Inoue, S. et al. Molecular and serological epidemiology of Japanese encephalitis virus (JEV) in a remote island of western Japan: an implication of JEV migration over the East China Sea. Trop Med Health 44, 8 (2016). https://doi.org/10.1186/s41182-016-0010-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41182-016-0010-0