Abstract
Background
The grasshopper family Lentulidae is endemic to eastern and southern Africa, with its center of diversity situated in South Africa, the highest diversity being found in the Cape Floristic Region, which is one of the global biodiversity hotspots. The family consists of 35 genera sorted in two subfamilies. This study provides first insights into the phylogeny of Lentulidae. Two mitochondrial genes (12S and NDS) were sequenced and the phylogeny was inferred through Maximum Likelihood and Bayesian Inference.
Results
Our results indicate that the current classification into the subfamilies Lentulinae and Shelforditinae may be incorrect as Uvarovidium, Leatettix (Shelforditinae) and Devylderia (Lentulinae) clustered together in one main clade, while Betiscoides, Basutacris and Gymnidium (all Lentulinae) formed the second main clade. The genera Uvarovidium and Leatettix, which had been assigned to the Acrididae (subfamily Hemiacridinae) in the past, grouped within the Lentulidae, confirming their current assignment to this family. The East African Usambilla group is likely to represent a sister clade to the south African Lentula and Eremidium. Diversification patterns in the genus Devylderia and Betiscoides suggest a higher number of species than currently known.
Conclusions
Our phylogeny is not in line with the current systematics of Lentulidae, suggesting that a broader sampling and a study of the genitalia would be useful to clarify the taxonomy. Furthermore, some genera (particularly Betiscoides and Devylderia) are in need of taxonomic revision, as the number of species within these genera is likely to be higher than the current taxonomy suggests.
Similar content being viewed by others
Background
The Cape Floristic Region in South Africa is one of the global biodiversity hotspots [1]. Numerous studies have dealt with the enormous plant diversity and its origin and a number of phylogenetic studies on certain plant families aimed at unravelling the drivers for diversification in this region [2–4]. The fauna of the Cape Floristic Region, though not less unique, has not been investigated as intensely as the flora, especially research on invertebrate diversity remains still scarce [5]. Recent evidence suggests that invertebrate diversity and endemism in the Cape Floristic Region might be comparable to the pattern found in plants [5, 6]. The Cape Floristic Region also maintains a variety of endemic families, subfamilies and genera, suggesting that it provided refugia also over a longer time-span. One of these subfamilies endemic to the Cape Floristic Region is the subfamily Shelforditinae Ritchie, 1982 within the family Lentulidae Dirsh, 1956. Within the family Lentulidae as a whole, the majority of taxa (70 %) are also endemic to South Africa [7], but some genera occur in East Africa, particularly the Usambilla group, which shows a radiation in the East African mountain systems [8].
All Lentulidae are completely wingless, and therefore expected to show high levels of genetic differentiation at a small geographic scale, as they have a low mobility and are often adapted to certain vegetation structures. For example, the Lentulidae genus Betiscoides Sjöstedt, 1923 is adapted to Restionaceae, which also show a high level of differentiation in the Cape Floristic Region [9]. Studies on the phylogenetics of grasshoppers from the Cape Floristic Region are lacking so far. However, several recent taxonomic studies suggest a high level of differentiation of flightless grasshopper species in the Cape Floristic Region with the identification of tens of hitherto undescribed species [10, 11]. Given the enormous number of unidentified species in times of increasing efforts to halt the loss of biodiversity, there have been approaches to record and assess biodiversity parameters also on higher taxonomic level [12]. Thus, it is of increasing interest to investigate the systematic relationships of endemic taxa to understand the importance of biodiversity hotspots.
The aim of our study was to investigate and reconstruct the phylogenetic relationships among genera within the family Lentulidae. We were particularly interested in the systematic position of the genera Leatettix Dirsh, 1956 and Uvarovidium Dirsh, 1956, which are currently assigned to the subfamily Shelforditinae. Furthermore, we aimed to clarify the phylogenetic relationships of the East African Usambilla group (here represented by the genera Usambilla Sjöstedt, 1910 and Rhainopomma Jago, 1981) with respect to the South African Lentulidae. We present the results of a first phylogenetic study on Lentulidae based upon two mitochondrial genes (12S rRNA and NDS – a fragment containing parts of the 16S rRNA, t-Leu and ND1), covering ten genera from both subfamilies, in order to provide new insights in evolutionary history of this family.
Results
The complete alignment contained 808–823 bp. The 12S fragment was between 318 and 327 bp (when including the outgroups between 316 and 327 bp) and NDS (consisting of a fragment of 16S, t-Leu and ND1) between 490 and 497 bp long (with outgroup: 487–497). NDS had more variable sites (254) compared to 12S with 178 variable sites (see Table 1), but the percentage of variable sites was slightly larger in 12S (54 % vs. 51 %). The highest genetic distance (p-Distance for the 12S data set) between genera within Lentulidae was detected between Usambilla and Uvarovidium with p = 0.224, whereas the lowest was found between Basutacris and Betiscoides (p = 0.057, Table 2).
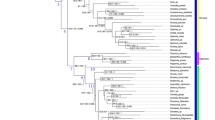
The phylogenetic reconstruction showed similar results for both methods, i.e. Maximum Likelihood and Bayesian Inference. Thus, only the results derived from the Bayesian Inference are presented and discussed here in detail. The Maximum Likelihood tree is provided in Additional file 1: Figure S1. The consensus tree from the Bayesian analysis showed a monophyly of the Lentulidae with respect to the chosen outgroups Sphingonotus rubescens and Frontifissia elegans and F. laevata (Fig. 1). Within the family Lentulidae, a clear basal split between two main clades was revealed, which was supported by a BPP value of 0.98. The first main clade consisted of the genera Gymnidium, Basutacris, Betiscoides, Lentula, Eremidium, Rhainopomma and Usambilla. Specimens collected in the same region clustered together, e.g. Table Mountain (Betiscoides), Hottentots Holland (Betiscoides), Jonaskop (Betiscoides), Baviaanskloof (Betiscoides), and Kogelberg (Gymnidium). The East African Usambilla group (genera Usambilla and Rhainopomma) formed a monophyletic group, related to the South African genera Eremidium and Lentula.
The second main clade, consisting of the genera Devylderia, Uvarovidium and Leatettix, revealed a rather distinct pattern regarding their geographical distribution. Leatettix specimens from Cederberg clustered in a single clade, strongly separated from the clades in Swartberg and Baviaanskloof, as a sister clade to Uvarovidium. However, this relationship was supported by a BPP value of only 0.84. Devylderia formed four subclades, one including individuals from Jonaskop, one from Cederberg and Groot Winterhoek, one represented in Jonaskop, Table Mountain, Limietberg, Hottentots Holland, Groot Winterhoek and De Hoop, and the fourth subclade included only one specimen from Cederberg.
Discussion
The results provide first insights into the phylogeny of the family Lentulidae. The clear split between the two main clades was highly supported. However, this split does not reflect the current subdivision into the two subfamilies Lentulinae and Shelforditinae. Even though the two Shelforditinae genera Leatettix and Uvarovidium clustered as a monophylum, they showed a clear sister group relationship to the genus Devylderia (Lentulinae). This could mean that either Devylderia belongs to Shelforditinae as well or that the current taxonomy within the complete family needs to be revised. A broader taxon sampling across all described lentulid genera would provide more precise information on the systematic relationships of the genera and clarify the assignments to the respective subfamilies. The East African Usambilla group is likely to represent a sister clade to Lentula and Eremidium, confirming their systematic position within the subfamily Lentulinae. This also suggests that the Lentulidae as a whole originated in South Africa and started to diversify here, before one group (the ancestor of the Usambilla group) spread to the north to radiate within the East African Mountain systems.
Although our sampling only covers a small fraction of the Lentulidae genera, a strong genetic differentiation between and within genera becomes already evident. In some cases, our phylogeny does not reflect the current taxonomy. This is the case for the genus Leatettix, which does not represent a monophylum, but a paraphyletic group. Otte [11] mentioned in his revision of Leatettix that further division of Leatettix species into different genera might be necessary. As Leatettix emota from Cederberg are closer related to U. peninsulare than to Leatettix moraki from Swartberg and Baviaanskloof, his suggestion seems to be supported by our findings. Further morphological and genetic research is required to clarify these findings.
Recent revisions of several lentulid genera from South Africa showed that the number of Lentulidae species is much higher than currently known. In total, 44 new species and even three new genera have recently been described (Armstrongium, Tanquata, Tsautettix, [13]). The center of species richness of Lentulidae is in South Africa with 103 of the 146 described species occurring in the Cape Floristic Region [7]. Recent studies show that insect species diversity in the Cape Floristic Region is generally much higher than current taxonomy suggests [10, 14]. This seems to be particularly true for the genus Betiscoides (Matenaar et al. unpubl.), which is also confirmed in our study. Although this genus represents a monophylum, specimens from different localities showed a high genetic differentiation. All specimens used in our study would morphologically be assigned to Betiscoides meridionalis. However, the results indicate four clades of this species with high genetic differentiation, suggesting the existence of cryptic species (which is confirmed by first morphological inspection).
The reasons for the unique diversity in the Cape Floristic Region and potential drivers of differentiation and speciation have been discussed before [2, 3, 15]. Concerning insect diversity, there is a consensus about the long-term isolation of populations starting in the early Miocene [16–18]. The Cape Floristic Region experienced climatic changes during late Mio- and Pliocene and repeated orographic changes through the uplift of the Cape Fold Belt as well as oceanic regression. These changing environmental conditions probably triggered and influenced dispersal as well as survival of insect taxa in refugia. Coastal regions and mountains of the Cape Floristic Region probably functioned as refugia during unfavorable periods, whereas ocean regression repeatedly enabled taxa to disperse into lowland habitats and the interior of the Cape Floristic Region [19]. Climatic stability in mountain ranges throughout the Pleistocene is believed to have had positive effects on diversity as it supported the origin of new species while keeping extinction rates low [15, 18]. As a result, species were able to persist in montane or coastal refugia. The lack of gene flow caused by low dispersal capabilities led to high genetic differentiation within genera. As all Lentulidae are flightless, they are likely to have limited dispersal capabilities, as has been shown in the genus Betiscoides [9]. Consequently, this might also explain diversity patterns within the Lentulidae in general, even though the initial splits into several main clades or genera probably occurred much earlier. Several palaeorelictual insects of the Cape Floristic Region are known [5], suggesting that parts of the Cape Floristic Region may have served as refugia for quite a long time. Some represent ancient Gondwanan lineages. Some Lentulidae genera (Betisocoides, Devylderia) seem to be adapted to plants of typical fynbos plants, which radiated 70 my ago. The phylogeny of Orthoptera indicates that Lentulidae started radiating during that period as well [20]. It is thus unlikely that Lentulidae include ancient Gondwanan genera despite their high endemicity.
Conclusions
Our results provide first insights in the phylogeny of the grasshopper family Lentulidae, indicating that the current subdivision into two subfamilies needs to be revised and that the East African Usambilla group represents a sister clade to the genera Lentula and Eremidium. Furthermore, species diversity within the genera seems to be higher than the current taxonomy suggests. Further research is needed including a broader taxon sampling and sequencing of additional genes.
Methods
Study objects
Members of the family Lentulidae are flightless and lack tympana. Further morphological features, such as shape of head or antenna, vary within this family. Lentulidae occur in South and East Africa with the center of diversity situated in South Africa. In total, 70 % of the Lentulidae are endemic to South Africa. The family is grouped into two subfamilies Lentulinae and Shelforditinae, the latter one only occurring in South Africa. Lentulinae currently comprise 88 species within 25 genera, Shelforditinae consist of ten genera with 44 species. Genera such as Leatettix and Uvarovidium had been classified as Hemiacridinae (family Acrididae) for some time, but were assigned to the Shelforditinae by Ritchie (1982). The East African Usambilla group belongs to the subfamily Lentulinae.
Sampling
A total of 50 Lentulidae specimens from six genera (Basutacris, Betiscoides, Devylderia, Gymnidium, Leatettix and Uvarovidium) were collected during four field trips from February 2012 to December 2013. Most specimens were collected in the eight reserves forming the UNESCO World Heritage Site “Cape Floral Region Protected Areas” and some in a private nature reserve “Jonaskop”. If possible, 3–5 specimens of each species were collected at each locality. Specimens were caught by sweep net or by hand and killed in a freezer. Afterwards they were stored in 99 % ethanol p.a. or dried and pinned. Information on specimens and localities and GenBank reference numbers is given in Fig. 2 and Table 3. Specimens are stored at the State Museum of Natural History in Stuttgart. Sphingonotus rubescens (Acrididae: Oedipodinae), Frontifissia elegans and F. laevata (Acrididae; Catantopinae) were chosen as outgroups.
Sequences for 12S from the genera Eremidium nr equuleus (AY569277.1), Lentula callani (NC_020774.1) Lentula obtusa (AY569276.1), Usambilla sagonai (AY569279.1) and Rhainopomma montanum (Z97601.1) were obtained from Genbank for an extended data set for 12S. The data sets for 12S and NDS were concatenated, data for NDS was coded missing when sequences were not available (Table 3).
DNA analysis
DNA was isolated from the insects’ hind leg muscles using the Qiagen DNeasy Blood and Tissue Kit (Qiagen GmbH, Hilden, Germany). We amplified two mitochondrial gene fragments: 12S rRNA and the gene fragment NDS (a combination of 16S rRNA, t-Leu and NADH-Dehydrogenase subunit 1 (ND1); see Table 4 for primer sequences). The gene fragment 12S was chosen as it typically amplifies well also in old samples (i.e. the pinned specimens used), whereas NDS was used because it is more variable and typically shows a better resolution than 12S. Two different polymerases were used for PCR reactions (HotMasterMix by 5Prime and HotStarTaq Master Mix by Qiagen). The reactions for both polymerases were compiled as follows: 26 μl of diH2O, 20 μl of HotStarMasterMix or HotStarTaq Master Mix respectively, 0.3 pmol of each primer and 50 ng of DNA template (total Vol. = 50 μl). HotStarMasterMix was used for amplifying 12S under the following conditions: 94 °C for 2 min., 37 Cycles of denaturation at 94 °C for 30 s., annealing at 45 °C for 30 s and elongation at 65 °C for 1 min and a final elongation step for 10 min at 65 °C. NDS was amplified with the HotStarTaq Master Mix under the following conditions: Initial denaturation at 96 °C for 20 min., 33 cycles of denaturation at 96 °C, annealing at 45 °C for and elongation at 68 °C for 1.30 min each following a final elongation step at 65 °C for 3 min. PCR products were visualized on an 1 % agarose gel stained with SYBR Green I (Biozym, Hessisch, Oldendorf, Germany). The products were purified using Roche High Pure PCR Product Purification Kit (Roche Deutschland Holding GmbH, Germany) and sequenced afterwards at MacroGen Cooperation (Amsterdam, The Netherlands) or on a MEGABACE 1000 automated sequencer at the University of Trier with the DYEnamic ET Terminator Cycle Sequencing Premixkit (GE Healthcare, Munich, Germany).
Sequence analysis
Sequences were inspected in MEGA 6.0 [21] and aligned using ClustalW. The Gap opening penalty was set to 15 for pairwise and multiple alignments respectively and the gap extension penalty was set to 6.66 for both pairwise and multiple alignment. IUB was chosen as DNA Weight Matrix with a transition weight of 0.5. Delay Divergent cutoff was set to 30 %. The genetic distance (p-Distance) between the genera was calculated only for the 12S sequences, as NDS was missing in GenBank for the genera Lentula, Eremidium, Rhainopomma and Usambilla. For tree reconstruction with Bayesian Inference we first used PartitionFinder in order to detect possible partitions in the data set and to find the best-fitting substitution models applicable in MrBayes [22]. Subsets were defined before Partitionfinder was run by dividing NDS in the components 16S (including t-Leu) and ND1 as well as the coding gene ND1 into its coding positions. In total, five different partitions with different substitution models were calculated. The model GTR + I + G was the best-fitting model for 12S, 16S and first coding position of ND1; HKY + I was calculated for the second coding position of ND1 and HKY + G for the third coding position of ND1.
We performed a Bayesian analyses on the datasets using MrBayes v.3.1.2 [23, 24]. The analysis was run for 20 million generations, sampling trees every 2000 generations. The first 2500 trees were discarded before a consensus tree was calculated and visualized in FIGTREE v. 1.4.2 [25].
The Maximum Likelihood tree reconstruction was run in MEGA 6.0. In addition to PartitionFinder, we applied the goodness-of-fit test for each nucleotide model in order to find the best substitution model applicable in MEGA [26]. Based on the BIC (Bayesian Information Criterion) and AICc (corrected Akaike Information Criterion), we chose the best-fitting models. The best fitting model for the Maximum likelihood analysis was GTR + G (BIC: 6153.091). The Bootstrap method was chosen as phylogenetic test and the number of replications of bootstraps was set to 500. Gaps or missing data were treated with partial deletion with a cutoff of 95 %. The heuristic method was set to Nearest Neighbor Interchange with the first tree being calculated automatically. The Branch swap filter was set to very strong.
References
Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GA, Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;403:853–8. doi:10.1038/35002501.
Goldblatt P, Savolainen V, Porteous O, Sostaric I, Powell M, Reeves G, et al. Radiation in the Cape flora and the phylogeny of peacock irises Moraea (Iridaceae) based on four plastid DNA regions. Mol Phylogenet Evol. 2002;25:341–60. doi:10.1016/S1055-7903(02)00235-X.
Linder HP. The radiation of the Cape flora, southern Africa. Biol Rev. 2003;78:597–638. doi:10.1017/S1464793103006171.
Richardson JE, Weitz FM, Fay MF, Cronk QC, Linder HP, Reeves G, et al. Rapid and recent origin of species richness in the Cape flora of South Africa. Nature. 2001;412:181–3. doi:10.1038/35084067.
Colville JF, Potts AJ, Bradshaw PL, Measey GJ, Snijman D, Picker MD, et al. Floristic and faunal Cape biochoria: do they exist? In: Allsopp N, Colville JF, Verboom GA, editors. Fynbos: Ecology, evolution, and conservation of a megadiverse region. Oxford, New York: Oxford University Press; 2014. p. 73–92.
Matenaar D, Bazelet CS, Hochkirch A. Simple tools for the evaluation of protected areas for the conservation of grasshoppers. Biol Conserv. 2015;192:192–9. doi:10.1016/j.biocon.2015.09.023.
Eades DC, Otte D, Cigliano MM, Braun H. Orthoptera Species File. Version 5.0/5.0; 2015.
Jago ND. A revision of the genus Usambilla Sjostedt (Orthoptera: Acridoidea) and its allies. Bulletin of the British Museum. Natural History. Entomology. 1981;43:1–38.
Matenaar D, Bröder L, Bazelet CS, Hochkirch A. Persisting in a windy habitat: population ecology and behavioral adaptations of two endemic grasshopper species in the Cape region (South Africa). J Insect Conserv. 2014;18:447–56. doi:10.1007/s10841-014-9654-4.
Spearman LA. Taxonomic Revision of the South African Grasshopper Genus Euloryma (Orthoptera: Acrididae). 2013;139:1–111. doi:10.3157/061.139.0101.
Otte D. Twenty-Six New Species of Grasshoppers from the Southern African Arid Zone (Orthoptera Acridoidea:Lentulidae). Trans Am Entomol Soc. 2014;140:293–347. doi:10.3157/061.140.0116.
Rach J, Desalle R, Sarkar IN, Schierwater B, Hadrys H. Character-based DNA barcoding allows discrimination of genera, species and populations in Odonata. Proc Biol Sci. 2008;275:237–47. doi:10.1098/rspb.2007.1290.
Brown HD. Armstrongium - an unusual new sylvan genus of Lentulidae (Orthoptera: Acridoidea) from KwaZulu-Natal Province, South Africa. Annals of the Ditsong National Museum of Natural History. 2014;49:33–50.
Switala AK, Sole CL, Scholtz CH. Phylogeny, historical biogeography and divergence time estimates of the genus Colophon Gray (Coleoptera: Lucanidae). Invert Systematics. 2014;28:326. doi:10.1071/IS13054.
Ellis AG, Anthony Verboom G, van der Niet T, Johnson SD, Peter LH. Speciation and extinction in the Greater Cape Floristic Region. In: Allsopp N, Colville JF, Verboom GA, editors. Fynbos: Ecology, evolution, and conservation of a megadiverse region. Oxford, New York: Oxford University Press; 2014. p. 119–41. doi:10.1093/acprof:oso/9780199679584.003.0006.
Daniels SR, Picker MD, Cowlin RM, Hammer ML. Unravelling evolutionary lineages among South African velvet worms (Onychophora: Peripatopsis) provides evidence for widespread cryptic speciation. Biol J Linn Soc. 2009;97:200–16. doi:10.1111/j.1095-8312.2009.01205.x.
de Jager ML, Ellis AG. The influence of pollinator phylogeography and mate preference on floral divergence in a sexually deceptive daisy. Evolution. 2013;67:1706–14. doi:10.1111/evo.12070.
Tolley K, Bowie R, Measey GJ, Price BJ, Forest F. The shifting landscapes of genes since the Pliocene: terrestrial phylogeography in the Greater Cape Floristic Region. In: Allsopp N, Colville JF, Verboom GA, editors. Fynbos: Ecology, evolution, and conservation of a megadiverse region. Oxford, New York: Oxford University Press; 2014. p. 142–63.
Allsopp N, Colville JF, Verboom GA, editors. Fynbos: Ecology, evolution, and conservation of a megadiverse region. Oxford, New York: Oxford University Press; 2014.
Song H, Amédégnato C, Cigliano MM, Desutter-Grandcolas L, Heads SW, Huang Y, et al. 300 million years of diversification: Elucidating the patterns of orthopteran evolution based on comprehensive taxon and gene sampling. Cladistics. 2015:1–31. doi:10.1111/cla.12116.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 2013;30:2725–9. doi:10.1093/molbev/mst197.
Lanfear R, Calcott B, Ho SYW, Guindon S. Partitionfinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 2012;29:1695–701. doi:10.1093/molbev/mss020.
Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. 2001;17:754–5.
Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–4. doi:10.1093/bioinformatics/btg180.
Rambaut A. FigTree 1.4.2. 2015. http://tree.bio.ed.ac.uk/software/figtree; 2015.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. doi:10.1093/molbev/msr121.
Kocher TD, Thomas WK, Meyer A, Edwards SV, Pääbo S, Villablanca FX, et al. Dynamics of mitochondrial DNA evolution in animals: Amplification and sequencing with conserved primers. Proc Natl Acad Sci. 1989;86:6196–200.
Vogler R, Desalle R. Phylogeographic Patterns in Coastal North American Tiger Beetles (Cicindela dorsalis Say) Inferred from Mitochondrial DNA Sequences. Evolution. 1993;47:1192–201.
Pashley D, Ke LD. Sequence evolution in the ribosomal and ND-1 genes in Lepidoptera: implications for phylogenetic analyses. Mol Biol Evol. 1992;9:1061–75.
Acknowledgements
We would like to thank SANParks, Eastern Cape Parks and Tourism Agency and CapeNature for providing the permits to work in the Cape Floral Region Protected Areas. In particular we thank the managers of the reserves for their continuous support during our studies. We are also grateful to Eva Heym for her helpful assistance in the field. This study was funded by German Research Foundation (DFG, GRK 1319) as part of the PhD of D.M.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
DM, LB and AH conceived the ideas; DM and LB conducted the field work and assessed the data; DM and AH wrote the manuscript. All authors read and approved the final manuscript.
Additional file
Additional file 1: Figure S1.
Phylogeny of the analyzed genera belonging to Lentulidae. Maximum likelihood tree for the genes 12S and NDS. Frontifissia and Sphingonotus were defined as outgroups. (JPG 1183 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Matenaar, D., Bröder, L. & Hochkirch, A. A preliminary phylogeny of the South African Lentulidae. Hereditas 153, 1 (2016). https://doi.org/10.1186/s41065-015-0005-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41065-015-0005-6