Abstract
While it is well established that cortical morphology differs in relation to a variety of inter-individual factors, it is often characterized using estimates of volume, thickness, surface area, or gyrification. Here we developed a computational approach for estimating sulcal width and depth that relies on cortical surface reconstructions output by FreeSurfer. While other approaches for estimating sulcal morphology exist, studies often require the use of multiple brain morphology programs that have been shown to differ in their approaches to localize sulcal landmarks, yielding morphological estimates based on inconsistent boundaries. To demonstrate the approach, sulcal morphology was estimated in three large sample of adults across the lifespan, in relation to aging. A fourth sample is additionally used to estimate test–retest reliability of the approach. This toolbox is now made freely available as supplemental to this paper: https://cmadan.github.io/calcSulc/.
Similar content being viewed by others
1 Introduction
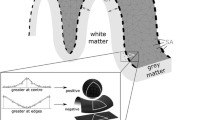
Cortical structure differs between individuals. It is well known that cortical thickness generally decreases with age [1,2,3,4,5,6,7,8,9,10,11]; however, a more visually prominent difference is the widening of sulci, sometimes described as ‘sulcal prominence’ [12,13,14,15,16,17]. In the literature, this measure has been referred to using a variety of names, including sulcal width, span, dilation, and enlargement, as well as fold opening. With respect to aging and brain morphology, sulcal width has been assessed qualitatively by clinicians as an index of cortical atrophy [12, 13, 15, 16, 18, 19]. An illustration of age-related differences in sulcal morphology is shown in Fig. 1.
Using quantitative approaches, sulcal width has been shown to increase with age [20,21,22,23] likely relating to subsequent findings of age-related decreases in cortical gyrification [2, 5,6,7, 24]. Sulcal widening has also been shown to be associated with decreases in cognitive abilities [25] and physical activity [26]. With respect to clinical conditions, increased sulcal width has been found in dementia patients relative to healthy controls [27,28,29,30,31,32,33], as well as with schizophrenia patients [34,35,36] and mood disorders [37].
One of the most common programs for conducting cortical surface analyses is FreeSurfer [38]. Unfortunately, though FreeSurfer reconstructs cortical surfaces, it does not estimate sulcal width or depth, leading researchers to use FreeSurfer along with another surface analysis program, BrainVISA [39,40,41,42], to characterize cortical thickness along with sulcal morphology (e.g, [22, 25, 26, 43, 44]). While this combination allows for the estimation of sulcal morphology in addition to standard measures such as cortical thickness, FreeSurfer and BrainVISA rely on different anatomical landmarks [45] which can yield differences in their resulting cortical surface reconstructions [46]. Admittedly, determining the boundaries for an individual sulcus and incorporating individual cortical variability is difficult [45, 47,48,49,50,51,52]. While an enumerate amount of other methods have already been proposed to identify and characterize sulcal morphology (e.g., [53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74]), ultimately these all are again using different landmarks than FreeSurfer uses for cortical parcellations (i.e., volume, thickness, surface area, gyrification). Note that, though FreeSurfer itself does compute sulcal maps, these are computed as normalized depths, not in real-world units (e.g., [75]); furthermore, these are also independent of sulcal width information.
Here we describe a procedure for estimating sulcal morphology and report age-related differences in sulcal width and depth using three large samples of adults across the lifespan: two of these datasets are from Western samples, Dallas Lifespan Brain Study (DLBS) and Open Access Series of Imaging Studies (OASIS), as well as one East Asian sample, Southwest University Adult Lifespan (SALD), as potential differences between populations have been relatively understudied [76, 77]. To further validate the method, test–retest reliability was also assessed using a sample of young adults who were scanned ten times within the span of a month [78, 79]. All four of these datasets are open-access and have sufficient sample sizes to be suitable for brain morphology research [77]. This procedure has been implemented as a MATLAB toolbox, calcSulc, that calculates sulcal morphology—both width and depth—using files generated as part of the standard FreeSurfer cortical reconstruction and parcellation pipeline. This toolbox is now made freely available as supplemental to this paper: https://cmadan.github.io/calcSulc/.
2 Materials and methods
2.1 Datasets
2.1.1 OASIS
This dataset consisted of 314 healthy adults (196 females), aged 18–94 (see Fig. 2), from the Open Access Series of Imaging Studies (OASIS) cross-sectional dataset (http://www.oasis-brains.org) [80]. Participants were recruited from a database of individuals who had (a) previously participated in MRI studies at Washington University, (b) were part of the Washington University Community, or (c) were from the longitudinal pool of the Washington University Alzheimer Disease Research Center. Participants were screened for neurological and psychiatric issues; the Mini-Mental State Examination (MMSE) and Clinical Dementia Rating (CDR) were administered to participants aged 60 and older. To only include healthy adults, participants with a CDR above zero were excluded; all remaining participants scored 25 or above on the MMSE. Multiple T1 volumes were acquired using a Siemens Vision 1.5 T with a MPRAGE sequence; only the first volume was used here. Scan parameters were: TR \(=\) 9.7 ms; TE \(=\) 4.0 ms; \(\hbox {flip angle} = 10^{\circ }\); \(\hbox {voxel size}=1.25 \times 1 \times 1 \, \hbox {mm}\). Age-related comparisons for volumetric and fractal dimensionality measures from the OASIS dataset were previously reported [7, 81, 82].Footnote 1
2.1.2 DLBS
This dataset consisted of 315 healthy adults (198 females), aged 20–89 (see Fig. 2), from wave 1 of the Dallas Lifespan Brain Study (DLBS), made available through the International Neuroimaging Data-sharing Initiative (INDI) [83] and hosted on the Neuroimaging Informatics Tools and Resources Clearinghouse (NITRC) [84] (http://fcon_1000.projects.nitrc.org/indi/retro/dlbs.html). Participants were screened for neurological and psychiatric issues. No participants in this dataset were excluded a priori. All participants scored 26 or above on the MMSE. T1 volumes were acquired using a Philips Achieva 3 T with a MPRAGE sequence. Scan parameters were: TR \(=\) 8.1 ms; TE \(=\) 3.7 ms; \(\hbox {flip angle} = 12 ^{\circ }\); \(\hbox {voxel size} = 1 \times 1 \times 1 \, \hbox {mm}\). See Kennedy et al. [85] and Chan et al. [86] for further details about the dataset. Age-related comparisons for volumetric and fractal dimensionality measures from the DLBS dataset were previously reported [7, 81, 82].\(^{1}\)
2.1.3 SALD
This dataset consisted of 483 healthy adults (303 females), aged 19–80 (see Fig. 2), from the Southwest University Adult Lifespan Dataset (SALD) [87], also made available through INDI and hosted on NITRC (http://fcon_1000.projects.nitrc.org/indi/retro/sald.html). No participants in this dataset were excluded a priori. T1 volumes were acquired using a Siemens Trio 3 T with a MPRAGE sequence. Scan parameters were: TR \(=\) 1.9 s; TE \(=\) 2.52 ms; \(\hbox {flip angle} = 9 ^{\circ }\); \(\hbox {voxel size} = 1 \times 1 \times 1 \, \hbox {mm}\).
2.1.4 CCBD
This dataset consisted of 30 healthy adults (15 females), aged 20–30, from the Center for Cognition and Brain Disorders (CCBD) at Hangzhou Normal University [78]. Each participant was scanned for 10 sessions, occurring 2–3 days apart over a 1-month period. No participants in this dataset were excluded a priori. T1 volumes were acquired using a SCANNER with a FSPGR sequence . Scan parameters were: TR \(=\) 8.06 ms; TE \(=\) 3.1 ms; \(\hbox {flip angle} = 8 ^{\circ }\); \(\hbox {voxel size}: 1 \times 1 \times 1 \, \hbox {mm}\). This dataset is included as part of the Consortium for Reliability and Reproducibility (CoRR) [88] as HNU1. Test–retest comparisons for volumetric and fractal dimensionality measures from the CCBD dataset were previously reported [79].\(^{1}\)
2.2 Procedure
Data were analyzed using FreeSurfer v6.0 (https://surfer.nmr.mgh.harvard.edu) on a machine running Red Hat Enterprise Linux (RHEL) v7.4. FreeSurfer was used to automatically volumetrically segment and parcellate cortical and subcortical structures from the T1-weighted images [38, 89] FreeSurfer’s standard pipeline was used (i.e., recon-all). No manual edits were made to the surface meshes, but surfaces were visually inspected. Cortical thickness is calculated as the distance between the white matter surface (white–gray interface) and pial surface (gray-CSF interface). Gyrification was also calculated using FreeSurfer, as described in Schaer et al. [90]. Cortical regions were parcellated based on the Destrieux et al. [91] atlas, also part of the standard FreeSurfer analysis pipeline.
3 Calculation
Here we outline a novel, simple yet robust, automated approach for estimating sulcal width and depth, based on intermediate files generated as part of the standard FreeSurfer analysis pipeline. This procedure and functionality has been implemented in an accompanying MATLAB toolbox, calcSulc. The toolbox is supplemental material to this paper and is made freely available: https://cmadan.github.io/calcSulc/.
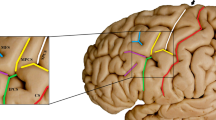
For each individual sulcus (for each hemisphere and participant), the following approach was used to characterize the sulcal morphology. The procedure has been validated and is supported for the following sulci: central, post-central, superior frontal, inferior frontal, parieto-occipital, occipito-temporal, middle occipital and lunate, and marginal part of the cingulate (S_central, S_postcentral, S_front_sup, S_front_inf, S_parieto_occipital, S_oc-temp_med&Lingual, S_oc_middle&Lunatus, S_cingul-Marginalis). All of the sulci are labeled in Fig. 3. An overview of the approach is illustrated in Fig. 4.
First the pial surface and Destrieux et al. [91] parcellation labels were read into MATLAB by using the FreeSurfer-MATLAB toolbox provided alongside FreeSurfer (calcSulc_load), this consists of the ?h.pial (FreeSurfer cortical surface mesh) ?h.aparc.a2009s.annot (FreeSurfer parcellation annotation) files. Using this, the faces associated with the individual sulcus were isolated as a 3D mesh (calcSulc_isolate).
The width of each sulcus (calcSulc_width) was calculated by determining which vertices lay on the boundary of the sulcus and the adjacent gyrus. An iterative procedure was then used to determine the ‘chain’ of edges that would form a contiguous edge loop that encircle the sulcal region (calcSulc_getEdgeLoop). This provided an exhaustive list of all vertices that were mid-way between the peak of the respective adjacent gyri and depth of the sulcus itself. For each vertex in this edge-loop, the nearest point in 3D space that was not neighboring in the loop was determined, with the goal of finding the nearest vertex in the edge that was on the opposite side of the sulcus—i.e., a line between these two vertices would ‘bridge’ across the sulcus. Since these nearest vertices in the edge loop are not necessarily the nearest vertex along the opposite sulcus wall, an exhaustive search (walk) was performed, moving up to a 4 edges from the initially determined nearest vertex (configurable as options.setWidthWalk). The sulcal width was then taken as the median of these distances that bridged across the sulcus (see Fig. 4).
The depth of each sulcus (calcSulc_depth) additionally used FreeSurfer’s sulcal maps (?h.sulc) to determine the relative inflections in the surface mesh, which would be in alignment with the gyral crown. The deepest points of the sulcus, i.e., the sulcal fundus, were taken as the 100 vertices within the sulcus with the lowest values in the sulcal map. For these 100 vertices, the shortest (i.e., Euclidean) distance to the smoothed enclosing surface was calculated (generated by FreeSurfer’s built-in gyrification analysis [?h.pial-outer-smoothed], [90]), and the median of these was then taken as the sulcal depth. While the use of a Euclidean distance here underestimates the true sulcal depth, it is nonetheless robust (as demonstrated in the present work) and does not markedly differ from other algorithmic approaches for estimating sulcal depth for much of the cortex (see [74] for a comparison).
Sulcal morphology, width and depth, was estimated for eight major sulci in each hemisphere: central, post-central, superior frontal, inferior frontal, parieto-occipital, occipito-temporal, middle occipital and lunate, and marginal part of the cingulate. Preliminary analyses additionally included superior and inferior temporal sulci and intraparietal sulcus, but these were removed from further analysis when the sulci width estimation was found to fail to determine a closed boundary edge-loop at an unacceptable rate (\(>10\%\)) for at least one hemisphere. This edge boundary determination failed when parcellated regions were labeled by FreeSurfer to comprise at least two discontinuous regions, such that they could not be identified using a single edge loop. Nonetheless, sulcal measures failed to be estimated for some participants, resulting in final samples of 310 adults from the OASIS dataset, 312 adults from the DLBS dataset, 481 adults from the SALD dataset, and 30 adults from the CCBD dataset (see Fig. 2).
3.1 Test–retest reliability
Test–retest reliability was assessed as intraclass correlation coefficient (ICC), which can be used to quantify the relationship between multiple measurements [79, 92,93,94,95,96,97,98]. McGraw and Wong [99] provide a comprehensive review of the various ICC formulas and their applicability to different research questions. ICC was calculated as the one-way random effects model for the consistency of single measurements, i.e., ICC(1, 1). As a general guideline, ICC values between .75 and 1.00 are considered ‘excellent,’ .60–.74 is ‘good,’ .40–.59 is ‘fair,’ and below .40 is ‘poor’ [100].
4 Results and discussion
4.1 Age-related differences in sulcal morphology
Scatter plots showing the relationships between each individual sulcal width and depth and age, for the OASIS dataset, are shown in Fig. 5; the corresponding correlations for all datasets are shown in Tables 1 and 2. The width and depth of the central and post-central sulci appear to be particularly correlated with age, with wider and shallower sulci in older adults. Age-related differences in sulcal width and depth and generally present in other sulci as well, but are generally weaker.
Age-related relationships for each sulcus were relatively consistent between the two Western lifespan datasets (OASIS and DLBS), but age-related differences in sulcal width (but not depth) were markedly weaker in the East Asian lifespan dataset (SALD). This finding will need to be studied further, but may be related to gross differences in anatomical structure [101,102,103]—and motivates the need to aging in samples that vary in ethnicity/race and are otherwise not of a so-called WEIRD (Western, Educated, Industrialized, Rich, and Democratic) demographic [77]. Additionally, there did not appear to be a significant influence of field strength (i.e., 1.5 T for the OASIS dataset vs. 3 T for the DLBS dataset) on estimates of sulcal morphology. Importantly, test–retest reliability, ICC(1, 1), was particularly good for the sulcal depth across individual sulci.
To obtain a coarse summary measure across sulci, we averaged the sulcal width across the 16 individual sulci for each individual, and with each dataset, and examined the relationship between mean sulcal width with age. These correlations, shown in Table 1, indicate that the mean sulcal width was generally a better indicator of age-related differences in sulcal morphology than individual sulci, and had increased test–retest reliability. Mean sulcal depth was similarly more sensitive to age-related differences than for an individual sulcus (e.g., it is unclear why the relationship between age and width of the central sulcus differed between samples) and the magnitude of this relationship was more consistent across datasets. Reliability was even higher for mean sulcal depth than mean sulcal width.
4.2 Comparison with other age-related structural differences
Within each dataset, mean sulcal depth and width correlated with age, as shown in Tables 1 and 2. Of course, other measures of brain morphology also differ with age, such as mean (global) cortical thickness [OASIS: \(r(308)=-.793\), \(p<.001\); DLBS: \(r(310)=-.759\), \(p<.001\); SALD: \(r(479)=-.642\), \(p<.001\)]. and volume of the third ventricle (ICV-corrected) [OASIS: \(r(308)=.665\), \(p<.001\); DLBS: \(r(310)=.677\), \(p<.001\); SALD: \(r(479)=.328\), \(p<.001\)]. Previous studies have demonstrated that both of these measures are robust estimates of age-related differences in brain structure [1,2,3,4,5,6, 8,9,10,11, 81, 104].
To test if these mean sulcal measures served as distinct measures of age-related differences in brain morphology, beyond those provided by other measures, such as mean cortical thickness and volume of the third ventricle, we conducted partial correlations that controlled for these two other measures of age-related atrophy. Mean sulcal width [OASIS: \(r_p(306)=.188\), \(p<.001\); DLBS: \(r_p(308)=.177\), \(p=.002\); SALD: \(r(477)=.003\), \(p=.96\)] and depth [OASIS: \(r_p(306)=-.443\), \(p<.001\); DLBS: \(r_p(308)=-.397\), \(p<.001\); SALD: \(r_p(477)=-.534\), \(p<.001\)] both explained unique variance in relation to age. Thus, even though more established measures of age-related differences in brain morphology were replicated here, the additional sulcal measures captured aspects of aging that are not accounted for by these extant measures, indicating that these sulcal measures are worth pursuing further and are not redundant with other measures of brain structure. Providing additional support for this, mean sulcal width and depth were only weakly related to each other [OASIS: \(r(308)=-.192\), \(p<.001\); DLBS: \(r(310)=.092\), \(p=.104\); SALD: \(r(479)=.119\), \(p=.009\)].
As with the individual sulci measures, we did observe a difference between samples where some age-related measures were less sensitive in the East Asian lifespan sample (SALD), here in the ventricle volume correlation and the unsurprisingly weaker age relationship in the partial correlation using sulcal width. These sample differences are puzzling, though there is a general correspondence between the two Western samples. Given that much of the literature is also based on Western samples, we think further research with East Asian samples, and particularly comparing samples with the same analysis pipeline, is necessary to shed further light on this initial finding.
5 Conclusion
Differences in sulcal width and depth are quite visually prominent, but are not often quantified when examining individual differences in cortical structure. Here we examined age-related differences in both sulcal measures as a proof-of-principle to demonstrate the utility of the calcSulc toolbox that accompanies this paper and is designed to closely compliments the standard FreeSurfer pipeline. This allows for the additional measurement of sulcal morphology, to add to the extant measures of brain morphology such as cortical thickness, area, and gyrification. Critically, this approach uses the same landmarks and boundaries as in the Destrieux et al. [91] parcellation atlas, in contrast to all previous approaches to characterize sulcal features. This toolbox is now made freely available as supplemental to this paper: https://cmadan.github.io/calcSulc/.
Using this approach, here we demonstrate age-related differences in sulcal width and depth, as well as high test–retest reliability. Since individual differences in sulcal morphology are sufficiently distinct from those characterized by other brain morphology measures, this approach should complement extant work of investigating factors that influence brain morphology, e.g., see Fig. 3 of Madan and Kensinger [7]. Given the flexibility in the methodological approach, these measures can be readily applied to other samples after being initially processed with FreeSurfer .
Notes
Note that analyses reported in these previous papers were based on preprocessing in FreeSurfer v5.3.0, rather than FreeSurfer v6.0.
References
Fjell AM, Westlye LT, Amlien I, Espeseth T, Reinvang I, Raz N et al (2009) High consistency of regional cortical thinning in aging across multiple samples. Cereb Cortex 19:2001–2012. https://doi.org/10.1093/cercor/bhn232
Hogstrom LJ, Westlye LT, Walhovd KB, Fjell AM (2013) The structure of the cerebral cortex across adult life: age-related patterns of surface area, thickness, and gyrification. Cereb Cortex 23:2521–2530. https://doi.org/10.1093/cercor/bhs231
Hutton C, Draganski B, Ashburner J, Weiskopf N (2009) A comparison between voxel-based cortical thickness and voxel-based morphometry in normal aging. NeuroImage 48:371–380. https://doi.org/10.1016/j.neuroimage.2009.06.043
Lemaitre H, Goldman AL, Sambataro F, Verchinski BA, Meyer-Lindenberg A, Weinberger DR, Mattay VS (2012) Normal age-related brain morphometric changes: nonuniformity across cortical thickness, surface area and gray matter volume? Neurobiol Aging 33:617.e1–617.e9. https://doi.org/10.1016/j.neurobiolaging.2010.07.013
Madan CR (2018) Age differences in head motion and estimates of cortical morphology. PeerJ 6:e5176. https://doi.org/10.7717/peerj.5176
Madan CR, Kensinger EA (2016) Cortical complexity as a measure of age-related brain atrophy. NeuroImage 134:617–629. https://doi.org/10.1016/j.neuroimage.2016.04.029
Madan CR, Kensinger EA (2018) Predicting age from cortical structure across the lifespan. Eur J Neurosci 47:399–416. https://doi.org/10.1111/ejn.13835
McKay DR, Knowles EEM, Winkler AAM, Sprooten E, Kochunov P, Olvera RL et al (2014) Influence of age, sex and genetic factors on the human brain. Brain Imaging Behav 8:143–152. https://doi.org/10.1007/s11682-013-9277-5
Salat DH, Buckner RL, Snyder AZ, Greve DN, Desikan RSR, Busa E et al (2004) Thinning of the cerebral cortex in aging. Cereb Cortex 14:721–730. https://doi.org/10.1093/cercor/bhh032
Sowell ER, Peterson BS, Kan E, Woods RP, Yoshii J, Bansal R et al (2007) Sex differences in cortical thickness mapped in 176 healthy individuals between 7 and 87 years of age. Cereb Cortex 17:1550–1560. https://doi.org/10.1093/cercor/bhl066
Sowell ER, Peterson BS, Thompson PM, Welcome SE, Henkenius AL, Toga AW (2003) Mapping cortical change across the human life span. Nat Neurosci 6:309–315. https://doi.org/10.1038/nn1008
Coffey CE, Wilkinson WE, Parashos L, Soady S, Sullivan RJ, Patterson LJ et al (1992) Quantitative cerebral anatomy of the aging human brain: a cross-sectional study using magnetic resonance imaging. Neurology 42(3):527–527. https://doi.org/10.1212/wnl.42.3.527
Drayer BP (1988) Imaging of the aging brain. Part I. Normal findings. Radiology 166(3):785–796. https://doi.org/10.1148/radiology.166.3.3277247
Jacoby RJ, Levy R, Dawson JM (1980) Computed tomography in the elderly: I. The normal population. Br J Psychiatry 136:249–255. https://doi.org/10.1192/bjp.136.3.249
Laffey PA, Peyster RG, Nathan R, Haskin ME, McGinley JA (1984) Computed tomography and aging: results in a normal elderly population. Neuroradiology 26:273–278. https://doi.org/10.1007/BF00339770
Tomlinson B, Blessed G, Roth M (1968) Observations on the brains of non-demented old people. J Neurol Sci 7:331–356. https://doi.org/10.1016/0022-510x(68)90154-8
Yue NC, Arnold AM, Longstreth WT, Elster AD, Jungreis CA, O’Leary DH et al (1997) Sulcal, ventricular, and white matter changes at MR imaging in the aging brain: data from the cardiovascular health study. Radiology 202:33–39. https://doi.org/10.1148/radiology.202.1.8988189
Pasquier F, Leys D, Weerts JG, Mounier-Vehier F, Barkhof F, Scheltens P (1996) Inter-and intraobserver reproducibility of cerebral atrophy assessment on MRI scans with hemispheric infarcts. Eur Neurol 36:268–272. https://doi.org/10.1159/000117270
Scheltens P, Pasquier F, Weerts JG, Barkhof F, Leys D (1997) Qualitative assessment of cerebral atrophy on MRI: inter- and intra-observer reproducibility in dementia and normal aging. Eur Neurol 37:95–99. https://doi.org/10.1159/000117417
Kochunov P, Mangin J-F, Coyle T, Lancaster J, Thompson P, Rivière D et al (2005) Age-related morphology trends of cortical sulci. Hum Brain Mapp 26:210–220. https://doi.org/10.1002/hbm.20198
Kochunov P, Thompson PM, Coyle TR, Lancaster JL, Kochunov V, Royall D et al (2008) Relationship among neuroimaging indices of cerebral health during normal aging. Hum Brain Mapp 29:36–45. https://doi.org/10.1002/hbm.20369
Liu T, Sachdev PS, Lipnicki DM, Jiang J, Geng G, Zhu W et al (2013) Limited relationships between two-year changes in sulcal morphology and other common neuroimaging indices in the elderly. NeuroImage 83:12–17. https://doi.org/10.1016/j.neuroimage.2013.06.058
Liu T, Wen W, Zhu W, Trollor J, Reppermund S, Crawford J et al (2010) The effects of age and sex on cortical sulci in the elderly. NeuroImage 51:19–27. https://doi.org/10.1016/j.neuroimage.2010.02.016
Cao B, Mwangi B, Passos IC, Wu M-J, Keser Z, Zunta-Soares GB et al (2017) Lifespan gyrification trajectories of human brain in healthy individuals and patients with major psychiatric disorders. Sci Rep 7:511. https://doi.org/10.1038/s41598-017-00582-1
Liu T, Wen W, Zhu W, Kochan NA, Trollor JN, Reppermund S et al (2011) The relationship between cortical sulcal variability and cognitive performance in the elderly. NeuroImage 56:865–873. https://doi.org/10.1016/j.neuroimage.2011.03.015
Lamont AJ, Mortby ME, Anstey KJ, Sachdev PS, Cherbuin N (2014) Using sulcal and gyral measures of brain structure to investigate benefits of an active lifestyle. NeuroImage 91:353–359. https://doi.org/10.1016/j.neuroimage.2014.01.008
Andersen SK, Jakobsen CE, Pedersen CH, Rasmussen AM, Plocharski M, Østergaard LR (2015) Classification of Alzheimer’s disease from MRI using sulcal morphology. In: Scandinavian conference on image analysis (SCIA): image analysis. Springer, pp 103–113. https://doi.org/10.1007/978-3-319-19665-7_9
Hamelin L, Bertoux M, Bottlaender M, Corne H, Lagarde J, Hahn V et al (2015) Sulcal morphology as a new imaging marker for the diagnosis of early onset Alzheimer’s disease. Neurobiol Aging 36(11):2932–2939. https://doi.org/10.1016/j.neurobiolaging.2015.04.019
Huckman MS, Fox J, Topel J (1975) The validity of criteria for the evaluation of cerebral atrophy by computed tomography. Radiology 116:85–92. https://doi.org/10.1148/116.1.85
Liu T, Lipnicki DM, Zhu W, Tao D, Zhang C, Cui Y et al (2012) Cortical gyrification and sulcal spans in early stage Alzheimer’s disease. PLoS ONE 7:e31083. https://doi.org/10.1371/journal.pone.0031083
Ming J, Harms MP, Morris JC, Beg MF, Wang L (2015) Integrated cortical structural marker for Alzheimer’s disease. Neurobiol Aging 36:S53–S59. https://doi.org/10.1016/j.neurobiolaging.2014.03.042
Plocharski M, Østergaard LR (2016) Extraction of sulcal medial surface and classification of Alzheimer’s disease using sulcal features. Comput Methods Progr Biomed 133:35–44. https://doi.org/10.1016/j.cmpb.2016.05.009
Reiner P, Jouvent E, Duchesnay E, Cuingnet R, Mangin J-F, Chabriat H (2012) Sulcal span in Alzheimer’s disease, amnestic mild cognitive impairment, and healthy controls. J Alzheimer’s Dis 29:605–613. https://doi.org/10.3233/JAD-2012-111622
Largen JW, Smith RC, Calderon M, Baumgartner R, Lu RB, Schoolar JC, Ravichandran GK (1984) Abnormalities of brain structure and density in schizophrenia. Biol Psychiatry 19:991–1013
Palaniyappan L, Park B, Balain V, Dangi R, Liddle P (2015) Abnormalities in structural covariance of cortical gyrification in schizophrenia. Brain Struct Funct 220(4):2059–2071. https://doi.org/10.1007/s00429-014-0772-2
Rieder RO, Donnelly EF, Herdt JR, Waldman IN (1979) Sulcal prominence in young chronic schizophrenic patients: CT scan findings associated with impairment on neuropsychological tests. Psychiatry Res 1(1):1–8. https://doi.org/10.1016/0165-1781(79)90021-0
Elkis H, Friedman L, Wise A, Meltzer HY (1995) Meta-analyses of studies of ventricular enlargement and cortical sulcal prominence in mood disorders. Arch Gen Psychiatry 52(9):735–746. https://doi.org/10.1001/archpsyc.1995.03950210029008
Fischl B (2012) FreeSurfer. NeuroImage 62:774–781. https://doi.org/10.1016/j.neuroimage.2012.01.021
Kochunov P, Rogers W, Mangin J-F, Lancaster J (2012) A library of cortical morphology analysis tools to study development, aging and genetics of cerebral cortex. Neuroinformatics 10:81–96. https://doi.org/10.1007/s12021-011-9127-9
Mangin J-F, Riviere D, Cachia A, Duchesnay E, Cointepas Y, Papadopoulos-Orfanos D et al (2004) Object-based morphometry of the cerebral cortex. IEEE Trans Med Imaging 23:968–982. https://doi.org/10.1109/tmi.2004.831204
Mangin J-F, Rivière D, Coulon O, Poupon C, Cachia A, Cointepas Y et al (2004) Coordinate-based versus structural approaches to brain image analysis. Artif Intell Med 30:177–197. https://doi.org/10.1016/s0933-3657(03)00064-2
Rivière D, Mangin J-F, Papadopoulos-Orfanos D, Martinez J-M, Frouin V, Régis J (2002) Automatic recognition of cortical sulci of the human brain using a congregation of neural networks. Med Image Anal 6:77–92. https://doi.org/10.1016/s1361-8415(02)00052-x
Cai K, Xu H, Guan H, Zhu W, Jiang J, Cui Y et al (2017) Identification of early-stage Alzheimer’s disease using sulcal morphology and other common neuroimaging indices. PLoS ONE 12:e0170875. https://doi.org/10.1371/journal.pone.0170875
Pizzagalli F, Auzias G, Kochunov P, Faskowitz JI, Thompson PM, Jahanshad N (2017) The core genetic network underlying sulcal morphometry. In: Romero E, Lepore N, Brieva J, Larrabide I (eds) International symposium on medical information processing and analysis. SPIE. https://doi.org/10.1117/12.2256959
Mikhael S, Hoogendoorn C, Valdes-Hernandez M, Pernet C (2018) A critical analysis of neuroanatomical software protocols reveals clinically relevant differences in parcellation schemes. NeuroImage 170:348–364. https://doi.org/10.1016/j.neuroimage.2017.02.082
Lee JK, Lee J-M, Kim JS, Kim IY, Evans AC, Kim SI (2006) A novel quantitative cross-validation of different cortical surface reconstruction algorithms using MRI phantom. NeuroImage 31:572–584. https://doi.org/10.1016/j.neuroimage.2005.12.044
Campero A, Ajler P, Emmerich J, Goldschmidt E, Martins C, Rhoton A (2014) Brain sulci and gyri: a practical anatomical review. J Clin Neurosci 21:2219–2225. https://doi.org/10.1016/j.jocn.2014.02.024
John JP, Wang L, Moffitt AJ, Singh HK, Gado MH, Csernansky JG (2006) Inter-rater reliability of manual segmentation of the superior, inferior and middle frontal gyri. Psychiatry Res Neuroimaging 148(2–3):151–163. https://doi.org/10.1016/j.pscychresns.2006.05.006
Ono M, Kubick S, Abernathey CD (1990) Atlas of the cerebral sulci. Thieme, Stuttgart
Rhoton AL (2007) The cerebrum. Neurosurgery 61(suppl1):SHC-37–SHC-119. https://doi.org/10.1227/01.neu.0000255490.88321.ce
ten Donkelaar HJ, Tzourio-Mazoyer N, Mai JK (2018) Toward a common terminology for the gyri and sulci of the human cerebral cortex. Front Neuroanat 12:93. https://doi.org/10.3389/fnana.2018.00093
Welker W (1990) Why does cerebral cortex fissure and fold? In: Jones EG, Peters A (eds) Cerebral cortex, vol 8B. Springer, Berlin, pp 3–136. https://doi.org/10.1007/978-1-4615-3824-0_1
Andreasen NC, Harris G, Cizadlo T, Arndt S, O’Leary DS, Swayze V, Flaum M (1994) Techniques for measuring sulcal/gyral patterns in the brain as visualized through magnetic resonance scanning: BRAINPLOT and BRAINMAP. Proc Natl Acad Sci 91(1):93–97. https://doi.org/10.1073/pnas.91.1.93
Auzias G, Brun L, Deruelle C, Coulon O (2015) Deep sulcal landmarks: algorithmic and conceptual improvements in the definition and extraction of sulcal pits. NeuroImage 111:12–25. https://doi.org/10.1016/j.neuroimage.2015.02.008
Beeston CJ, Taylor CJ (2000) Automatic landmarking of cortical sulci. In: Medical image computing and computer-assisted intervention–MICCAI 2000. Springer, Berlin, pp 125–133. https://doi.org/10.1007/978-3-540-40899-4_13
Behnke KJ, Rettmann ME, Pham DL, Shen D, Resnick SM, Davatzikos C, Prince JL (2003) Automatic classification of sulcal regions of the human brain cortex using pattern recognition. In: Sonka M, Fitzpatrick JM (eds) Medical imaging 2003: image processing. SPIE, pp 1499–1510. https://doi.org/10.1117/12.480834
Eskildsen SF, Uldahl M, Ostergaard LR (2005) Extraction of the cerebral cortical boundaries from MRI for measurement of cortical thickness. In: Fitzpatrick JM, Reinhardt JM (eds) Medical imaging 2005: image processing. SPIE, Bellingham. https://doi.org/10.1117/12.595145
Im K, Jo HJ, Mangin J-F, Evans AC, Kim SI, Lee J-M (2010) Spatial distribution of deep sulcal landmarks and hemispherical asymmetry on the cortical surface. Cereb Cortex 20(3):602–611. https://doi.org/10.1093/cercor/bhp127
Jones SE, Buchbinder BR, Aharon I (2000) Three-dimensional mapping of cortical thickness using Laplace’s equation. Hum Brain Mapp 11(1):12–32. https://doi.org/10.1002/1097-0193(200009)11:1<12::AID-HBM20>3.0.CO;2-K
Le Goualher G, Barillot C, Bizais YJ, Scarabin J-M (1996) Three-dimensional segmentation of cortical sulci using active models. In: Loew MH, Hanson KM (eds) Medical imaging 1996: image processing. SPIE, pp 254–263. https://doi.org/10.1117/12.237928
Le Goualher G, Collins DL, Barillot C, Evans AC (1998) Automatic identificaiton of cortical sulci using a 3d probabilistic atlas. In: Medical image computing and computer-assisted intervention—MICCAI’98. Springer, Berlin, pp 509–518. https://doi.org/10.1007/bfb0056236
Le Troter A, Auzias G, Coulon O (2012) Automatic sulcal line extraction on cortical surfaces using geodesic path density maps. NeuroImage 61(4):941–949. https://doi.org/10.1016/j.neuroimage.2012.04.021
Li G, Guo L, Nie J, Liu T (2010) An automated pipeline for cortical sulcal fundi extraction. Med Image Anal 14(3):343–359. https://doi.org/10.1016/j.media.2010.01.005
Li G, Shen D (2011) Consistent sulcal parcellation of longitudinal cortical surfaces. NeuroImage 57:76–88. https://doi.org/10.1016/j.neuroimage.2011.03.064
Lohmann G, von Cramon DY (2000) Automatic labelling of the human cortical surface using sulcal basins. Med Image Anal 4(3):179–188. https://doi.org/10.1016/s1361-8415(00)00024-4
Lohmann G, von Cramon DY, Colchester ACF (2008) Deep sulcal landmarks provide an organizing framework for human cortical folding. Cereb Cortex 18(6):1415–1420. https://doi.org/10.1093/cercor/bhm174
Nowinski WL, Raphel JK, Nguyen BT (1996) Atlas-based identification of cortical sulci. In: Kim Y (ed) Medical imaging 1996: image display. SPIE, Bellingham, pp 64–74. https://doi.org/10.1117/12.238488
Oguz I, Cates J, Fletcher T, Whitaker R, Cool D, Aylward S, Styner M (2008) Cortical correspondence using entropy-based particle systems and local features. In: 2008 5th IEEE international symposium on biomedical imaging: from nano to macro. IEEE, pp 1637–1640. https://doi.org/10.1109/isbi.2008.4541327
Perrot M, Rivière D, Mangin J-F (2011) Cortical sulci recognition and spatial normalization. Med Image Anal 15(4):529–550. https://doi.org/10.1016/j.media.2011.02.008
Royackkers N, Desvignes M, Fawal H, Revenu M (1999) Detection and statistical analysis of human cortical sulci. NeuroImage 10(6):625–641. https://doi.org/10.1006/nimg.1999.0512
Thompson PM, Schwartz C, Lin RT, Khan AA, Toga AW (1996) Three-dimensional statistical analysis of sulcal variability in the human brain. J Neurosci 16(13):4261–4274. https://doi.org/10.1523/jneurosci.16-13-04261.1996
Vaillant M, Davatzikos C (1997) Finding parametric representations of the cortical sulci using an active contour model. Med Image Anal 1(4):295–315. https://doi.org/10.1016/s1361-8415(97)85003-7
Yang F, Kruggel F (2008) Automatic segmentation of human brain sulci. Med Image Anal 12:442–451. https://doi.org/10.1016/j.media.2008.01.003
Yun HJ, Im K, Yang J-J, Yoon U, Lee J-M (2013) Automated sulcal depth measurement on cortical surface reflecting geometrical properties of sulci. PLoS ONE 8(2):e55977. https://doi.org/10.1371/journal.pone.0055977
Kippenhan JS, Olsen RK, Mervis CB, Morris CA, Kohn P, Meyer-Lindenberg A, Berman KF (2005) Genetic contributions to human gyrification: sulcal morphometry in Williams syndrome. J Neurosci 25(34):7840–7846. https://doi.org/10.1523/jneurosci.1722-05.2005
Leong RL, Lo JC, Sim SK, Zheng H, Tandi J, Zhou J, Chee MW (2017) Longitudinal brain structure and cognitive changes over 8 years in an east asian cohort. NeuroImage 147:852–860. https://doi.org/10.1016/j.neuroimage.2016.10.016
Madan CR (2017) Advances in studying brain morphology: the benefits of open-access data. Front Hum Neurosci 11:405. https://doi.org/10.3389/fnhum.2017.00405
Chen B, Xu T, Zhou C, Wang L, Yang N, Wang Z et al (2015) Individual variability and test–retest reliability revealed by ten repeated resting-state brain scans over one month. PLoS ONE 10:e0144963. https://doi.org/10.1371/journal.pone.0144963
Madan CR, Kensinger EA (2017b) Test–retest reliability of brain morphology estimates. Brain Inform 4:107–121. https://doi.org/10.1007/s40708-016-0060-4
Marcus DS, Wang TH, Parker J, Csernansky JG, Morris JC, Buckner RL (2007) Open access series of imaging studies (OASIS): cross-sectional MRI data in young, middle aged, nondemented, and demented older adults. J Cognit Neurosci 19:1498–1507. https://doi.org/10.1162/jocn.2007.19.9.1498
Madan CR, Kensinger EA (2017a) Age-related differences in the structural complexity of subcortical and ventricular structures. Neurobiol Aging 50:87–95. https://doi.org/10.1016/j.neurobiolaging.2016.10.023
Madan CR (2019) Shape-related characteristics of age-related differences in subcortical structures. Aging Mental Health 23:800–810. https://doi.org/10.1080/13607863.2017.1421613
Mennes M, Biswal BB, Castellanos FX, Milham MP (2013) Making data sharing work: the FCP/INDI experience. NeuroImage 82:683–691. https://doi.org/10.1016/j.neuroimage.2012.10.064
Kennedy DN, Haselgrove C, Riehl J, Preuss N, Buccigrossi R (2016) The NITRC image repository. NeuroImage 124:1069–1073. https://doi.org/10.1016/j.neuroimage.2015.05.074
Kennedy KM, Rodrigue KM, Bischof GN, Hebrank AC, Reuter-Lorenz PA, Park DC (2015) Age trajectories of functional activation under conditions of low and high processing demands: an adult lifespan fMRI study of the aging brain. NeuroImage 104:21–34. https://doi.org/10.1016/j.neuroimage.2014.09.056
Chan MY, Park DC, Savalia NK, Petersen SE, Wig GS (2014) Decreased segregation of brain systems across the healthy adult lifespan. Proc Natl Acad Sci USA 111:E4997–E5006. https://doi.org/10.1073/pnas.1415122111
Wei D, Zhuang K, Ai L, Chen Q, Yang W, Liu W et al (2018) Structural and functional brain scans from the cross-sectional southwest university adult lifespan dataset. Sci Data 5:180134. https://doi.org/10.1038/sdata.2018.134
Zuo X-N, Anderson JS, Bellec P, Birn RM, Biswal BB, Blautzik J et al (2014) An open science resource for establishing reliability and reproducibility in functional connectomics. Sci Data 1:140049. https://doi.org/10.1038/sdata.2014.49
Fischl B, Dale AM (2000) Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci USA 97:11050–11055. https://doi.org/10.1073/pnas.200033797
Schaer M, Cuadra MB, Schmansky N, Fischl B, Thiran J-P, Eliez S (2012) How to measure cortical folding from MR images: a step-by-step tutorial to compute local gyrification index. J Vis Exp 59:e3417. https://doi.org/10.3791/3417
Destrieux C, Fischl B, Dale A, Halgren E (2010) Automatic parcellation of human cortical gyri and sulci using standard anatomical nomenclature. NeuroImage 53:1–15. https://doi.org/10.1016/j.neuroimage.2010.06.010
Asendorpf J, Wallbott HG (1979) Maße der Beobachterübereinstimmung: ein systematischer Vergleich. Zeitschrift für Sozialpsychologie. 10:243–252
Bartko JJ (1966) The intraclass correlation coefficient as a measure of reliability. Psychol Rep 19:3–11. https://doi.org/10.2466/pr0.1966.19.1.3
Chen G, Taylor PA, Haller SP, Kircanski K, Stoddard J, Pine DS et al (2018) Intraclass correlation: improved modeling approaches and applications for neuroimaging. Hum Brain Mapp 39:1187–1206. https://doi.org/10.1002/hbm.23909
Hallgren KA (2012) Computing inter-rater reliability for observational data: an overview and tutorial. Tutor Quant Methods Psychol 8:23–34. https://doi.org/10.20982/tqmp.08.1.p023
Koo TK, Li MY (2016) A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J Chiropr Med 15:155–163. https://doi.org/10.1016/j.jcm.2016.02.012
Rajaratnam N (1960) Reliability formulas for independent decision data when reliability data are matched. Psychometrika 25:261–271. https://doi.org/10.1007/bf02289730
Shrout PE, Fleiss JL (1979) Intraclass correlations: uses in assessing rater reliability. Psychol Bull 86:420–428. https://doi.org/10.1037/0033-2909.86.2.420
McGraw KO, Wong SP (1996) Forming inferences about some intraclass correlation coefficients. Psychol Methods 1:30–46. https://doi.org/10.1037/1082-989x.1.1.30
Cicchetti DV (1994) Guidelines, criteria, and rules of thumb for evaluating normed and standardized assessment instruments in psychology. Psychol Assess 6:284–290. https://doi.org/10.1037/1040-3590.6.4.284
Kochunov P, Fox P, Lancaster J, Tan LH, Amunts K, Zilles K et al (2003) Localized morphological brain differences between english-speaking caucasians and chinese-speaking asians: new evidence of anatomical plasticity. NeuroReport 14(7):961–964. https://doi.org/10.1097/01.wnr.0000075417.59944.00
Longstreth WT, Arnold AM, Manolio TA, Burke GL, Bryan N, Jungreis CA et al (2000) Clinical correlates of ventricular and sulcal size on cranial magnetic resonance imaging of 3301 elderly people. Neuroepidemiology 19:30–42. https://doi.org/10.1159/000026235
Tang Y, Hojatkashani C, Dinov ID, Sun B, Fan L, Lin X et al (2010) The construction of a Chinese MRI brain atlas: a morphometric comparison study between Chinese and Caucasian cohorts. NeuroImage 51(1):33–41. https://doi.org/10.1016/j.neuroimage.2010.01.111
Walhovd KB, Westlye LT, Amlien I, Espeseth T, Reinvang I, Raz N et al (2011) Consistent neuroanatomical age-related volume differences across multiple samples. Neurobiol Aging 32:916–932. https://doi.org/10.1016/j.neurobiolaging.2009.05.013
Acknowledgements
MRI data used in the preparation of this article were obtained from several sources, data were provided in part by: (1) the Open Access Series of Imaging Studies (OASIS) [80]; (2) wave 1 of the Dallas Lifespan Brain Study (DLBS) led by Dr. Denise Park and distributed through INDI [83] and NITRC [84]; (3) the Southwest University Adult Lifespan Dataset (SALD) [87], also made available through INDI and hosted on NITRC; and (4) the Center for Cognition and Brain Disorders (CCBD) [78] as dataset HNU1 in the Consortium for Reliability and Reproducibility (CoRR) [88].
Author information
Authors and Affiliations
Contributions
CM conceptualised the study, developed the analysis approach and implemented it in software, processed the data, and wrote the manuscript. The author read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The author declares that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Madan, C.R. Robust estimation of sulcal morphology. Brain Inf. 6, 5 (2019). https://doi.org/10.1186/s40708-019-0098-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40708-019-0098-1