Abstract
Background
Helicobacter pylori is detected by pathogen recognition receptors including toll-like receptors (TLR) and nucleotide-binding oligomerization domain (NOD)-like receptors, eliciting an innate immune response against this bacteria. The aim of this study was to assess if polymorphisms of TLR2, TLR4, TLR5, NOD1 and NOD2 genes are associated with gastric cancer, in particular in individuals infected with H. pylori.
Results
A case-control study of 297 gastric cancer patients and 300 controls was performed to assess the association of 17 polymorphisms. Analyses performed under the allele model did not find association with gastric cancer. However, NOD1 rs2075820 (p.E266K) showed association with intestinal-type gastric cancer among H. pylori infected subjects (OR = 2.69, 95% CI 1.41–5.13, p = 0.0026). The association was not statistically significant in diffuse-type gastric cancer cases (OR = 1.26, 95% CI 0.63–2.52, p = 0.51). When the analyses were performed in patients carrying H. pylori strains harboring the cag pathogenicity island (cagPAI), we noticed significant association with NOD1 rs2075820 (OR = 4.90, 95% CI 1.80–3.36, p = 0.0019), in particular for intestinal-type gastric cancer cases (OR = 7.16, 95% CI 2.40–21.33, p = 4.1 × 10− 4) but not among diffuse-type gastric cancer cases (OR = 3.39, 95% CI 1.13–0.10, p = 0.03).
Conclusions
NOD1 rs2075820 increases the risk of intestinal-type gastric cancer among individuals infected with H. pylori, particularly in those harboring the cagPAI.
Similar content being viewed by others
Background
The burden of gastric cancer varies according to the region of the world, with incidence being higher in East Asia (22.4/100,000) followed by Central and Eastern Europe (11.4/100,000 and 9.5/100,000 respectively), and lower in Africa and North America (near to 4.0/100,000) [1]. Risk factors for gastric cancer include, among others, Helicobacter pylori infection and genetic factors [2]. H. pylori is a bacterium that colonizes the stomach, with a prevalence of 60% in Latin America [3]. This pathogen is associated with peptic ulcers (reported in 1–10% of infected subjects), gastric cancer (0.1–3%) and other extra-gastric diseases [4]. According to the IARC, H. pylori is considered as carcinogenic to humans (Group 1). A systematic review and meta-analysis [5] of published studies assessing the association of genetic factors with gastric cancer, described nine polymorphisms with significant association. Many of them were found using a candidate gene approach. Some of these polymorphisms were found to be associated in Asians and others in Caucasians.
The Lauren classification recognizes two histological types of gastric adenocarcinoma: intestinal and diffuse. For the first, Correa has described a precancerous cascade starting with atrophic gastritis, intestinal metaplasia and dysplasia [6]. In fact, atrophic gastritis is related with H. pylori and associated with gastric cancer risk [7]. This cascade emphasizes the role of inflammation in the pathogenesis of intestinal-type gastric cancer [8]. In contrast, diffuse-type gastric cancer is poorly differentiated and the underlying biological mechanisms leading to this type of gastric cancer are not fully understood [9]. H. pylori is detected by pathogen recognition receptors to elicit an innate immune response against the bacteria [10]. The main pathogen recognition receptors involved in this response include toll-like receptors (TLR) and nucleotide-binding oligomerization domain (NOD)-like receptors.
Ten toll-like receptors have been identified in humans, with TLR2, TLR4 and TLR5 being those involved in the recognition of H. pylori (reviewed by [11]). TLR2 and TLR4 recognize bacterial lipopolysaccharide (LPS) resulting in the activation of NF-kB [11]. Flagellin is recognized by TLR5, which induces the activation of a MyD88-dependent pathway [12]. The NOD-like receptor family include intracellular proteins that mediate an innate immune response [13]. NOD1 and NOD2 are NOD-like receptors expressed in epithelial and antigen-presenting cells and recognize H. pylori peptidoglycan [14] which enters to the host cell through the type IV secretion system of H. pylori [15]. The proteins that compose this bacterial apparatus are encoded by the cag pathogenicity island (cagPAI), a 40 kb DNA element of the H. pylori genome containing approximately 32 genes [16].
Some studies assessing the role of polymorphisms in TLR2, TLR4, TLR5, NOD1 and NOD2 in gastric cancer have been published. The associations of TLR4 rs4986790 (Asp299Gly) (minor allele frequency-MAF = 0.06) and rs4986791 (MAF = 0.04) were found to be statistically significant in meta-analyses [5]. The evidence for association of polymorphisms in TLR5 [17], NOD1 and NOD2 is scarce [18, 19]. Almost all studies were in Eastern countries, mainly China.
The aim of our study was to assess the association of common polymorphisms in TLR2, TLR4, TLR5, NOD1 and NOD2 with gastric cancer in H. pylori infected subjects. We found that a polymorphism in NOD1 (rs2075820 p.Glu266Lys) increases the risk of intestinal-type gastric cancer among individuals infected with H. pylori, particularly harboring cagPAI.
Results
The genotype count of the analyzed polymorphisms in 297 gastric cancer cases and 300 controls are shown in Additional file 1: Table S1. None of the 17 polymorphisms are associated with gastric cancer under the allele model (Additional file 1: Table S1).
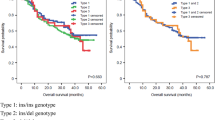
To study whether the association of the polymorphisms depends on the H. pylori status of the subject, we performed the analysis on a subgroup of infected patients. One hundred and two cases (39.4%) and 102 controls (49.3%) resulted positive for H. pylori infection. In the group of 102 gastric cancer cases and 102 controls we did not observe significant associations (Additional file 2). Nevertheless, after stratification according to histological type, the NOD1 rs2075820 (p.E266K) polymorphism was associated only among cases with intestinal-type gastric cancer in the unadjusted analysis (OR = 2.69, 95% CI 1.41–5.13, P = 2.6 × 10− 3). The association was not statistically significant in diffuse-type gastric cancer cases (OR = 1.26, 95% CI 0.63–2.52, P = 0.51).
Helicobacter pylori peptidoglycan is delivered to the cytoplasm of the epithelial cell through the T4SS injection apparatus encoded by the cagPAI, and is recognized by intracellular pathogen recognition receptors such as NOD1 and NOD2. To assess if polymorphisms modify their risk depending on the presence of the cagPAI, we carried out the association analysis in 87 gastric cancer patients (85.3%) and 43 controls (42.2%) infected with cagPAI-positive strains of H. pylori. We noticed significant association with NOD1 rs2075820 (OR = 4.90, 95% CI 1.80–13.36, P = 1.9 × 10− 3) (Additional file 2), in particular in intestinal-type gastric cancer cases (OR = 7.16, 95% CI 2.40–21.33, P = 4.1 × 10− 4) but no among diffuse-type gastric cancer cases (OR = 3.39, 95% CI 1.13–10.10, P = 0.03). Table 1 summarizes the results of the stratified analyses of NOD1 rs2075820. Therefore, the results suggest that this polymorphism confers increased risk for gastric cancer only among patients infected with H. pylori, in particular those infected with cagPAI-positive strains.
NOD1 rs2075820 is a non-synonymous polymorphism that replaces glutamic acid, a negatively charged amino acid, with lysine, a positively charged amino acid, at residue 266 of NOD1 protein (p.E266K). Additional file 3: Figure S1 shows a multiple sequence alignment of NOD1 from different species. Position 266 is relatively conserved, and glutamic acid or aspartic acid is present at this residue in almost all species, suggesting that this residue corresponds to a negatively-charged amino acid. SIFT and PolyPhen2 were used to assess if the polymorphism affects the function of the encoded protein. The score was 0.043 for SIFT, which is considered “damaging”, and 0.897 for PolyPhen2 classified as “possibly damaging”. Since the crystal structure of NOD1 is not available, we modeled ab initio its structure to infer whether the polymorphism alters the structure of the protein. Residue 266 lies in a alpha helix (Additional file 4: Figure S2). According to the ab initio modeled protein (Additional file 4: Figure S2), lysine at residue 266 does not modify the structure of the alpha helix compared to the protein with aspartic acid at 266. Nevertheless, a detailed inspection of the structure reveals a change in the orientation of lateral chains at residues 266 and 267 (Additional file 4: Figure S2). Taken together, it is possible that this polymorphism affects the function of NOD1.
Discussion
In the present study we assessed the role of polymorphisms in genes encoding pathogen recognition receptors in gastric cancer risk. We also aimed to analyze whether the risk changes among patients infected with H. pylori, in particular those with strains harbouring the cagPAI. The NOD1 rs2075820 polymorphism (p.E266K) was not associated with gastric cancer, nevertheless, the risk conferred by this SNP was evident only among patients carrying cagPAI positive H. pylori strains.
Little is known about the association of NOD1 rs2075820 with gastric cancer. Wang et al. [19] did not find association with gastric cancer in Chinese population. However, they found an increased risk to diffuse-type gastric cancer among individuals infected with H. pylori in the recessive model (OR = 1.89 [95% CI 1.07–3.32]) but the risk was not statistically significant in the dominant model (OR = 0.60 [95% CI 0.21–1.70]). The association with intestinal-type gastric cancer was not reported. We were not able to replicate a statistically significant result for the recessive model because just one case homozygous for the A allele was observed in diffuse-type gastric cancer cases (n = 52) and again only one case among the controls (n = 102) infected with H. pylori. Atrophic gastritis and intestinal metaplasia are premalignant lesions for gastric cancer. Among cagA seropositive patients, Kara et al. [20] found that carriers of allele A of NOD1 rs2075820 had a significantly increased risk for antral atrophy and metaplasia. In the study by Kim et al. [21], healthy subjects infected with H. pylori and homozygous for the risk allele (A) of NOD1 rs2075820 had a high gastritis score compared with carriers of the G allele. This effect was not observed among non-infected subjects. In addition, the IL-8 mRNA level of the AA genotype was significantly higher than for GA and GG genotypes among subjects infected with H. pylori harboring intact cagPAI [21]. Those studies suggest that H. pylori influence the association of NOD1 rs2075820 with preneoplasic lesions of intestinal-type gastric cancer.
Some studies have shed light on the role of NOD1 in gastric carcinogenesis. In an in vivo experiment, intestinal metaplasia was more frequently seen in tissue from NOD1-deficient mice (−/−) infected with H. pylori as compared to NOD1-intact mice [22]. The proportion of mice with intestinal metaplasia was low in non infected mice. They demonstrated in vitro that expression of cdx2, a protein involved in the normal differentiation of the intestinal epithelium, is higher in NOD1-deficient infected mice than in NOD1-intact infected mice. Uninfected mice do not express cdx2. In vitro analyses suggest that NOD1 is a negative regulator of cdx2 expression [22]. In another in vivo study in mice, the preactivation of NOD1 previous to infection by H. pylori reduces the frequency of adenocarcinoma and inflammation score [23]. A recently published article [24] shows that the prevalence of gastric dysplasia in INS-GAS NOD1 −/− infected mice was significantly higher than INS-GAS NOD1 +/+ mice. The above studies suggest that a diminished function of NOD1 contributes to the development of intestinal-type gastric cancer in infected subjects. In the case of NOD1 rs2075820, we hypothesize that it partially reduces the function of the NOD1 protein.
Our in silico analysis of the possible consequence of the amino acid exchange at the residue 266 induced by the NOD1 rs2075820 polymorphism was not fully conclusive. This residue lies in the central nucleotide-binding domain termed “NACHT” [25]. It is hypothesized that a crucial step in NOD1 activation is the oligomerization of this domain that acts as a platform for binding of adaptor molecules and effector proteins, resulting in an inflammatory response [26]. We propose that the change of a negatively-charged amino-acid to a positively-charged one, together with the possible change in the orientation of lateral chains at residues 266 and 277, could affect the interaction of effector proteins and adaptor molecules with the NACHT domain. The consequence of NOD1 rs2075820 at the functional and structural level requires further investigation.
The present study has some limitations. It does not necessarily represent the general population of Chile since it is a multi-centric hospital-based study. The sample size is small. Data regarding environmental risk factors were not available, which did not allow us to analyze gene–environment interactions. We did not adjust for age due to the low prevalence of gastric cancer in this study [27].
Conclusions
Our results indicate that NOD1 rs2075820 increases the risk of intestinal-type gastric cancer among individuals infected with H. pylori, particularly for those carrying the cagPAI. This could begin to explain the complex interaction between host genetic factors and H. pylori infection.
Methods
Subjects
The study included 299 cases (195 men and 104 women) diagnosed with gastric adenocarcinoma according to the histopathological report. The median age of diagnosis was 66 years, and ranged from 25 to 93. Patients were recruited at the time of surgical resection between 2001 and 2018 from the following hospitals in Santiago, Chile: University of Chile Clinical Hospital and Biobanco de Tejidos y Fluidos de la Universidad de Chile (BTUCH), Salvador Hospital, Barros Luco Trudeau Hospital, San Juan de Dios Hospital and Military Hospital of Santiago. Tumors were classified as intestinal-type or diffuse-type according to the Lauren’s classification. The control group was composed of 301 individuals (179 men and 122 women, median age 53 years, ranging from 18 to 82) with no personal history of cancer. Blood samples were collected in EDTA vacutainers for all participants. A sample of gastric mucosa was obtained from 259 cases and 207 controls. For cases, the sample was taken from corpus distant to the tumor, immediately from the resected stomach prior to the histopathological procedure. In the case of controls, a fresh sample was obtained from antrum and corpus, in patients who underwent an upper gastrointestinal endoscopy prescribed by a physician. This study was approved by the institutional review board of University of Chile School of Medicine (#045/2015) and was performed in accordance with the Declaration of Helsinki. All participants gave their written informed consent.
Genotyping
Genomic DNA was isolated from blood samples using the salting out and Proteinase K method, or according to the protocol described by Chomczynski and Sacchi [28] was further purified using Monarch PCR and DNA cleanup columns (New England Biolabs, USA). Single nucleotide polymorphisms (SNPs) were genotyped at the Human Genomics Facility (HuGe-F) in Erasmus MC, Netherlands, by Infinium Global Screening Array-24 BeadChip (Illumina, USA) according to the manufacturer’s protocol. Quality control of the genotyping data was performed following the guidelines in Anderson et al. [29] using PLINK 1.9 [30]. All studied samples and analyzed SNPs passed the quality control test. SNPs of TLR2, TLR4, TLR5, NOD1 and NOD2 genes were selected according to the following criteria: (1) positioned from 5 kb upstream from the transcription start site to 5 kb downstream from the stop triplet according to the GRCh37 assembly of human genome; (2) minor allele frequency (MAF) > 0.10 and (3) with no departure from Hardy-Weinberg equilibrium in the studied population (p < 0.01). We excluded rs5030728, rs4722988, rs2111234, rs2076756, rs4785224 and rs5743266 because they are in linkage disequilibrium (LD) (r2 > 0.8) with rs2770150, rs62447420, rs8057341, rs17313265, rs11647841 and rs2066842, respectively. Additional file 5: Table S2 describes the list of 17 polymorphisms analyzed in this study.
Detection of H. pylori and cag pathogenicity island
Analyses were performed using DNA from gastric mucosa isolated with FavorPrep Tissue DNA Extraction Mini Kit (Favorgen Biotech Corp, Taiwan, China). Samples were assessed for the presence or absence of H. pylori DNA by amplification of the 16S rRNA gene by 5′ exonuclease assay as described by Kobayashi et al. [31]. cagE gene was used as a marker of the presence of the cagPAI, and its detection was performed by 5′exonuclease assay. Primers were: F: 5′TGTGCTTGTAGCTCTTGGATTC-3′ and R: 5′-TCATGAACGCTTTGTTTTTCAC-3′ and the probe corresponds to 5′-[FAM] CTTTATCAAAGAATGGAGCGAGCGATG-3′ [BHQ1]. Primers and probes were synthesized by Macrogen Inc. (Korea). The 5′exonuclease assay was carried out using 5X HOT FIREPol Probe qPCR Mix Plus (ROX) (Solis BioDyne, Estonia) according to the manufacturer’s directions in a StepOne Real Time PCR system (Applied Biosystems, USA). Samples with a negative result in the cagE gene assay were subject to further detection of cagPAI empty site by PCR using the primers: 5′-ACATTTTGGCTAAATAAACRYTG-3′ and 5′-CACGCATTTTCCCTTRATC-3′, which amplify a fragment of 532 bp for cagPAI-negative samples. The reaction mixture was set up using RBC TaqDNA Polymerase (RBC Bioscience, Taiwan, China) and PCR reagents according to the manufacturer’s instructions. Reaction conditions for PCR were as follows: initial denaturation at 94 °C for 3 min, 40 cycles of denaturation at 94 °C for 30 s, annealing at 58 °C for 30 s, elongation at 72 °C for 45 s, and final elongation at 72° for 2 min. The product was analyzed in 1.5% agarose gel electrophoresis.
Prediction of functional consequences of NOD1 rs2075820 polymorphism
The multiple sequence alignment for NOD1 from different species was performed using PRALINE (http://www.ibi.vu.nl/programs/pralinewww/). Sequences were retrieved from UniProt with the following accession codes: Homo Sapiens (NP_006083.1), Callithrix jacchus (F7GYY3), Heterocephalus glaber (G5BRM7), Equus caballus (F7AF18), Rattus norvegicus (D4ADT7), Mus musculus (Q8BHB0), Sus scrofa (B0FSM7), Canis lupus familiaris (E2R9L3), Bos Taurus (E1B7V7), Loxodonta africana (G3T781), Ailuropoda menaloleuca (G1LJ87), Myotis lucifugus (G1NWX5), and Oryctolagus cuniculus (G1SNK2). SIFT [32] and PolyPhen-2 [33] were used to predict the possible impact of the amino acid substitution. The structure of NOD1 was predicted using the I-TASSER server [34] using the protein sequence with accession number NP_006083, and the resulting .pdb file was submitted to Missense3D [35] to model the structure of the mutant protein.
Statistical analyses
Statistical analyses were performed using PLINK 1.9 [30]. The exact test was used to detect departures from the Hardy-Weinberg equilibrium. The set of autosomal genotypes, obtained from Infinium Global Screening Array, was pruned from extended regions of high LD (r2 > 0.2) using indep-pairwise to obtain a set of 184,909 autosome SNPs. This set was submitted for principal component analysis (PCA) using pca. Principal component (PC) 1 and PC2 were used as estimates of population stratification [36]. The plot of the distribution of PC1 and PC2 among cases and controls is shown in Additional file 6: Fig. S3. A logistic regression analysis was performed to assess association of SNPs under the allele model, unadjusted or adjusting for sex, PC1 and PC2. Fisher’s exact test of independence was used to compare genotype distribution between cases and controls. A p-value < 2.9 × 10− 3 was considered statistically significant according to Bonferroni’s correction for multiple comparisons.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- TLR:
-
Toll-like receptor
- NOD:
-
Nucleotide-binding oligomerization domain
- OR:
-
Odds ratio
- cagPAI:
-
cag pathogenicity island
- LPS:
-
Lipopolysaccharide
- MAF:
-
Minor allele frequency
- SNP:
-
Single nucleotide polymorphism
- PC1:
-
Principal component 1
- PC2:
-
Principal component 2
References
CANCER TODAY [Internet]. [cited 2019 Dec 21]. Available from: http://gco.iarc.fr/today/home.
Guggenheim DE, Shah MA. Gastric cancer epidemiology and risk factors. J Surg Oncol. 2013;107:230–6.
Curado MP, de Oliveira MM, de Araújo Fagundes M. Prevalence of Helicobacter pylori infection in Latin America and the Caribbean populations: a systematic review and meta-analysis. Cancer Epidemiol. 2019;60:141–8.
McColl KEL. Helicobacter pylori Infection. N Engl J Med. 2010;362:1597–604.
Jie T, Guanchu L, Chunjian Z, Caiyang L, Wanlun H, Huanwen C. Genetic polymorphisms and gastric cancer risk: a comprehensive review synopsis from meta-analysis and genome-wide association studies. Cancer Biol Med. 2019;16:361.
Correa P, Camargo MC, Piazuelo MB. Overview and pathology of gastric cancer. In: Wang T, Fox J, Giraud A, editors. The biology of gastric cancer. New York: Springer; 2009. p. 1–24.
Hall CR. Pathology of gastric cancer. In: Gore RM, editor. Gastric cancer. Cambridge: Cambridge University Press; 2010. pp. 22–41.
Wu M-S. Host-Environment, interactions. Their impact on progression from gastric inflammation to carcinogenesis and on development of new approaches to prevent and treat gastric cancer. Cancer Epidemiol Biomark Prev. 2005;14:1878–82.
Riquelme I, Saavedra K, Espinoza JA, Weber H, García P, Nervi B, et al. Molecular classification of gastric cancer: Towards a pathway-driven targeted therapy. Oncotarget. 2015;6:24750–79.
Rudnicka K, Backert S, Chmiela M. Genetic polymorphisms in inflammatory and other regulators in gastric cancer: risks and clinical consequences. In: Backert S, editor. Molecular mechanisms of inflammation: induction, resolution and escape by Helicobacter pylori. Switzerland: Springer Nature; 2019. p. 56–76.
Meliț LE, Mărginean CO, Mărginean CD, Mărginean MO. The Relationship between Toll-like Receptors and Helicobacter pylori -Related Gastropathies: Still a Controversial Topic. Journal of Immunology Research. 2019;2019:1–10.
Pachathundikandi SK, Lind J, Tegtmeyer N, El-Omar EM, Backert S. Interplay of the gastric pathogen Helicobacter pylori with toll-like receptors. Biomed Res Int. 2015;2015:1–12.
Platnich JM, Muruve DA. NOD-like receptors and inflammasomes: A review of their canonical and non-canonical signaling pathways. Arch Biochem Biophys. 2019;670:4–14.
Castaño-Rodríguez N, Kaakoush NO, Mitchell HM. Pattern-recognition receptors and gastric cancer. Front Immunol. 2014;5:336.
Viala J, Chaput C, Boneca IG, Cardona A, Girardin SE, Moran AP, et al. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat Immunol. 2004;5:1166–74.
Noto JM, Peek RM. The Helicobacter pylori cag Pathogenicity Island. Methods Mol Biol. 2012;921:41–50.
Xu T, Fu D, Ren Y, Dai Y, Lin J, Tang L, et al. Genetic variations of TLR5 gene interacted with Helicobacter pylori infection among carcinogenesis of gastric cancer. Oncotarget. 2017;8:31016–22.
Li Z-X, Wang Y-M, Tang F-B, Zhang L, Zhang Y, Ma J-L, et al. NOD1 and NOD2 genetic variants in association with risk of gastric cancer and its precursors in a Chinese population. PLOS ONE. 2015;10:e0124949.
Wang P. Association of NOD1 and NOD2 genes polymorphisms with Helicobacter pylori related gastric cancer in a Chinese population. World J Gastroenterol. 2012;18:2112.
Kara B, Akkiz H, Doran F, Bayram S, Erken E, Gumurdullu Y, et al. The significance of E266K polymorphism in the NOD1 gene on Helicobacter pylori infection: an effective force on pathogenesis? Clin Exp Med. 2010;10:107–12.
Kim EJ, Lee JR, Chung WC, Jung SH, Sung HJ, Lee YW, et al. Association between genetic polymorphisms of NOD 1 and Helicobacter pylori-induced gastric mucosal inflammation in healthy Korean population. Helicobacter. 2013;18:143–50.
Asano N, Imatani A, Watanabe T, Fushiya J, Kondo Y, Jin X, et al. Cdx2 expression and intestinal metaplasia induced by H. pylori infection of gastric cells is regulated by NOD1-mediated innate immune responses. Cancer Res. 2016;76:1135–45.
Suarez G, Romero-Gallo J, Piazuelo MB, Wang G, Maier RJ, Forsberg LS, et al. Modification of Helicobacter pylori peptidoglycan enhances NOD1 activation and promotes cancer of the stomach. Cancer Res. 2015;75:1749–59.
Suarez G, Romero-Gallo J, Piazuelo MB, Sierra JC, Delgado AG, Washington MK, et al. Nod1 imprints inflammatory and carcinogenic responses toward the gastric pathogen Helicobacter pylori. Cancer Res. 2019;79:1600–11.
Boyle JP, Mayle S, Parkhouse R, Monie TP. Comparative genomic and sequence analysis provides insight into the molecular functionality of NOD1 and NOD2. Front Immunol. 2013;4:317.
Proell M, Riedl SJ, Fritz JH, Rojas AM, Schwarzenbacher R. The nod-like receptor (NLR) family: a tale of similarities and differences. PLoS ONE. 2008;3:e2119.
Pirinen M, Donnelly P, Spencer CCA. Including known covariates can reduce power to detect genetic effects in case-control studies. Nat Genet. 2012;44:848–51.
Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–9.
Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, Zondervan KT. Data quality control in genetic case-control association studies. Nat Protoc. 2010;5:1564–73.
Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaSci. 2015;4:7.
Kobayashi D, Eishi Y, Ohkusa T, Ishige null, Suzuki T, Minami J, et al. Gastric mucosal density of Helicobacter pylori estimated by real-time PCR compared with results of urea breath test and histological grading. J Med Microbiol. 2002;51:305–11.
Vaser R, Adusumalli S, Leng SN, Sikic M, Ng PC. SIFT missense predictions for genomes. Nat Protoc. 2016;11:1–9.
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9.
Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat Methods. 2015;12:7–8.
Ittisoponpisan S, Islam SA, Khanna T, Alhuzimi E, David A, Sternberg MJE. Can predicted protein 3D structures provide reliable insights into whether Missense variants are disease associated? J Mol Biol. 2019;431:2197–212.
Price AL, Zaitlen NA, Reich D, Patterson N. New approaches to population stratification in genome-wide association studies. Nat Rev Genet. 2010;11:459–63.
Acknowledgements
The authors would like to acknowledge BSc Tracy Wormwood for her help in proofreading the manuscript, and Susana Escandar for her assistance in collecting samples from control group.
Funding
This work was partially supported by Fondo Nacional de Desarrollo Científico y Tecnológico -Chile [FONDECYT 1151015].
Author information
Authors and Affiliations
Contributions
PG-H and DP conceived, designed the analysis, performed statistical analyses and wrote the draft manuscript; MM, MB, JSt, RP, HV, EL, HC, JSu, LAQ, NMV and ZB collected samples and clinicopathological data, MM cured and analyzed clinicopathological data, DP, LJ and VGC: Performed and contributed to laboratory procedures; PG-H supervised. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the institutional review board of University of Chile School of Medicine (#045/2015) and was performed in accordance with the Declaration of Helsinki. All participants gave their written informed consent.
Consent to publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
Genotype count of studied polymorphisms among gastric cancer cases and controls.
Additional file 2.
Association analysis of the studied SNPs.
Additional file 3.
Multiple sequence alignment of NOD1 from different species.
Additional file 4.
Analysis of the consequences of p.E266K on NOD1 protein.
Additional file 5.
Description of the 17 SNPs analyzed in this study.
Additional file 6.
Distribution of Principal Component 1 and Principal Component 2 among gastric cancer cases and controls.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Gonzalez-Hormazabal, P., Pelaez, D., Musleh, M. et al. NOD1 rs2075820 (p.E266K) polymorphism is associated with gastric cancer among individuals infected with cagPAI-positive H. pylori. Biol Res 54, 13 (2021). https://doi.org/10.1186/s40659-021-00336-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40659-021-00336-4