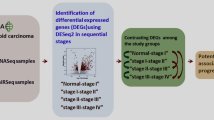
Abstract
Objective
In this study, crucial genes and microRNAs (miRNAs) associated with the progression, staging, and prognosis of papillary thyroid cancer (PTC) were identified.
Methods
Four PTC datasets, including our own mRNA-sequencing (mRNA-seq) dataset and three public datasets downloaded from Gene Expression Omnibus and The Cancer Genome Atlas, were used to analyze differentially expressed genes (DEGs) and miRNAs (DEMs) between PTC tumor tissues and paired normal tissues (control). Gene ontology (GO) terms and pathways associated with these DEGs were identified, and protein–protein interactions (PPIs) were analyzed. Additionally, an miRNA-mRNA regulatory network was constructed and the functions of DEMs were explored. Finally, miRNAs/mRNAs associated with tumor staging and prognosis were identified. The expression levels of several key genes and miRNAs were validated by qRT-PCR.
Results
Numerous DEGs and DEMs were identified between tumor and control groups in four datasets. The DEGs were significantly enriched in cell adhesion and cancer-related GO terms and pathways. In the constructed PPI network, ITGA2, FN1, ICAM1, TIMP1 and CDH2 were hub proteins. In the miRNA-mRNA negative regulatory networks, miR-204-5p regulated the largest number of target genes, such as TNFRSF12A. miR-146b, miR-204, miR-7-2, and FN1 were associated with tumor stage in PTC, and TNFRSF12A and CLDN1 were related to prognosis.
Conclusions
Our results suggested the important roles of ITGA2, FN1, ICAM1, TIMP1 and CDH2 in the progression of PTC. miR-204-5p, miR-7-2, and miR-146b are potential biomarkers for PTC staging and FN1, CLDN1, and TNFRSF12A may serve as markers of prognosis in PTC.
Similar content being viewed by others
Background
Thyroid cancer is the most common endocrine neoplasm, accounting for approximately 1.7% of all human malignancies [1]. Thyroid cancer is usually classified into four types: papillary thyroid cancer (PTC), anaplastic thyroid cancer, follicular thyroid cancer, and medullary thyroid cancer [2]. Among the four types, PTC is the most common, accounting for 75–85% of all thyroid cancer cases [3]. PTC is usually curable and has a 5-year survival rate of over 95% [4]. However, PTC occasionally dedifferentiates into more aggressive and lethal thyroid cancers. Additionally, recurrences are observed in approximately 30% of patients with PTC. Therefore, further analyses of the molecular characteristics of this disease are necessary [5].
Genetic mutations have been identified in the majority of PTC cases and are believed to be responsible for PTC initiation [6]. A high frequency of activating somatic alterations in genes encoding effectors in the MAPK (mitogen-activated protein kinase) signaling pathway, such as BRAF, RAS, and RET, has been found in PTC [7,8,9]; these mutations act as drivers in approximately 70% of PTC cases [10]. Additionally, galectin-3 (LGALS3), platelet-derived growth factor (PDGF), and epithelial muctin-1 (MUC-1) are also potential biomarkers of PTC [11, 12]. A recent study has demonstrated that numerous microRNAs (miRNAs) are transcriptionally up-regulated in PTC compared with normal tissues [13]. miR-181b, miR-221, and miR-222 have been suggested to be overexpressed in most thyroid tissues originating from patients affected by PTC [14]. Although numerous molecular markers associated with the carcinogenesis of PTC have been identified, significant uncertainty remains regarding its molecular mechanisms. Furthermore, prognosis- or staging-related molecular targets need to be explored.
Microarrays have made it possible to analyze genome-wide miRNA or mRNA expression [15]. In this study, we combined our mRNA-sequencing (mRNA-seq) data with three public datasets (miRNA-seq and mRNA-seq) downloaded from Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and The Cancer Genome Atlas (TCGA, https://tcga-data.nci.nih.gov/) to analyze differentially expressed genes (DEGs) and miRNAs (DEMs) between PTC tumor tissues and normal tissue samples (control). Common miRNAs and mRNAs between our mRNA-seq dataset and public datasets were selected for subsequent analyses, including analyses of gene ontology (GO) biological processes, pathway enrichment, and protein–protein interactions (PPIs), the construction of miRNA-mRNA regulatory networks, as well as the identification of miRNAs and mRNAs associated with staging and prognosis. Moreover, the expression levels of several key mRNAs and miRNAs identified in these analyses were confirmed by qRT-PCR. Loci identified in this study may act as important therapeutic targets or prognostic markers in PTC.
Methods
Tissue samples and mRNA-seq data collection
Five PTC tumor samples (P1–P5) and paired peritumoral thyroid tissue samples as controls (N1–N5) were obtained from patients with PTC who underwent thyroidectomy. The tumor samples and paired peritumoral thyroid tissue samples were obtained by thyroidectomy and stored in RNA store Reagent DP408-02 (Tiangen Biotech Co., Ltd., Beijing, China) at 4 °C in a refrigerator. Tumor stages were determined based on the tumor-node-metastasis (TNM) criteria. Detailed patient information is provided in Table 1. Informed consent was obtained from all patients. This study was approved by the local Research Ethics Committee.
After tissue collection, total RNAs were extracted using the RNAprep Pure Tissue Kit (Tiangen Biotech Co., Ltd.) according to the manufacturer’s protocol and was then converted to Tru-Seq libraries for sequencing using the Illumina HiSeq2000 platform (Illumina, San Diego, CA, USA). After sequencing, RNA-seq data were preprocessed, including the removal of the adaptor sequence from raw reads and the filtering of reads with a high N content and low quality. The detailed sequencing and preprocessing procedures are described in our previous study [16].
Public data collection
miRNA microarray data (accession GSE57780) were downloaded from the GEO database. A total of 9 samples were available, i.e., 3 PTC tissues, 3 normal tissues, and 3 nodal metastases. In this study, 3 PTC and 3 normal tissues were selected for further analysis. The platform used to obtain these data was the Illumina HiSeq2000.
Additionally, PTC-associated mRNA-seq, miRNA-seq, and clinical data were downloaded from TCGA using the TCGAbiolinks R package. In total, 552 samples were included.
Differential expression analysis
Using our own mRNA-seq data, gene expression levels were calculated using the RPKM method. DEGs between the PTC and control groups were detected using the edgeR [17] tool (version 3.4, http://www.bioconductor.org/packages/release/bioc/html/edgeR.html), and the significance of DEGs was calculated based on the negative binomial model. The obtained P-values were adjusted using the Benjamini and Hochberg method [5] to obtain the false discovery rate (FDR). The thresholds for DEG selection were FDR < 0.05 and |log2 fold change (FC)| > 2.
The mRNA-seq and miRNA-seq data obtained from TCGA and GEO were also preprocessed using edgeR [17] (version 3.4) in R. The raw counts were converted to log2-counts-per-million (logCPM) followed by linear modeling and the mean–variance relationship was modeled with precision weights using the voom [18] method in limma. The differential expression analysis (tumor vs. control) was performed using t-tests implemented in the limma package [19]. The P-values were adjusted using the Benjamini and Hochberg method. The cut-off values were FDR < 0.05 and |log2 FC| > 2 for DEGs and FDR < 0.05 and |log2 FC| > 1.5 for DEMs.
Functional enrichment analysis
For the identified DEGs, gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed using multifaceted analysis tool for human transcriptome (MATHT) (www.biocloudservice.com). The GO analysis included biological process, cellular component, and molecular function categories. P < 0.05 was used as a threshold for the identification of significant GO terms and pathways.
PPI network analysis
Based on the PPIs in the search tool for the retrieval of interacting genes/proteins (STRING, http://string-db.org/) [20], a PPI analysis of the DEGs was performed with threshold required confidence (combined score) of > 0.4. The obtained PPI pairs were then visualized to construct a PPI network using Cytoscape (http://www.cytoscape.org/) [21]. The topological properties of nodes in the PPI network were analyzed using CytoNCA [22] plugin (version 2.1.6, http://apps.cytoscape.org/apps/cytonca). Based on estimates of degree centrality, betweenness centrality, and closeness centrality of nodes, key nodes involved in the PPI network, named hub proteins, were identified [23].
miRNA–mRNA regulatory network analysis
The target genes of DEMs were predicted using the miRWalk 2.0 tool [24, 25] (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/). The prediction criterion was that the target gene must be identified in three or more of the following prediction databases: miRWalk (http://mirwalk.uni-hd.de/), miRanda (http://www.microrna.org/miranda.html), RNA22 (http://cm.jefferson.edu/rna22v1.0/), and Targetscan (http://www.targetscan.org). Then, target genes were combined with DEGs to select DEM-differentially expressed target gene pairs. Subsequently, the Pearson correlation coefficients were calculated for DEM-differentially expressed target genes pairs and pairs with r < − 0.4 (negative regulation) were used to construct the miRNA-mRNA negative regulatory network. The key miRNAs and genes were identified based on a network topological property analysis.
miRNA function analysis
For genes involved in the miRNA-mRNA negative regulatory network, GO and KEGG pathway enrichment analyses were performed using clusterProfiler [26] in R. These GO terms and KEGG pathways were interpreted as functions of miRNAs involved in the miRNA-mRNA regulatory network.
Associations between miRNAs/mRNAs and tumor stage
According to the clinical stage and metastasis information obtained from TCGA, samples were divided into the following groups: stage III–IV vs. stage I–II and M1 vs. M0. Differential analyses of stage III–IV vs. stage I–II and M1 vs. M0 were performed to identify DEMs and DEMs associated with stage and metastasis status following the same methods described above “Differential expression analysis”. The cut-off values for DEG selection were FDR < 0.05 and |log2 FC| > 0.585, and for DEM selection were FDR < 0.05 and |log2 FC| > 1.
Survival analysis for DEGs and DEMs
Clinical information related to prognosis, including overall survival and vital status, were obtained. For all DEMs and DEGs, samples were divided into high and low miRNA/mRNA expression groups based on the median expression values. Kaplan–Meier (KM) curves were then generated using survminer (https://cran.r-project.org/web/packages/survminer/index.html) in R and were analyzed using the log-rank test with a significance threshold of P < 0.05.
Verification of the expression levels of key mRNAs and miRNAs
An additional nine PTC tumor samples (P6–P14) and paired peritumoral normal thyroid tissues (N6–N14) were collected for qRT-PCR verification of the expression levels of key mRNAs and miRNAs. The detailed patient information is listed in Table 1. Total RNA was extracted from tissue samples using TRIzol reagent (9109; TaKaRa, Dalian, China). cDNA was generated with PrimeScript™ RT Master Mix (RR036A; TaKaRa) and the PrimeScript™ II 1st Strand cDNA Synthesis Kit (6210A; TaKaRa). PCR amplification was performed using the ABI ViiA7 real-time PCR system (Applied Biosystems, Foster City, CA, USA) using Power SYBR Green PCR Master (4367659; Thermo, Waltham, MA, USA). GAPDH and U6 were used as endogenous controls. The primers used for in the study are listed in Table 1. The relative gene expression levels were determined with the 2−ΔΔCT method.
Statistical analysis
Data are expressed as mean ± standard error of the mean (SEM). Differences between groups were analyzed using Student’s t-test implemented in SPSS 22.0 (SPSS, Inc., Chicago, IL, USA). P < 0.05 was considered statistically significant. Experiments were repeated three times.
Results
DEG and DEM identification
A total of 496 up-regulated and 440 down-regulated DEGs were identified using our own mRNA-seq data. For publicly available datasets, 12 up-regulated and 21 down-regulated DEMs were identified using TCGA miRNA-seq data, 491 up-regulated and 541 down-regulated DEGs were identified using TCGA mRNA-seq data, and 14 up-regulated and 15 down-regulated DEGs were identified using GSE57780 data. Subsequently, we constructed a Venn diagram for the DEGs and DEMs. As shown in Fig. 1, 10 intersecting DEMs (5 up- and 5 down-regulation), such as miR-222, miR-221, miR-146b, and miR-21 (Fig. 1a), and 548 intersecting DEGs, including 263 up- and 285 down-regulated DEGs, were identified (Fig. 1b).
Functional enrichment analysis
A functional enrichment analysis of the intersecting DEGs revealed that the up-regulated DEGs were significantly involved in cell adhesion, signal transduction, cell cycle, and cancer-related GO terms and pathways (Table 2 and Fig. 2a). The down-regulated DEGs were significantly enriched in bicarbonate transport, negative regulation of growth, tyrosine metabolism, and drug metabolism-associated GO terms and pathways (Table 2 and Fig. 2b).
PPI network analysis
The PPI network included 369 nodes and 841 interaction pairs (Fig. 3). A topological property analysis of nodes in the PPI network showed that integrin subunit alpha 2 (ITGA2), intercellular adhesion molecule 1 (ICAM1), fibronectin 1 (FN1), tissue inhibitor of metalloproteinases 1 (TIMP1), nitric oxide synthase 1 (NOS1), cadherin 2 (CDH2), and neural cell adhesion molecule 1 (NCAM1) had high scores, among other loci. The top 10 nodes with high topological property scores are shown in Table 3.
miRNA-mRNA regulatory network analysis
The constructed miRNA–mRNA negatively regulatory networks are shown in Fig. 4. The network included 175 nodes (102 up-regulated mRNAs, 57 down-regulated mRNAs, 8 up-regulated miRNAs, and 8 down-regulated mRNAs) and 272 interacting pairs. A topological property analysis showed that miR-204-5p regulated the largest number of target genes (n = 31), such as such as TNF receptor superfamily member 12A (TNFRSF12A), and surfactant protein B (SFTPB) was regulated by 6 miRNAs (the most one), such as miR-204-5p.
miRNA-mRNA negative regulatory networks of differentially expressed genes (DEGs) and miRNAs (DEMs). Each red circle represents an up-regulated DEG and each blue rhombus represents a down-regulated DEG; red triangles represent up-regulated DEMs and blue triangles represent down-regulated DEMs. Line thicknesses represents the Pearson correlation coefficient
miRNA function analysis
Based on the target genes of DEMs involved in the miRNA–mRNA negative regulatory networks, we investigated GO function and pathway enrichment for these DEMs. As shown in Fig. 5, miR-21 was associated with thyroid hormone synthesis and cytokine–cytokine receptor interactions; miR-222 was involved in the adipocytokine signaling pathway and Jak-STAT signaling pathway; miR-7-2 was associated with cytokine–cytokine receptor interactions and tight junctions.
Associations between miRNAs/mRNAs and tumor stage
Applying the thresholds indicated in “Methods”, 4 DEMs, i.e., miR-146b, miR-204, miR-1179, and miR-7-2, and 61 DEGs, such as FN1, baculoviral IAP repeat containing 7 (BIRC7), and matrix metallopeptidase 7 (MMP7), were identified for the comparison between stage III–IV and stage I–II (Table 4). Additionally, the expression trends of these miRNAs/mRNAs were in accord with those identified in tumor vs. normal tissues. Neither DEMs nor DEGs were identified in the M1 vs. M0 comparison.
Survival analysis of DEGs and DEMs
Using overall survival and vital status information, we performed a survival analysis for DEGs and DEMs. A total of 59 DEGs were associated with prognosis, such as TNFRSF12A (Fig. 6a) and claudin 1 (CLDN1) (Fig. 6b), while no prognosis-associated DEMs were identified. Based on the comparation of the 61 tumor stage and 59 prognosis related DEGs, Kallikrein-related peptidase 7 (KLK7) and KLK 10 were presented in both sets. The differences in survival probabilities between the high and low gene expression groups were significant (P < 0.05).
Expression levels of key mRNAs and miRNAs
The expression levels of seven mRNAs (ITGA2, FN1, ICAM1, CDH2, TIMP1, CLDN1, and TNFRSF12A) and three miRNAs (miR-204-5p, miR-146b, and miR-222) were evaluated by qRT-PCR. As shown in Fig. 7, in comparison with the control group, except for miR-204-5p, the expression levels of ITGA2, FN1, ICAM1, CDH2, TIMP1, CLDN1, TNFRSF12A, miR-146b, and miR-222 were all up-regulated in the tumor group, consistent with the results of the bioinformatics analyses. Additionally, significant differences were identified in miR-204-5p, ITGA2, FN1, ICAM1, CDH2, TIMP1, CLDN1, and TNFRSF12A expression between groups (P < 0.05 or 0.01).
Discussion
In the present study, numerous DEGs and DEMs between tumor and control groups were identified in four datasets. In a Venn diagram analysis, 10 intersecting DEMs, like miR-204-5p, miR-7-2, miR-146b, and miR-222, and 548 intersecting DEGs were identified. The intersecting DEGs were significantly enriched in cell adhesion and cancer-related GO terms and pathways. In the constructed PPI network, ITGA2, FN1, ICAM1, TIMP1 and CDH2 were identified as hub proteins. In the miRNA–mRNA negative regulatory networks, miR-204-5p regulated the largest number of target genes. Additionally, miR-146b, miR-204, miR-7-2, and FN1 were associated with the tumor stage of PTC, and TNFRSF12A and CLDN1 were related to the prognosis of PTC.
Cell adhesion determines cell polarity and is involved in tissue maintenance. Abnormalities in cell adhesion are a morphological hallmark of malignant tumors, and have effects on the biological characteristics of cancers [27]. The present results showed that up-regulated DEGs identified in multiple datasets, such as ICAM1 and CDH2, were significantly involved in cell adhesion-associated functions and pathways (GO: 0007155~cell adhesion, hsa04514: cell adhesion molecules). Specifically, ICAM1 and CDH2 were hub proteins in the PPI network. ICAM1, a transmembrane glycoprotein receptor, belongs to the immunoglobulin superfamily of adhesion molecules [28]. The expression of ICAM1 is elevated in many human cancers, including PTC [29]. ICAM1 can facilitate the spread of metastatic cancer cells to secondary sites by recruiting inflammatory cells that release angiogenic and growth factors stimulating angiogenesis, cell proliferation, and invasion [30]. CDH2 belongs to the cadherin gene family of cell–cell adhesion molecules and encodes N-cadherin. CDH2 is commonly up-regulated in various human cancers, including thyroid cancer [31], and its overexpression can promote tumor cell invasion [32]. In addition, CDH2 plays a significant role in epithelial-mesenchymal transition, a pivotal event for tumor cell acquisition of metastatic ability [33]. Importantly, ICAM1 and CDH2 were up-regulated in PTC tissues in a validation experiment. Taken together, the pathway and genes associated with cell adhesion may play important roles in PTC development.
Most complex networks, including PPI networks, are scale-free, containing a small number of highly connected nodes (hubs) and a large number of poorly connected nodes (non-hubs). A genome-wide analysis has shown that the deletion of a hub is more likely to be lethal than the deletion of a non-hub, suggesting the importance of hubs in network organization [23]. In this study, we identified several hub proteins in the PPI network. In addition to ICAM1 and CDH2 mentioned above, TIMP1, ITGA2 and FN1 had high degrees and accordingly may play important roles in PTC. Additionally, FN1 was identified to be a clinical stage-associated gene. ITGA2 is an important collagen receptor on epithelial cells, and its expression regulated during normal cell differentiation and altered during tumorigenesis [34]. FN1 is an extracellular matrix protein synthesized by fibroblasts, and is often associated with goiters [35]. FN1 was first reported to be overexpressed in thyroid cancer in 1988 [36]. It is the most strikingly up-regulated marker in intermediate-risk PTC and is considered a negative prognostic marker in patients with PTC [37]. In the present study, TIMP1, ITGA2 and FN1 were up-regulated in PTC tissues. Additionally, ITGA2 and FN1 were associated with GO terms and pathways related to extracellular matrix (ECM) organization and cancer (hsa05222: small cell lung cancer, hsa04512: ECM-receptor interaction, hsa05200: pathways in cancer). ECM is essential at various stages of the carcinogenic process and abnormal ECM is considered a hallmark of cancer [38]. Therefore, ITGA2 and FN1 may play key roles in PTC progression via these cancer-related pathways. TIMP1 is recognized as a multifunctional protein. Inhibiting matrix metalloproteinases and antiangiogenic activity are two important roles of TIMP1 [39, 40]. Study has demonstrated inhibition of TIMP1-mediated tumor invasion and metastasis in many types of tumor cells [41, 42]. Elevated serum and tissue levels of TIMP1 have been found in cancer patients [43]. In this study, TIMP1 was demonstrated to up-regulate in PTC tumor tissue, which was in consistent with the previous study. Thus, TIMP1 may serves as key biomarker of PTC.
miRNAs negatively regulate gene expression at the post-transcriptional level and have profound biological importance. The dysregulation of miRNA expression has been implicated in human cancers [44]. In the present study, several DEMs were identified, such as miR-204-5p, miR-7-2, miR-146b, and miR-222. miR-204-5p was a hub in the miRNA–mRNA regulatory network, regulating the most target genes. It was also associated with clinical stage. A recent study found that miR-204-5p was down-regulated in PTC and acted as a tumor suppressor [45]. In accordance with this finding, miR-204-5p was down-regulated in PTC tissues in our study and regulated TNFRSF12A, a prognosis-associated DEG. Besides, result of qRT-PCR also showed that miR-204-5p expression was significantly decreased while TNFRSF12A expression was significantly increased in tumor compared with control. Based on a GO functional enrichment analysis, TNFRSF12A was related to angiogenesis. A previous study has found that angiogenesis is essential for tumor growth and progression [46]. Importantly, Tanaka et al. [47] reported that the balance between angiogenic and antiangiogenic factors is correlated with the distinct invasion of PTC, suggesting the importance of angiogenesis in PTC.
miR-7-2 was another down-regulated DEM associated with clinical stage. Based on a functional enrichment analysis, the target genes of miR-7-2, such as CLDN1, were significantly enriched for hsa04530: tight junction pathway. CLDN1 encodes a member of the claudin family, a component of tight junction strands. Tight junctions are one mode of cell-to-cell adhesion in endothelial or epithelial cell sheets; they can alter cell polarity, cell fate, and cell migration, thereby impacting cancer progression. Alterations in tight junctions facilitate breast cancer initiation and progression [48]. Interestingly, CLDN1 was a prognosis-associated DEG in this study. Therefore, miR-7-2 and CLDN1 may be used as biomarkers of stage and prognosis in PTC.
miR-146b and miR-222 were up-regulated in the tumor group in this study, and miR-146b was a DEM associated with clinical stage. miR-146b has been implicated in several cancer types by microarray analyses, such as cervical cancers, breast cancer, and prostate cancers [49]. Moreover, miR-222 is a cancer-related miRNA and has been reported to promote cancer cell proliferation [50]. Importantly, miR-146b and miR-222 are overexpressed in PTC tissue and are significantly associated with extrathyroidal invasion [51]. Validation experiment also found that miR-146b and miR-222 were up-regulated in PTC tissue, although there was no significant difference. Therefore, our data were consistent with previous results, further supporting the important role of miR-146b and miR-222 in PTC.
By comparing the 61 tumor stage and 59 prognosis related DEGs, there were two members in the intersection: KLK7 and KLK 10. Kallikrein are serine proteases which have many physiological functions, containing the remodeling of ECM. They are able to function individually or in cascade pathway(s) and acted as diagnostic and prognostic biomarkers for tumors [52]. The expression of KLK7 and KLK10 in PTC were significantly upregulated, with a mean fold change of 28.8 and 27.8, reletive to matched normal tissue, suggesting that KLK7 and KLK10 might be useful biomarker for the diagnosis of PTC [53]. In this study, KLK7 and KLK10 were identified as the biomarkers of tumor stage and prognosis of PTC, which were consistent with the previous report. In addition, miR-1179 was another DEM related to tumor stage of PTC. A recently report showed that miR-1179 inhibited glioblastoma cell proliferation through targeting E2F transcription factor 5 [54]. In this study, miR-1179 was down-regulated in PTC patients, and the regulation mechanism of miR-1179 in the PTC progress was need further study. Although some genes and miRNAs markers were identified and validated, this study had some limitations. The sample size was small; therefore, we will collect additional samples in the future to further investigate the roles of these genes and miRNAs in the pathogenesis of PTC.
In conclusion, our results suggested the important roles of ITGA2, FN1, ICAM1, TIMP1 and CDH2 in the progression of PTC via various cancer-related pathways. Additionally, miR-204-5p, miR-7-2, miR-146b, and miR-222 may play critical roles in tumorigenesis of PTC. Furthermore, FN1, miR-204-5p, miR-7-2, and miR-146b may be used as biomarkers for PTC staging, and CLDN1 and TNFRSF12A may serve as prognostic markers.
References
Lodewijk L, Prins AM, Kist JW, Valk GD, Kranenburg O, Rinkes IH, et al. The value of miRNA in diagnosing thyroid cancer: a systematic review. Cancer Biomark. 2012;11(6):229–38.
Xing M. BRAF mutation in thyroid cancer. Endocr Relat Cancer. 2005;12(2):245–62.
Geraldo MV, Kimura ET. Integrated analysis of thyroid cancer public datasets reveals role of post-transcriptional regulation on tumor progression by targeting of immune system mediators. PLoS ONE. 2015;10(11):e0141726.
Hay ID, Thompson GB, Grant CS, Bergstralh EJ, Dvorak CE, Gorman CA, et al. Papillary thyroid carcinoma managed at the Mayo Clinic during six decades (1940–1999): temporal trends in initial therapy and long-term outcome in 2444 consecutively treated patients. World J Surg. 2002;26(8):879–85.
Esin YE, Duygu T, Serkan T, Ezel UF, Ugur G, Nejat D. microRNA expression profiles in papillary thyroid carcinoma, benign thyroid nodules and healthy controls. J Cancer. 2016;7(7):803–9.
Vasko V, Espinosa AV, Scouten W, He H, Auer H, Liyanarachchi S, et al. Gene expression and functional evidence of epithelial-to-mesenchymal transition in papillary thyroid carcinoma invasion. Proc Natl Acad Sci. 2007;104(8):2803–8.
Cohen Y, Xing M, Mambo E, Guo Z, Wu G, Trink B, et al. BRAF mutation in papillary thyroid carcinoma. J Natl Cancer Inst. 2003;95(8):625.
Kimura ET, Nikiforova MN, Zhu Z, Knauf JA, Nikiforov YE, Fagin JA. High prevalence of BRAF mutations in thyroid cancer: genetic evidence for constitutive activation of the RET/PTC-RAS-BRAF signaling pathway in papillary thyroid carcinoma. Can Res. 2003;63(7):1454.
Grieco M, Santoro M, Berlingieri MT, Melillo RM, Donghi R, Bongarzone I, et al. PTC is a novel rearranged form of the ret proto-oncogene and is frequently detected in vivo in human thyroid papillary carcinomas. Cell. 1990;60(4):557–63.
Eszlinger M, Paschke R. Molecular fine-needle aspiration biopsy diagnosis of thyroid nodules by tumor specific mutations and gene expression patterns. Mol Cell Endocrinol. 2010;322(1–2):29–37.
Yano Y, Uematsu N, Yashiro T, Hara H, Ueno E, Miwa M, et al. Gene expression profiling identifies platelet-derived growth factor as a diagnostic molecular marker for papillary thyroid carcinoma. Clin Cancer Res. 2004;10(6):2035–43.
Sheils O. Molecular classification and biomarker discovery in papillary thyroid carcinoma [J]. Expert Rev Mol Diagn. 2005;5(6):927–46.
Liu X, He M, Hou Y, Liang B, Zhao L, Ma S, et al. Expression profiles of microRNAs and their target genes in papillary thyroid carcinoma. Oncol Rep. 2013;29(4):1415–20.
Pallante P, Visone R, Ferracin M, Ferraro A, Berlingieri M, Troncone G, et al. MicroRNA deregulation in human thyroid papillary carcinomas. Endocr Relat Cancer. 2006;13(2):497–508.
Liang RQ, Li W, Li Y, Tan C, Li JX, Jin YX, et al. An oligonucleotide microarray for microRNA expression analysis based on labeling RNA with quantum dot and nanogold probe. Nucleic Acids Res. 2005;33(2):e17.
Qiu J, Zhang W, Xia Q, Liu F, Li L, Zhao S, et al. RNA sequencing identifies crucial genes in papillary thyroid carcinoma (PTC) progression. Exp Mol Pathol. 2015;100(1):151.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. https://doi.org/10.1093/bioinformatics/btp616.
Law CW, Chen Y, Shi W, Smyth GK. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15(2):R29.
Smyth GK. Limma: linear models for microarray data, in Bioinformatics and computational biology solutions using R and bioconductor. New York: Springer; 2005. p. 397–420.
Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–52. https://doi.org/10.1093/nar/gku1003.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504.
Tang Y, Li M, Wang J, Pan Y, Wu FX. CytoNCA: a cytoscape plugin for centrality analysis and evaluation of biological networks. BioSystems. 2014;11:005. https://doi.org/10.1016/j.biosystems.
He X, Zhang J. Why do hubs tend to be essential in protein networks? PLoS Genet. 2006;2(6):e88. https://doi.org/10.1371/journal.pgen.0020088.
Breuer K, Foroushani AK, Laird MR, Chen C, Sribnaia A, Lo R, et al. InnateDB: systems biology of innate immunity and beyond—recent updates and continuing curation. Nucleic Acids Res. 2013;41(Database issue):D1228–33. https://doi.org/10.1093/nar/gks1147.
Dweep H, Gretz N. miRWalk2. 0: a comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12(8):697.
Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–7.
Hirohashi S, Kanai Y. Cell adhesion system and human cancer morphogenesis. Cancer Sci. 2003;94(7):575–81.
Yang L, Froio RM, Sciuto TE, Dvorak AM, Alon R, Luscinskas FW. ICAM-1 regulates neutrophil adhesion and transcellular migration of TNF-alpha-activated vascular endothelium under flow. Blood. 2005;106(2):584.
Buitrago D, Keutgen X, Crowley M, Filicori F, Aldailami H, Hoda R, et al. Intercellular adhesion molecule-1 (ICAM-1) is upregulated in aggressive papillary thyroid carcinoma. Ann Surg Oncol. 2012;19(3):973–80.
Skelding KA, Barry RD, Shafren DR. Systemic targeting of metastatic human breast tumor xenografts by Coxsackievirus A21. Breast Cancer Res Treat. 2009;113(1):21–30.
Husmark J, Heldin NE, Nilsson M. N-cadherin-mediated adhesion and aberrant catenin expression in anaplastic thyroid-carcinoma cell lines. Int J Cancer. 1999;83(5):692–9.
Zhang X, Liu G, Kang Y, Dong Z, Qian Q, Ma X. N-cadherin expression is associated with acquisition of EMT phenotype and with enhanced invasion in erlotinib-resistant lung cancer cell lines. PLoS ONE. 2013;8(3):e57692.
Richards EJ, Zhang G, Li ZP, Permuth-Wey J, Challa S, Li Y, et al. Long non-coding RNAs (LncRNA) regulated by transforming growth factor (TGF) β: LncRNA-hit-mediated TGFβ-induced epithelial to mesenchymal transition in mammary epithelia. J Biol Chem. 2015;290(11):6857–67.
Ferraro A, Boni T, Pintzas A. EZH2 regulates cofilin activity and colon cancer cell migration by targeting ITGA2 gene. Plos ONE. 2014;9(12):e115276.
Prasad ML, Pellegata NS, Huang Y, Nagaraja HN, de la Chapell A, Kloos RT. Galectin-3, fibronectin-1, CITED-1, HBME1 and cytokeratin-19 immunohistochemistry is useful for the differential diagnosis of thyroid tumors. Mod Pathol. 2004;18(1):48–57.
Sugenoya A, Usuda N, Adachi W, Oohashi M, Nagata T, Iida F. Immunohistochemical studies on the localization of fibronectin in human thyroid neoplastic tissues. Endocrinol Jpn. 1988;35(1):111.
Sponziello M, Rosignolo F, Celano M, Maggisano V, Pecce V, De Rose RF, et al. Fibronectin-1 expression is increased in aggressive thyroid cancer and favors the migration and invasion of cancer cells. Mol Cell Endocrinol. 2016;431:123–32.
Lu P, Weaver VM, Werb Z. The extracellular matrix: a dynamic niche in cancer progression. J Cell Biol. 2012;196(4):395.
Martin DC, Sanchezsweatman OH, Ho AT, Inderdeo DS, Tsao MS, Khokha R. Transgenic TIMP-1 inhibits simian virus 40 T antigen-induced hepatocarcinogenesis by impairment of hepatocellular proliferation and tumor angiogenesis. Lab Invest. 1999;79(2):225–34.
Murphy G, Docherty AJP. The matrix metalloproteinases and their inhibitors. Am J Respir Cell Mol Biol. 1992;7(2):120.
Khokha R, Zimmer MJ, Graham CH, Lala PK, Waterhouse P. Suppression of invasion by inducible expression of tissue inhibitor of metalloproteinase-1 (TIMP-1) in B16-F10 melanoma cells. J Natl Cancer Inst. 1992;84(13):1017–22.
Tsuchiya Y, Sato H, Endo Y, Okada Y, Mai M, Sasaki T, et al. Tissue inhibitor of metalloproteinase 1 is a negative regulator of the metastatic ability of a human gastric cancer cell line, KKLS, in the chick embryo. Can Res. 1993;53(6):1397.
Talvensaari-Mattila A, Turpeenniemi-Hujanen T. High preoperative serum TIMP-1 is a prognostic indicator for survival in breast carcinoma. Breast Cancer Res Treat. 2005;89(1):29–34.
Tetzlaff MT, Liu A, Xu X, Master SR, Baldwin DA, Tobias JW, et al. Differential expression of miRNAs in papillary thyroid carcinoma compared to multinodular goiter using formalin fixed paraffin embedded tissues. Endocr Pathol. 2007;18(3):163–73.
Liu L, Wang J, Li X, Ma J, Shi C, Zhu H, et al. MiR-204-5p suppresses cell proliferation by inhibiting IGFBP5 in papillary thyroid carcinoma. Biochem Biophys Res Commun. 2015;457(4):621–6.
Carmeliet P, Jain RK. Angiogenesis in cancer and other diseases. Nature. 2000;407(6801):249–57.
Tanaka K, Sonoo H, Kurebayashi J, Nomura T, Ohkubo S, Yamamoto Y, et al. Inhibition of infiltration and angiogenesis by thrombospondin-1 in papillary thyroid carcinoma. Clin Cancer Res. 2002;8(5):1125–31.
Brennan K, Offiah G, Mcsherry EA, Hopkins AM. Tight junctions: a barrier to the initiation and progression of breast cancer? J Biomed Biotechnol. 2010;2010(1):460607.
Wang X, Tang S, Le SY, Lu R, Rader JS, Meyers C, et al. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS ONE. 2008;3(7):e2557.
Li N, Tang B, Zhu ED, Li BS, Zhuang Y, Yu S, et al. Increased miR-222 in H. pylori-associated gastric cancer correlated with tumor progression by promoting cancer cell proliferation and targeting RECK. FEBS Lett. 2012;586(6):722–8.
Chou CK, Chen RF, Chou FF, Chang HW, Chen YJ, Lee YF, et al. miR-146b is highly expressed in adult papillary thyroid carcinomas with high risk features including extrathyroidal invasion and the BRAF(V600E) mutation. Thyroid. 2010;20(5):489.
Chang WSW, Chou RH, Wu CW, Chang JY. Human tissue kallikreins as prognostic biomarkers and as potential targets for anticancer therapy. Expert Opin Ther Pat. 2007;17(10):1227–40.
Macewan M, Mehta V, Darr EA, Xie S, Nowicki TS, Iacob C, et al. Abstract 1502: increased expression of KLK7 and KLK10 in papillary thyroid cancer. Cancer Res. 2011;71(8 Supplement):1502.
Xu X, Cai N, Zhi T, Bao Z, Wang D, Liu Y, et al. MicroRNA-1179 inhibits glioblastoma cell proliferation and cell cycle progression via directly targeting E2F transcription factor 5. Am J Cancer Res. 2017;7(8):1680–92.
Authors’ contributions
Conception and design of the research: JQ, YS. Acquisition of data: JQ, WZ. Analysis and interpretation of data: JQ, CZ, XL. Statistical analysis: WZ, FL, RG. Drafting the manuscript: JQ, WZ. Revision of manuscript for important intellectual content: JQ, YS. All authors read and approved the final manuscript.
Acknowledgements
None.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
Not applicable.
Consent for publication
Not applicable.
Ethics approval and consent to participate
This study was approved by Ethics Committee of The affiliated Hospital of Qingdao University.
Funding
This work was supported by the Qingdao Municipal Science and Technology Bureau (Grant Number 13-1-4-166-jch).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Qiu, J., Zhang, W., Zang, C. et al. Identification of key genes and miRNAs markers of papillary thyroid cancer. Biol Res 51, 45 (2018). https://doi.org/10.1186/s40659-018-0188-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40659-018-0188-1