Abstract
Background
Diet analysis is essential to understanding the functional role of large bird species in food webs. Morphological analysis of regurgitated bird pellet contents is time intensive and may underestimate biodiversity. DNA metabarcoding has the ability to circumvent these issues, but has yet to be done.
Methods
We present a pilot study using DNA metabarcoding of MT-RNR1 and MT-CO1 markers to determine the species of origin and prey of 45 pellets collected in Qinghai and Gansu Provinces, China.
Results
We detected four raptor species [Eurasian Eagle Owl (Bubo bubo), Saker Falcon (Falco cherrug), Steppe Eagle (Aquila nipalensis), and Upland Buzzard (Buteo hemilasius)] and 11 unique prey species across 10 families and 4 classes. Mammals were the greatest detected prey class with Plateau Pika (Ochotona curzoniae) being the most frequent. Observed Shannon’s and Simpson’s diversity for Upland Buzzard were 1.089 and 0.479, respectively, while expected values were 1.312 ± 0.266 and 0.485 ± 0.086. For Eurasian Eagle Owl, observed values were 1.202 and 0.565, while expected values were 1.502 ± 0.340 and 0.580 ± 0.114. Interspecific dietary niche partitioning between the two species was not detected.
Conclusions
Our results demonstrate successful use of DNA metabarcoding for understanding diet via a novel noninvasive sample type to identify common and uncommon species. More work is needed to understand how raptor diets vary locally, and the mechanisms that enable exploitation of similar dietary resources. This approach has wide ranging applicability to other birds of prey, and demonstrates the power of using DNA metabarcoding to study species noninvasively.
Similar content being viewed by others
Background
Understanding predator–prey interactions is an important component of community ecology and management (Estes et al. 2011). Sympatric species with similar ecological demands must find ways to reduce competition. One way this is accomplished is through dietary niche partitioning (Schoener 1974). Understanding this overlap can discern how species allocate resources. Such knowledge is important for conservation planning as communities with lower niche overlap can support greater biodiversity (Pianka 1974). Unfortunately, food web dynamics are complex and require accurate information of items consumed (Pompanon et al. 2012). Understanding the trophic niche of birds is important as knowledge of prey composition plays an important role in shaping conservation policies (Grier 1982). Dietary assessment methods for avian species include direct observation (Margalida et al. 2005, 2009), camera placement at nest sites (Margalida et al. 2005; Tremblay et al. 2005), stomach pumping (Wilson 1984; Walter and O’Neill 1986), examination of stomach contents (Miller and McEwen 1995), digestive tract flushing (Moody 1970), forced vomiting (Valera et al. 1997), examination of fatty acid and isotope signatures (Iverson et al. 2007), molecular fecal analysis (Treves et al. 2016; Jedlicka et al. 2017; Trevelline et al. 2018a, b) and morphological investigation of pellets (Li et al. 2004).
Bird pellets are the accumulation of undigested prey that are regurgitated through the mouth in compact units (Taberlet and Fumagalli 1996). Examination of diet via pellets has historically involved collection and morphological assessment of contents (Ewins et al. 1994; Symondson 2002; Sándor and Ionescu 2009). This method has many shortcomings. Digestive processes may render samples unrecognizable (Symondson 2002; Galan et al. 2012), pellet appearance can vary widely based on life stage and sex (Galan et al. 2012), small to medium size prey are often overestimated and unusual prey items unrecorded (Marchesi et al. 2002), and taxonomic specialists are needed for wide ranging genera (Galan et al. 2012).
Molecular approaches involving the examination of DNA can circumvent these issues and increase the detectable prey spectrum relative to sampling and analysis effort (Oehm et al. 2017). DNA sequencing techniques have been called for since the early 2000s (Symondson 2002), but remain under used despite the stability of DNA in bird pellets (Taberlet and Fumagalli 1996), and the advent of Next-Generation Sequencing (NGS) technology which can identify species from a high volume of samples (Galan et al. 2012). In DNA metabarcoding, generalized primers target and amplify a segment, the DNA barcoding region, of a conserved gene (Pompanon et al. 2012). This gene must have low intra-species variation (Galan et al. 2012) and high inter-species variation for taxonomic classification (Simon et al. 1994). Degradation of longer DNA segments remains problematic. Thus, researchers rely on shorter segments known as “mini-barcodes” (Meusnier et al. 2008). Researchers also use mitochondrial genes, as they have higher copy numbers compared to nuclear genes and greater PCR amplification success (Freeland 2017).
MT-RNR1 is an RNA gene previously used in studies examining the identity and genetic relationships of animals (Riaz et al. 2011). However, MT-RNR1 lacks the genetic diversity to discern wild versus domestic goat (Capra hircus) and sheep species (Shehzad et al. 2012; Hacker et al. 2021). This is problematic for the discernment of domestic sheep and argali (Ovis ammon), as well as domestic goat and Siberian Ibex (Capra sibirica), in areas where they are sympatric (Reading et al. 2020). Thus, an additional marker is necessary. MT-COI is widely used for DNA barcoding as it has both conserved regions and segments with high divergence (Hebert et al. 2003). Recent work identified a segment of MT-COI capable of discerning wild and domestic goat and sheep taxa in Central Asia, making it an important addition for diet research of predators and scavengers distributed there (Hacker et al. 2021).
Numerous sympatric birds of prey are found on the Qinghai-Tibetan Plateau (QTP). They play vital roles in maintaining ecosystem balance (Xia et al. 1991) under the context of vast modernization and environmental shifts (Liu and Chen 2000; Foggin 2008). They face threats associated with climate change (Liu and Chen 2000) as well as anthropogenically-induced mortality, such as electrocution from power lines (Dixon et al. 2013) and pika poisoning (Badingqiuying et al. 2016). Raptor species include the Eurasian Eagle Owl (Bubo bubo) (Birdlife International 2017), Upland Buzzard (Buteo hemilasius), Northern Goshawk (Accipiter gentilis), Golden Eagle (Aquila chrysaetos), Saker Falcon (Falco cherrug) (Dixon et al. 2013; Birdlife International 2018a), Steppe Eagle (Aquila nipalensis), Himalayan Vulture (Gyps himalayensis), and Bearded Vulture (Gypaetus barbatus) (Schaller 1998; Cui et al. 2008; Birdlife International 2019). Understanding their diets is necessary for effective conservation action, and DNA metabarcoding of regurgitated pellets provides a method to do so noninvasively.
The aims of this study were to (1) determine applicability of DNA metabarcoding to species and prey identification of avian pellets; (2) examine metrics surrounding species presence and mechanisms of coexistence; and (3) make suggestions based on our results for conservation action planning.
Methods
Study site
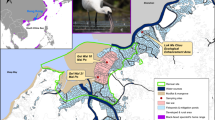
Samples were collected in the Qilian Shan Mountains (hereafter “Qilian Shan”) of Qinghai and Gansu Provinces, China, in the eastern Kunlan Mountains in Dulan County, Qinghai Province, China and in Zhiduo County, Yushu Prefecture, Qinghai Province, China (Fig. 1). Qilian Shan runs along the northeastern corner of the QTP (> 3000 m above sea level). It comprises three parallel subsidiary ranges—the Tuali Nanshan, Shule Nanshan, and Danghe Nanshan (Schaller et al. 1988). Qilian Shan is composed of deserts at lower elevations giving way to shrubs, grasses, and alpine meadows (Schaller et al. 1988). Yushu Prefecture is in the southwestern corner of Qinghai Province, and has alpine meadow vegetation with small rugged ranges surrounded by rolling grassland, with juniper forests along mountainsides (Schaller et al. 1988). The Kunlan Mountains are the longest mountain system in Asia with its eastern end south west of Qilian Shan (Miller and Bedunah 1994). The landscape of Dulan County is primarily rugged grassland with rock slopes (Liu 1993). Species found on the QTP include the Blue Sheep (Pseudois nayaur), Tibetan Gazelle (Procapra picticaudata), Argali (Ovis ammon), and White-lipped Deer (Cervus albirostris), and carnivores such as the Tibetan Wolf (Canis lupus), Red Fox (Vulpes vulpes), Snow Leopard (Panthera uncia), Eurasian Lynx (Lynx lynx), and Pallas’s Cat (Otocolobus manul), among others. Small mammals include voles (Neodon sp.), pikas (Ochotona sp.), zokors (Myospalax sp.), Woolly Hare (Lepus oiostolus), and Himalayan Marmots (Marmota himalyana) (Schaller et al. 1988; Jackson 2012).
Sample collection
Permits were obtained prior to sample collection. Pellets were collected opportunistically as part of a separate snow leopard study over seven sampling trips from September 2017 to July 2019. Sampling methods are described in Janečka et al. (2008, 2011).
DNA extraction
Samples were stored in a − 20 °C freezer. Pellets were defrosted and individually placed in a Petri dish. Tweezers were then used to remove material from the outside and inside the pellet to capture DNA of both the prey and host and placed in a 1.5 mL centrifuge tube. DNA was extracted using QIAamp DNA Stool Mini Kits (QIAGEN, Hilden, Germany) with two additional centrifugation steps to remove residual Buffer AW1 and AW2. Aliquots were quantified using a NanoDrop Lite Spectrophotometer.
PCR for species and diet analysis
A short segment (~ 100-bp) of MT-RNR1 (primers 12SV5F/12SV5R; Riaz et al. 2011) was used to discern all predator and prey species except those belonging to goats and sheep. The primers for MT-CO1 (MT-CO1-379F and MT-CO1-604Rd; Hacker et al. 2021) were designed to amplify a gene segment (330-bp) capable of differentiating closely related caprines. Each segment was amplified separately using a PCR reaction containing 1.5 µL of DNA template, 7.94 µL of KAPA HiFi HotStart ReadyMix (2 ×) (Kapa Biosystems, Wilmington, MA, USA), 0.16 µL of 20 µM forward primer, 0.16 µL of 20 µM reverse primer, and 5.2 µL of PCR grade water. PCR conditions consisted of 95 °C for 3 min, 35 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s, followed by a 5 min extension step at 72 °C and 4 °C hold.
Next-generation sequencing
Amplicons were mixed in equal ratios determined by measuring gel band brightness using GeneTools Analysis Software Version 4.03.05.0 (SynGene, Frederick, MD, USA). The E.Z.N.A.® Gel Extraction Kit (Omega Bio-Tek, Inc., Norcross, GA, USA) was used to isolate and purify products. The NEBNext® Ultra™ II DNA Library Prep Kit for Illumina® (New England Biolabs, MA, USA) was used to prepare sequencing libraries. Indexing oligonucleotides (Nextera XT, Illumina, San Diego, CA, USA) were incorporated and the pooled ampliconic library quantified using an Invitrogen Qubit® 2.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). Paired-end 250-bp sequencing was completed on an Illumina NovaSeq 6000 by Guangdong Magigene Biotechnology Co., Ltd. (Guangzhou, China).
Diet analysis
FASTQ sequences were demultiplexed, sequencing adapters removed, and reads imported into CLC Genomics Workbench v12.0 (CLC bio, QIAGEN, Aarhus, Denmark). Raw sequencing reads were trimmed using a quality score limit of 0.1. A reference FASTA file was created by downloading MT-CO1 and MT-RNR1 sequences for potential prey and host species, which were determined via a literature search and by consulting with local experts (Additional file 1). Sequencing reads were then mapped to the reference FASTA file using local alignment with the following parameters—mismatch cost: 2; insertion cost: 3; deletion cost: 3; length fraction: 0.9; similarity: 0.94; non-specific matches mapped randomly (Hacker et al. 2021). To identify the host species of each pellet, identification was made when the sequenced reads were mapped to a reference sequence with > 98% similarity. Unique haplotypes of the MT-RNR1 segment for each host species were determined in MEGA 7.0 by aligning sequences using MUSCLE (Edgar 2004). Prey identification was made by selecting the prey species from the reference database with the highest number of mapped reads and fewest number of mismatches. To ensure prey DNA sequences were correctly classified, the consensus sequence for the host and prey items were extracted and a blastn search performed against the nr/nt nucleotide collection GenBank databases using megablast for highly similar sequences (Additional file 2). In addition, collection sites were compared with known species distributions (CITES Red List range maps, https://www.iucnredlist.org/search/map).
Samples with a large proportion of unmapped reads (> 5% of total reads) were analyzed to rule out an incomplete reference file. This was done by performing a de novo assembly with the following parameters—minimum contig length of 100; mismatch cost: 2; insertion cost: 3; deletion cost: 3; length fraction: 0.9; similarity: 0.98. Consensus sequences were extracted for contigs with the highest number of mapped reads. At least 10,000 reads were required to generate a consensus sequence. Nucleotides in sites with conflicting reads were resolved via majority rule and ambiguous sites were coded with an “N”. Species were identified using the same blastn search described above.
Statistical analysis
All statistical analyses were performed in R version 3.5.2 (R Core Team 2018) using base R functions, vegan (Oksanen et al. 2013), iNEXT (Hsieh et al. 2016), and EcoSimR packages (Gotelli et al. 2015). The mean number of unique prey taxa per pellet was calculated by summing the number of detected prey species among each pellet and dividing this sum by the number of pellets analyzed. This calculation was also repeated for each bird species sampled for interspecific comparisons. Dietary frequency of occurrence was calculated by dividing the number of pellets in which a prey species was detected by the total number of pellets for each predator species. Observed dietary taxonomic richness, as well as Shannon’s index (Shannon and Weaver 1949) and Simpson’s index (Simpson 1949; Marchesi et al. 2002) were calculated using the “iNEXT” function (Hsieh et al. 2016). Effective taxonomic richness and diversity of predator diet was calculated using Hill numbers (Hill 1973) and the “iNEXT” function (Hsieh et al. 2016). This enabled comparison of dietary diversity between species with varying sample sizes (Hurlbert 1971; Heck et al. 1975) and allowed for evaluation of sampling completeness.
For the Upland Buzzard and the Eurasian Eagle Owl, Pianka’s metric of niche overlap (Pianka 1974) was calculated using the ‘niche_null_model’ function in the EcoSimR package (Gotelli et al. 2015) and compared to 999 null model simulations to determine if observed dietary niche overlap was higher or lower than expected. The occurrence of interspecific niche differentiation between these two species was determined by performing a permutational multivariate analysis of variance (PERMANOVA; Anderson 2017) using the “adonis” function and Jaccard distance matrix within the vegan R package (Oksanen et al. 2013). Additional information on statistical analyses can be found in Additional file 3.
Results
Predator species detected
Using the MT-RNR1 genetic marker, four host bird species were identified among 45 pellets—Upland Buzzard (n = 26, 23 from Qilian Shan and 3 from Yushu), Eurasian Eagle Owl (n = 15, 14 from Qilian Shan and 1 from Yushu), Steppe Eagle (n = 3, from Dulan County), and Saker Falcon (n = 1, from Qilian Shan) (Fig. 2). A total of 8 unique MT-RNR1 haplotypes were found for Upland Buzzard, 5 for Eurasian Eagle Owl, 1 for Saker Falcon, and 1 for Steppe Eagle (Additional file 4).
The host bird species identified via DNA-metabarcoding of bird pellets collected on the Qinghai-Tibetan Plateau. a Steppe Eagle (photo credit: Jia Li). b Upland Buzzard (photo credit: Charlotte Hacker). c Saker Falcon (photo credit: Charlotte Hacker). d Eurasian Eagle Owl (photo credit: Munib Khanyari). Figure made in Microsoft PowerPoint for Mac version 16.42
Predator diet composition
A total of 41 of the 45 collected pellets (91.1%) had prey DNA sequences discerned. A total of 11 unique prey taxa spanning 7 orders and 4 classes were detected (Table 1). The most frequently detected prey species, Plateau Pika (Ochotona curzoniae), was found in 77.8% (35/45) of all diets analyzed and was the only prey taxa detected in the Saker Falcon samples (Table 1). While Mammalia was the most taxonomically-rich prey class (7 species; Table 1), predator species in this study also consumed Ray-finned Fishes (Actinopterygii, 2 species), small birds (Aves), and one species of toad (Amphibia).
Dietary richness, diversity and overlap
The average number of unique prey taxa detected in a single pellet across all species was 1.16 (range 0–3), with Upland Buzzard and Eurasian Eagle Owl having an average of 1.16 and 1.13 unique prey taxa per pellet, respectively. We observed the average number of unique prey taxa per pellet for Saker Falcon and Steppe Eagle in our study to be 1 and 0.333, respectively. Our analysis detected 7 prey species within pellets collected from Upland Buzzard, 6 from Eurasian Eagle Owl, and one each for Steppe Eagle and Saker Falcon. Steppe Eagle (n = 3) and Saker Falcon (n = 1) were removed from further analyses due to limited sample sizes.
Though these data are preliminary, they provide an important starting point for understanding dietary overlap between two large bird species. The expected dietary richness of Upland Buzzard and Eurasian Eagle Owl was 14.692 ± 11.227 (mean ± SE) and 11.6 ± 6.594, respectively (Fig. 3). DNA barcoding detected 87.3% of the Upland Buzzard’s and 79.9% of the Eurasian Eagle Owl’s expected prey taxa. Observed Shannon’s and Simpson’s diversity for Upland Buzzard were 1.089 and 0.479, respectively, while expected values were 1.312 ± 0.266 and 0.485 ± 0.086. Observed Shannon’s and Simpson’s diversity for Eurasian Eagle Owl were 1.202 and 0.565, while expected values were 1.502 ± 0.340 and 0.580 ± 0.114, respectively. Our PERMANOVA results did not detect interspecific dietary niche partitioning between Upland Buzzard and Eurasian Eagle Owl (Pseudo-F(1, 36) = 1.86, p = 0.129) and Pianka’s index of dietary niche overlap was calculated to be 0.511, within the expected 95% confidence interval derived from null model simulations (CI range: 0.477–0.522).
Estimation of prey taxonomic richness with rarefaction and extrapolation. Using the iNEXT function, rarefaction curves were made to estimate sample completeness and expected taxonomic richness with additional sampling for the Eurasian Eagle Owl (n = 15) and Upland Buzzard (n = 26). Interpolation (solid lines) was performed using the sample sizes for each predator species while extrapolation (dotted lines) of species richness was performed on double the sample size. Figure made in R version 3.5.2
Discussion
Pika was the dominant consumed species in this study. Pikas are non-hibernating and diurnal, providing a year-long accessible food source (Badingqiuying et al. 2016). Pikas have been labeled as pests, with a poisoning program to control and eradicate them launched in 1958 (Smith et al. 1990; Smith and Foggin 1999). However, more recent research recognizes pikas as important environmental engineers that cause minimal damage to alpine grassland ecosystems (Smith and Foggin 1999; Wei et al. 2020). Poisoning them is more likely to have negative impacts (Lai and Smith 2003; Badingqiuying et al. 2016). In a study by Badingqiuying et al. (2016), recordings of Steppe Eagles, Saker Falcons, and Upland Buzzards were three times less frequent at sites with active poisoning programs compared to those without; though data were insufficient to make any conclusions about Eurasian Eagle Owls. Similarly, Lai and Smith (2003) reported that Upland Buzzards were found 11.2 times more frequently at non-poisoned versus poisoned pika sites.
The one pellet collected from a Saker Falcon revealed pika as the dietary item. In previous studies, pikas were found to comprise 90% of the food items a Saker Falcon pair fed to their young in the Chang Tang region (Schaller 2012). In this study, pikas were detected in 80% of pellets from Eurasian Eagle Owl and 84.6% of Upland Buzzard pellets, aligning with previous studies. Our results further substantiate the important role pika play as a primary food source for large bird species. However, some species may be able to adapt to reductions in pika populations by exploiting other prey species (Cui et al. 2008).
Dietary diversity assessments could only be done with Upland Buzzards and Eurasian Eagle Owls due to the low sample size of Saker Falcon and Steppe Eagle pellets. While the Upland Buzzard had one more unique species represented in their diet compared to the Eurasian Eagle Owl, the observed diversity indices suggested that Eurasian Eagle Owls exhibit greater dietary taxonomic diversity. The values resulting from the Shannon-Weiner diversity index and Simpson’s index between the two species were relatively similar with a difference of 0.113 for Shannon-Weiner and 0.086 for Simpson’s. Previous work by Cui et al. (2008), who compared the dietary diversity of Upland Buzzards and Eurasian Eagle Owls via morphological assessment of regurgitated pellets found that Eurasian Eagle Owls exhibited higher, but similar diversity to that of the Upland Buzzard with a difference between the two species of 0.17. When calculated to expected degrees of dietary diversity, differences between Upland Buzzard and Eurasian Eagle Owl in this study were a bit larger, with a difference of 0.19 expected for Shannon-Weiner and 0.095 for Simpson’s. Greater expected dietary diversity for Eurasian Eagle Owls is not unexpected as they are known to be generalists (Hiraldo et al. 1975).
Despite the relatively small number of pellets collected, the sampling effort in this study captured 87.3% of the expected prey taxa for the Upland Buzzard and 79.9% of expected prey taxa for the Eurasian Eagle Owl. Species that have been previously recorded as present in the diets of birds of prey living on the QTP but not found in this study include Zokors (Myospalax fontanierii; M. baileyi), Gansu Pika (Ochotona cansus), Root Voles (Micrtous oeconomus), Plateau Voles (Lasiopodomys fuscus), and Mountain Weasels (Mustela altaica) (Lai and Smith 2003; Li et al. 2004; Cui et al. 2008). The presence and frequency of food items in the diets of birds of prey are typically reflective of prey presence and frequency in their habitats (Bontzorlos et al. 2005). While data could reflect low numbers or absence of expected species that may spark concern surrounding conservation status of wildlife, species absence in this dataset is most likely due to low sample size or a result of variable methodology in comparison to previous studies.
We found no evidence of dietary niche partitioning between Upland Buzzard and Eurasian Eagle Owl, indicating that temporal or spatial partitioning is responsible for coexistence. Previous work by Cui et al. (2008) had similar findings. Temporally, Eurasian Eagle Owls are nocturnal while Upland Buzzards are diurnal (Lei 1995; Yang et al. 2000), and resource exploitation may occur at different parts of the day. Across a broader temporal scale, Upland Buzzards partially migrate, spending the breeding season in China, while Eurasian Eagle Owls are not migratory (Birdlife International 2017). However, with the exception of two samples collected in September, all remaining pellets were collected during the corresponding breeding season for Upland Buzzards (April through July) (Rasmussen and Anderson 2005), and both would be expected to be present. Eurasian Eagle Owls and Upland Buzzards also deploy different hunting strategies which may separate them behaviorally. Eurasian Eagle Owls are opportunistic (Hiraldo et al. 1975), swooping down on prey and departing whether the kill is successful or not, while Upland Buzzards deploy a sit-and-wait strategy (Cui et al. 2008). In addition, we observed ungulates (Domestic Goat and Blue Sheep) in two Upland Buzzard pellets, likely attributed to scavenging.
Unique and previously under-reported prey items were identified. It may be that birds at present are rapidly adapting to changes in prey base; however, it is also possible that traditional methods reliant on the morphological assessment of pellets may have missed rare or unexpected prey items. In this study, Przewalski’s Naked Carp (Gymnocyris przewalksii) was found in one Eurasian Eagle Owl pellet. Przewalski’s Naked Carp is found in Qinghai Lake and is classified as Endangered on the China Species Red List (Chen et al. 2009; Zhang et al. 2015). Loss of the species could cause severe negative consequences within the lake (Chen et al. 2009). A previous study examining the distribution of Przewalski’s Naked Carp in Qinghai Lake showed that the species was found ~ 2 m below the surface (Chen et al. 2009), where individuals moving upward could be caught by birds of prey.
Although DNA metabarcoding is sensitive enough to detect the presence of rare and elusive species (Granjon et al. 2002; Thiam et al. 2008), the degree to which they are being predated would be unknown due to the inability to count the number of individuals consumed within in a pellet (Symondson 2002; Emmrich and Düttmann 2011). This may be problematic as raw occurrence counts of dietary items can artificially inflate the occurrence of rare food taxa (Deagle et al. 2019). Previous research has suggested that the number of mapped reads may be quasi-indicative of the quantity of a dietary item within a sample (Kartzinel et al. 2015; Deagle et al. 2019; Lamb et al. 2019). We greatly caution against this assumption, as read mapping success can be influenced by other factors such as PCR amplification bias, sample quality, and reference sequence availability (Pompanon et al. 2012; Lamb et al. 2019). Other potential pitfalls of DNA metabarcoding which should be considered include the level of DNA degradation and whether the genetic marker used provides necessary taxonomic resolution.
In this study, four of the pellets collected were unable to have prey DNA amplified. This may be because the primer pairs used did not amplify the prey’s target gene, or could be a result of DNA degradation. The QTP is a dry, cold environment. Regardless, DNA degradation may make the rendering of sequences from some samples impossible (Guimaraes et al. 2016). Shorter diagnostic fragments are preferred as they are more likely to remain intact (Meusnier et al. 2008). However, use of shorter DNA segments warrants caution as this may increase difficulty in discerning species (Symondson 2002), especially for those belonging to taxonomically-rich groups.
Rodents represent 40% of all mammalian species globally (Musser and Carleton 2005), and were found in our dataset. While unmapped reads were examined to identify any species potentially missing from the reference file, it remains possible that the MT-RNR1 segment does not amplify all possible rodent species within our study area. Rodents are a food source for many predators on the QTP, extending beyond birds (Hacker et al. 2021), and thus an accurate assessment of their consumption would be of use in large-scale conservation planning. Sequences which cannot be discerned down to the species-level may be grouped into a “prey OTU”, as commonly done in dietary and microbial studies for identification and analysis purposes (Marchesi et al. 2002). Galan et al. (2012) designed a mini-barcode marker based on a 136-bp DNA segment of MT-CYB capable of discerning a vast array of Rodentia. While our study did not face intensive challenges associated with rodent discernment, our sample size was small and concentrated in two areas. Study questions tied to rich taxonomic groups as a resource in an area with high biodiversity may want to consider the addition of such a marker. Researchers should also consider the likelihood of false identifications which may result in the query sequence being assigned to the incorrect taxon and subsequently choose appropriate sequence based species identification methods to increase confidence, accuracy, and efficiency of data processing (Soergel et al. 2012; Boyer et al. 2016; Shi et al. 2018).
The MT-COI marker was used as a diagnostic marker to differentiate wild and domestic caprid species found in the study area (Hacker et al. 2021). Though ungulates were not necessarily expected in the diet based on previously available literature, it was surmised that past work using morphological analysis may have missed these larger species as bones and other hard parts would likely not be ingested by birds due to their size. Indeed, our study found two occurrences of caprids in the diet of the Upland Buzzard, one matching to Domestic Goat and one to Blue Sheep. Unfortunately, and as would be an issue with morphological analyses, DNA metabarcoding is unable to distinguish between scavenging versus legitimate kills (Symondson 2002). Larger birds of prey, such as the Golden Eagle have been observed taking larger prey such as domestic calves and sheep in North America (Avery and Cummings 2004). Upland Buzzards are considerably smaller in size, and would most likely not be able to kill caprids; with the exception of newborns. Scavenging on the carcass of an already deceased individual or placenta is more likely. Determining whether native raptors kill newborn livestock would be of particular interest to herders, and would present a further challenge for conservation as this could lead to conflict.
The genetic identification of pellet hosts allowed for accurate information of species presence in a particular area. Of the 45 pellets identified in this study, only 3 belonged to Steppe Eagle and 1 to Saker Falcon. China is breeding habitat for both species, corresponding to when samples were collected, but both birds are classified as Endangered by the IUCN and lack of representation in our dataset is consistent with these low population numbers (Birdlife International 2018a, 2019). In contrast, the Upland Buzzard and Eurasian Eagle Owl are listed as Least Concern (Birdlife International 2017, 2018b). More detailed studies may be able to apply noninvasive surveys using regurgitated pellets and DNA metabarcoding to map geographic distribution or shifts in temporal presence during breeding seasons and migration events, as well as habitat preference and overall population status. This can similarly be applied to prey in an area, though birds may be moving long distances and thus the pellets they regurgitate may not be indicative of immediate local wildlife presence per se.
Albeit a small sample size, our findings provide information important for conservation action on the QTP. First, results demonstrate that pikas are an important food source, thus large-scale policies to eradicate them should be reconsidered. Second, Eurasian Eagle Owls and Upland Buzzards depend on similar dietary items. Persistence of these prey will be necessary for survival of both species. Eurasian Eagle Owls may be better able to exploit a wider range of prey as they demonstrate the ability to increase dietary plasticity, while Upland Buzzards may lack the adaptability to quickly changing and resource poor landscapes. Third, birds of prey may contribute to conflict with humans in the form of livestock depredation or death. Reports of such from local herders, or findings of larger than expected mammals in pellets, should be taken seriously to build mitigation actions for reducing livestock loss.
Conclusions
Biodiversity tends to be richer and more quickly changing than acknowledged, and assessment methods must be reliable and comprehensive (Pimm et al. 1995). DNA metabarcoding is a conservation technology tool capable of providing such information. We show the utility of this technique in determining dietary items from regurgitated bird pellets, and subsequently identified 11 unique prey species for 4 raptor species with pika confirmed as an important food resource, with high dietary niche overlap between Eurasian Eagle Owl and Upland Buzzard. While we focus on bird of prey species on the QTP, our methodology could be applied to pellet analysis for species worldwide. Our work also demonstrates the power of DNA metabarcoding with noninvasive samples, encouraging other sample types to also be explored. We also showed that sampling efforts may not need to be as intensive as previously assumed, given the relatively high number of observed taxa captured compared to the expected number of taxa despite limited sample sizes. This could decrease effort, time, and costs associated with fieldwork. However, we also contend that the application of this technique has great potential to answer complex questions surrounding birds of prey that will require rigorous and large-scale study design.
Availability of data and materials
The datasets generated during the current study are available via email to the corresponding authors.
References
Anderson MJ. Permutational multivariate analysis of variance (PERMANOVA). Wiley StatsRef Stat Refer Online. 2017. https://doi.org/10.1002/9781118445112.stat07841.
Avery ML, Cummings JL. Livestock depredations by black vultures and golden eagles. Sheep Goat Res J. 2004;19:58–63.
Badingqiuying, Smith AT, Senko J, Siladan MU. Plateau pika Ochotona curzoniae poisoning campaign reduces carnivore abundance in southern Qinghai, China. Mammal Study. 2016;41:1–8.
Birdlife International. Bubo bubo (amended version of 2016 assessment). The IUCN Red List of Threatened Species 2017: e.T22688927A113569670. 2017. https://doi.org/10.2305/IUCN.UK.2017-1.RLTS.T22688927A113569670.en.
Birdlife International. Falco cherrug (amended version of 2016 assessment). The IUCN Red List of Threatened Species 2017: e.T22696495A110525916. 2018a. https://doi.org/10.2305/IUCN.UK.2017-1.RLTS.T22696495A110525916.en.
Birdlife International. Buteo hemilasius. The IUCN Red List of Threatened Species 2018: e.T22695967A131937792. 2018b. https://doi.org/10.2305/IUCN.UK.2018-2.RLTS.T22695967A131937792.en.
Birdlife International. Aquila nipalensis (amended version of 2017 assessment). The IUCN Red List of Threatened Species 2019: e.T22696038A155419092. 2019. https://doi.org/10.2305/IUCN.UK.2019-3.RLTS.T22696038A155419092.en.
Bontzorlos VA, Peris SJ, Vlachos CG, Bakaloudis DE. The diet of barn owl in the agricultural landscapes of central Greece. Folia Zool. 2005;54:99–110.
Boyer F, Mercier C, Bonin A, Le Bras Y, Taberlet P, Coissac E. Obitools: a unix-inspired software package for DNA metabarcoding. Mol Ecol Resour. 2016;16:176–82.
Chen D, Zhang X, Tan X, Wang K, Qiao Y, Chang Y. Hydroacoustic study of spatial and temporal distribution of Gymnocypris przewalskii (Kessler, 1876) in Qinghai Lake, China. Environ Biol Fish. 2009;84:231–9.
Cui Q, Su J, Jiang Z. Summer diet of two sympatric species of raptors Upland Buzzard (Buteo hemilasius) and Eurasian Eagle Owl (Bubo bubo) in alpine meadow: problem of coexistence. Pol J Ecol. 2008;56:173–9.
Deagle BE, Thomas AC, McInnes JC, Clarke LJ, Vesterinen EJ, Clare EL, et al. Counting with DNA in metabarcoding studies: how should we convert sequence reads to dietary data? Mol Ecol. 2019;28:391–406.
Dixon A, Maming R, Gunga A, Purev-Ochir G, Batbayar N. The problem of raptor electrocution in Asia: case studies from Mongolia and China. Bird Conserv Int. 2013;23:520–9.
Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7.
Emmrich M, Düttmann H. Seasonal shifts in diet composition of Great Cormorants Phalacrocorax carbo sinensis foraging at a shallow eutrophic inland lake. Ardea. 2011;99:207–16.
Estes JA, Terborgh J, Brashares JS, Power ME, Berger J, Bond WJ, et al. Trophic downgrading of planet earth. Science. 2011;333:301–6.
Ewins PJ, Weseloh DV, Groom JH, Dobos RZ, Mineau P. The diet of Herring Gulls (Larus argentatus) during winter and early spring on the lower Great Lakes. Hydrobiologia. 1994;279(280):39–55.
Foggin JM. Depopulating the Tibetan grasslands: national policies and perspectives for the future of Tibetan herders in Qinghai Province, China. Mt Res Dev. 2008;28:26–31.
Freeland JR. The importance of molecular markers and primer design when characterizing biodiversity from environmental DNA. Genome. 2017;60:358–74.
Galan M, Pagès M, Cosson JF. Next-generation sequencing for rodent barcoding: species identification from fresh, degraded and environmental samples. PLoS ONE. 2012;7:e48374.
Gotelli NJ, Hart EM, Ellison AM. EcoSimR: null model analysis for ecological data. R Package version 0.1.0. 2015. https://doi.org/10.5281/zenodo.16522.
Granjon L, Bruderer C, Cosson JF, Dia AT, Colas F. The small mammal community of a coastal site of south-west Mauritania. Afr J Ecol. 2002;40:10–7.
Grier JW. Ban of DDT and subsequent recovery of reproduction in bald eagles. Science. 1982;218:1232–5.
Guimaraes S, Fernandez-Jalvo Y, Stoetzel E, Gorgé O, Bennett EA, Denys C, et al. Owl pellets: a wise DNA source for small mammal genetics. J Zool. 2016;298:64–74.
Hacker CE, Jevit M, Hussain S, Muhammad G, Munkhtsog B, Munkhtsog B, et al. Regional comparison of snow leopard (Panthera uncia) diet using DNA metabarcoding. Biodivers Conserv. 2021;30:797–817.
Hebert PDN, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proc R Soc B Biol Sci. 2003;270:313–21.
Heck KL, van Belle G, Simberloff D. Explicit calculation of the rarefaction diversity measurement and the determination of sufficient sample size. Ecology. 1975;56:1459–61.
Hill MO. Diversity and evenness: a unifying notation and its consequences. Ecology. 1973;54:427–32.
Hiraldo F, Andrada J, Parreño F. Diet of the eagle owl (Bubo bubo) in Mediterranean Spain. Doñana Acta Vertebr. 1975;2:161–77.
Hsieh TC, Ma KH, Chao A. iNEXT: an R package for rarefaction and extrapolation of species diversity (Hill numbers). Method Ecol Evol. 2016;7:1451–6.
Hurlbert SH. The nonconcept of species diversity: a critique and alternative parameters. Ecology. 1971;52:577–86.
Iverson SJ, Springer AM, Kitaysky AS. Seabirds as indicators of food web structure and ecosystem variability: qualitative and quantitative diet analyses using fatty acids. Mar Ecol Prog Ser. 2007;352:235–44.
Jackson R. Fostering community-based stewardship of wildlife in Central Asia: transforming snow leopards from pests into valued assets. In: Squires VR, editor. Rangeland stewardship in Central Asia. Dordrecht: Springer; 2012. p. 357–80.
Janečka JE, Jackson R, Yuquang Z, Diqiang L, Munkhtsog B, Buckley-Beason V, et al. Population monitoring of snow leopards using noninvasive collection of scat samples: a pilot study. Anim Conserv. 2008;11:401–11.
Janečka JE, Munkhtsog B, Jackson RM, Naranbaatar G, Mallon DP, Murphy WJ. Comparison of noninvasive genetic and camera-trapping techniques for surveying snow leopards. J Mammal. 2011;92:771–83.
Jedlicka JA, Vo ATE, Almeida RPP. Molecular scatology and high-throughput sequencing reveal predominately herbivorous insects in the diets of adult and nestling Western Bluebirds (Sialia mexicana) in California vineyards. Auk. 2017;134:116–27.
Kartzinel TR, Chen PA, Coverdale TC, Erickson DL, Kress WJ, Kuzmina ML, et al. DNA metabarcoding illuminates dietary niche partitioning by African large herbivores. Proc Natl Acad Sci USA. 2015;112:8019–24.
Lai CH, Smith AT. Keystone status of plateau pikas (Ochotona curzoniae): effect of control on biodiversity of native birds. Biodivers Conserv. 2003;12:1901–12.
Lamb PD, Hunter E, Pinnegar JK, Creer S, Davies RG, Taylor MI. How quantitative is metabarcoding: a meta-analytical approach. Mol Ecol. 2019;28:420–30.
Lei F. A study on diet of the Little Owl (Athene noctua plumipes) in Qishan, Shanxi Province, China. Wuyi Sci J. 1995;12:136–42 (in Chinese).
Li L, Yi X, Li M, Zhang X. Diet composition of upland buzzard: analysis on stomach content and food pellet. Zool Res. 2004;25:162–5 (in Chinese).
Liu X, Chen B. Climatic warming in the Tibetan Plateau during recent decades. Int J Climatol. 2000;20:1729–42.
Liu Y. International hunting and the involvement of local people, Dulan, Qinghai, People’s Republic of China. Master’s Thesis. Missoula: University of Montana; 1993.
Marchesi L, Pedrini P, Sergio F. Biases associated with diet study methods in the Eurasian Eagle-Owl. J Raptor Res. 2002;36:11–6.
Margalida A, Bertran J, Boudet J. Assessing the diet of nestling Bearded Vultures: a comparison between direct observation methods. J Field Ornithol. 2005;76:40–5.
Margalida A, Bertran J, Heredia R. Diet and food preferences of the endangered Bearded Vulture Gypaetus barbatus: a basis for their conservation. Ibis. 2009;151:235–43.
Meusnier I, Singer GAC, Landry JF, Hickey DA, Hebert PDN, Hajibabaei M. A universal DNA mini-barcode for biodiversity analysis. BMC Genomics. 2008;9:214.
Miller C, McEwen L. Diet of nesting Savannah Sparrows in interior Alaska. J Field Ornithol. 1995;66:152–8.
Miller DJ, Bedunah DJ. Rangelands of the Kunlun Mountains in Western China. Rangelands. 1994;16:71–6.
Moody T. A method for obtaining food samples from insectivorous birds. Auk. 1970;87:579.
Musser GG, Carleton MD. Superfamily Muroidea. In: Wilson D, Reeder D, editors. Mammal species of the world: a taxonomic and geographic reference. 3rd ed. Baltimore: John Hopkins University; 2005. p. 894–1531.
Oehm J, Thalinger B, Eisenkölbl S, Traugott M. Diet analysis in piscivorous birds: what can the addition of molecular tools offer? Ecol Evol. 2017;7:1984–95.
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, et al. Package vegan: community ecology package. R Package. version 2.3-1. 2013.
Pianka ER. Niche overlap and diffuse competition. Proc Natl Acad Sci USA. 1974;71:2141–5.
Pimm SL, Russell GJ, Gittleman JL, Brooks TM. The future of biodiversity. Science. 1995;269:347–50.
Pompanon F, Deagle BE, Symondson WOC, Brown DS, Jarman SN, Taberlet P. Who is eating what: diet assessment using next generation sequencing. Mol Ecol. 2012;21:1931–50.
Rasmussen P, Anderson J. Birds of South Asia: the ripley guide. Washington, DC, and Barcelona: Smithsonian Institution and Lynx Edicions; 2005.
Reading R, Michel S, Amgalanbaatar S. Ovis ammon. The IUCN Red List of Threatened Species 2020: e.T15733A22146397. 2020. https://doi.org/10.2305/IUCN.UK.2020-2.RLTS.T15733A22146397.en.
Riaz T, Shehzad W, Viari A, Pompanon F, Taberlet P, Coissac E. ecoPrimers: inference of new DNA barcode markers from whole genome sequence analysis. Nucleic Acids Res. 2011;39:e145.
Sándor AD, Ionescu DT. Diet of the eagle owl (Bubo bubo) in Braşov, Romania. North West J Zool. 2009;5:170–8.
Schaller GB. Wildlife of the Tibetan Steppe. Chicago: University of Chicago Press; 1998.
Schaller GB. Tibet Wild. Washington: Island Press; 2012.
Schaller GB, Junrang R, Mingjiang Q. Status of the snow Leopard Panthera uncia in Qinghai and Gansu Provinces, China. Biol Conserv. 1988;45:179–94.
Schoener TW. Competition and the form of habitat shift. Theor Popul Biol. 1974;6:265–307.
Shannon C, Weaver W. The mathematical theory of communication. Urbana: University of Illinois Press; 1949.
Shehzad W, McCarthy TM, Pompanon F, Purevjav L, Coissac E, Riaz T, et al. Prey preference of snow leopard (Panthera uncia) in South Gobi, Mongolia. PLoS ONE. 2012;7:e32104.
Shi ZY, Yang CQ, Hao MD, Wang XY, Ward RD, Zhang AB. FuzzyID2: a software package for large data set species identification via barcoding and metabarcoding using hidden Markov models and fuzzy set methods. Mol Ecol Resour. 2018;18:666–75.
Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann Entomol Soc Am. 1994;87:651–701.
Simpson EH. Measurement of diversity. Nature. 1949;163:688.
Smith AT, Formozov NA, Hoffman RS, Zeng C, Erbajeva MA. The pikas. In: Chapman J, Flux J, editors. Rabbits, hares and pikas. Status survery and conservation action plan. Chapter 3. Gland, Switzerland: IUCN; 1990. pp. 14–60.
Smith AT, Foggin JM. The plateau pika (Ochotona curzoniae) is a keystone species for biodiversity on the Tibetan plateau. Anim Conserv. 1999;2:235–40.
Soergel DAW, Dey N, Knight R, Brenner SE. Selection of primers for optimal taxonomic classification of environmental 16S rRNA gene sequences. ISME J. 2012;6:1440–4.
Symondson WOC. Molecular identification of prey in predator diets. Mol Ecol. 2002;11:627–41.
Taberlet P, Fumagalli L. Owl pellets as a source of DNA for genetic studies of small mammals. Mol Ecol. 1996;5:301–5.
Thiam M, Bâ K, Duplantier JM. Impacts of climatic changes on small mammal communities in the Sahel (West Africa) as evidenced by owl pellet analysis. African Zool. 2008;43:135–43.
Tremblay I, Thomas D, Blondel J, Perret P, Lambrechts MM. The effect of habitat quality on foraging patterns, provisioning rate and nestling growth in Corsican Blue Tits Parus caeruleus. Ibis. 2005;147:17–24.
Trevelline BK, Nuttle T, Porter BA, Brouwer N, Hoenig BD, Steffensmeier ZD, Latta SC. Stream acidification and reduced migratory prey availablity are associated with dietary shifts in an obligate riparian Neotropical migratory songbird. PeerJ. 2018a;16:e5141.
Trevelline BK, Nuttle T, Hoenig BD, Brouwer NL, Porter BA, Latta SC. DNA metabarcoding of nestling feces reveals provisioning of aquatic prey and resource partitioning among Neotropical migratory songbirds in a riparian habitat. Oecologia. 2018b;187:85–98.
Treves A, Krofel M, McManus J. Predator control should not be a shot in the dark. Front Ecol Environ. 2016;14:380–8.
Valera F, Gutiérrez JE, Barrios R. Effectiveness, biases and mortality in the use of apomorphine for determining the diet of granivorous passerines. Condor. 1997;99:765–72.
Walter CB, O’Neill E. Electrophoresis in the study of diets and digestive rates of seabirds. Comp Biochem Physiol A Comp Physiol. 1986;84:763–5.
Wei WR, He JD, Zheng QY. Plateau pikas (Ochotona curzoniae) at low densities have no destructive effect on winter pasture in alpine meadows. Rangel J. 2020;42:55–61.
Wilson RP. An improved stomach pump for penguins and other seabirds. J Field Ornithol. 1984;55:109–12.
Xia W, Zhou X, Liu J, Zhang X. The bio-community in the region of alpine meadow. In: Liu J, Wang Z, editors. Alpine meadow ecosystem. Beijing, China: Science Press; 1991. p. 1–7.
Yang Z, Gong M, Huang X, Li C, Liu X. Biodiversity of birds of prey in Shiqu county. J Sichuan Teach Coll. 2000;21:137–40 (in Chinese).
Zhang R, Ludwig A, Zhang C, Tong C, Li G, Tang Y, et al. Local adaptation of Gymnocypris przewalskii (Cyprinidae) on the Tibetan Plateau. Sci Rep. 2015;5:9780.
Acknowledgements
We would like to thank Mr. Yun Ma, Jiong He, and Rou Bao for their generous assistance in the field surveys.
Funding
This project was funded by the National Key Technology R&D Program of China (2019YFA0607103), Snow Leopard Conservancy (G1900014, G2000019), Cleveland Metroparks Zoo (G1800082), Panthera Corporation and The Andrew Sabin Family Foundation (G1900011, G2000017), and Welfare Project of the National Scientific Research Institution (CAFYBB2019ZE003).
Author information
Authors and Affiliations
Contributions
CEH formulated study concept. BDH performed statistical analyses. LW led field teams. CEH, LW, WC, YD, YL, JL, YX, YZ, HC, and YZ collected study samples. CEH, WC, JY, and YJ performed DNA extractions. CEH, WC, and JY performed sequence read mapping and determination of dietary items. CEH, YZ, and JEJ acquired funding for the study. CEH wrote the study manuscript. CEH, BDH, YZ, and JEJ interpreted study results. BDH, YZ, and JEJ edited the manuscript and provided additional expertise. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Supplementary Information
Additional file 1.
The FASTA file of reference sequences used for read mapping of possible host and prey species.
Additional file 2.
Results of subject sequences blasted to reference sequences of determined host and prey items.
Additional file 3.
The r-script used in the statistical analyses of the dataset.
Additional file 4.
Unique haplotypes for a segment of MT-RNR1 amplified by primers (12SV5F/12SV5R, Riaz et al. 2011) by species.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Hacker, C.E., Hoenig, B.D., Wu, L. et al. Use of DNA metabarcoding of bird pellets in understanding raptor diet on the Qinghai-Tibetan Plateau of China. Avian Res 12, 42 (2021). https://doi.org/10.1186/s40657-021-00276-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40657-021-00276-3