Abstract
In current years, natural pigments are facing a fast-growing global market due to the increase of people’s awareness of health and the discovery of novel pharmacological effects of various natural pigments, e.g., carotenoids, flavonoids, and curcuminoids. However, the traditional production approaches are source-dependent and generally subject to the low contents of target pigment compounds. In order to scale-up industrial production, many efforts have been devoted to increasing pigment production from natural producers, via development of both in vitro plant cell/tissue culture systems, as well as optimization of microbial cultivation approaches. Moreover, synthetic biology has opened the door for heterologous biosynthesis of pigments via design and re-construction of novel biological modules as well as biological systems in bio-platforms. In this review, the innovative methods and strategies for optimization and engineering of both native and heterologous producers of natural pigments are comprehensively summarized. Current progress in the production of several representative high-value natural pigments is also presented; and the remaining challenges and future perspectives are discussed.
Graphical Abstract

Similar content being viewed by others
Introduction
Pigments are defined as finely divided and usually water-insoluble colorants, absorbing and reflecting the visible light to show different colors (Rapp 2009). They are of larger molecular weight, less water-soluble, and less transparent than dyes. The usage of pigments has a long history, dating back to the beginning of ancient civilizations (ancient China, India, and Egypt), whereby natural plants, insects, and minerals were used to dye textiles, color foods, paint, color the body in religious ceremonies, and more. In 1856, English chemist William Henry Perkin discovered the first synthesized organic dye from coal tar distillate (Chandler 2001), which triggered the commercial flourishing of synthetic pigments. However, they are derived from chemical reactions, causing heavy environment stress. More notably, many serious health hazards such as toxicity, oncogenicity, and teratogenicity were found associated with synthetic pigments (Li and Tian 2017).
With the increase in public health awareness, replacement of synthetic pigments with safer and more ecofriendly natural pigments has become the current marketing trend. According to the report by GRAND VIEW RESEARCH, the global market for natural pigments in food industry alone is estimated to reach 2.5 billion USD by 2025 (https://www.grandviewresearch.com/industry-analysis/food-colorants-market). Besides usage as colorants, many of natural pigments show great potential in pharmaceutical, nutrition, and skincare industries, due to their health benefits (Amalraj et al. 2017; Chen and Zhong 2015; Yamagata et al. 2015). For instance, as one of the most bioactive xanthophyll pigments that mainly presents in higher plants and microalgae, lutein can prevent and treat retinal damage, including age-related macular degeneration, glaucoma and diabetic retinopathy (Krinsky et al. 2003; Seddon et al. 1994; Sun et al. 2011; Zhang et al. 2016a). It has also been shown to prevent cardiovascular diseases, atherosclerosis and cancers (Astorg 1997; James et al. 2001). The global market for lutein is fast growing, which is expected to reach 357.7 million USD and 2121.2 tons by 2022 (Markets 2017).
According to their structural characteristics, natural pigments can be classified into tetrapyrroles, carotenoids, flavonoids, curcuminoids, betalains, quinones, and others. They are widely distributed across all the natural kingdoms, e.g., plants, animals, and microorganisms. However, each natural source has its own limitations and currently cannot compete economically with synthetic counterparts. Taking astaxanthin (a typical carotenoid compound) as an example, its extraction from green microalga is subject to the lengthy autotrophic cultivation in open freshwater ponds (10–15 days) and the requirement of cell wall disruption to release the carotenoids (Mota et al. 2021); the frequently reported astaxanthin contents from crustaceans, salmon, and trout sources are less than 1 mg/g (Ahmadkelayeh and Hawboldt 2020); the yield of astaxanthin in most wild strains of the microorganism Phaffia rhodozyma can only reach 0.3–0.9% while in bioreactor high cell density and production titers can be achieved in much shorter times (Wan et al. 2021). Therefore, improving biotechnological productivity towards achieving large-scale production is presently an important challenge.
To date, great efforts have been devoted to in vitro production of natural pigments via plant cell/tissue culture and optimization of microbial cultivation. Moreover, rational engineering via pathway mining and genetic engineering has been proposed and applied to improve the yield of various natural pigments. Although considerable achievements have been obtained from biotechnological modification of natural producers, in most cases their productivity still cannot meet the requirements for industrial-scale production.
At the beginning of the twenty-first century, the emergence of “omics” techniques greatly improved our understanding of natural biological systems. Based on these knowledge treasures, synthetic biology came into the forefront, whereby engineering principles were employed to design and reconstruct novel biological modules as well as biological systems (Osbourn et al. 2012). By constructing artificial metabolic pathways using designed parts in well-known biological systems, unlimited supply of previously expensive or unfeasible products could be realized. Currently, model microorganisms like Escherichia coli and Saccharomyces cerevisiae are regarded as the ideal chassis for engineering, owing to their genetic tractability, short cycle of life, and mature technology of high-density fermentation. One classic example is the production of crocins (Fangyu et al. 2018; Liu et al. 2020), which demonstrates the potential of synthetic biology in realizing large-scale production of rare and high-value natural pigments. The simultaneous blooming of genome sequencing, genetic editing, and computer modeling techniques further accelerated progress in heterologous biosynthesis, ushering in the dawn of industrial production of natural pigments.
During the past two decades, several reviews (Begum et al. 2016; Lagashetti et al. 2019; Manivasagan et al. 2018; Rodriguez-Amaya 2016; Sigurdson et al. 2017) have been published on the topic of natural pigments. These articles mainly focused on the characteristics, biosynthesis, regulation, functions and applications of pigments, while comparatively little information on the biotechnological progresses in their production was given. More recently, certain pigments (mainly carotenoids) have been well reviewed, covering their potential use, chemistry and biosynthesis, economic importance, and strategies to enhance production (Heider et al. 2014; Hu et al. 2018; Mussagy et al. 2019b; Saini et al. 2020; Saini and Keum 2019; Venil et al. 2013; Wang et al. 2021a). However, readers cannot get a full picture of the progresses and challenges in industrial application of natural pigments due to the limited categories of pigments and producers (like microalgae) covered in those reviews. Furthermore, the biotechnological engineering methods and strategies are rarely systematically analyzed and summarized. In this review, we present a systematic introduction on the progresses in biosynthesis of natural pigments with the aim to provide a more complete picture of natural pigments development. Specifically, part 2 introduces the current progresses in improving pigment production from natural producers; part 3 summarizes the technical developments towards efficient heterologous biosynthesis of pigments in non-pigment producing organisms; part 4 focuses on representative high-value products among natural pigments, and comprehensively reviews the efforts made in promoting their biosynthesis; lastly, part 5 discusses the challenges and future perspectives in industrial application of natural pigments.
Biosynthesis of pigments from natural producers
Plant cell culture and tissue culture
Plants are the most abundant source of natural pigments, including chlorophyll, anthocyanin, carotenoids, betalains, etc. Traditionally, most of the commercial pigments are derived from natural sources, either foraging or large-scale plant field cultivation. Due to the low accumulated contents of desired biochemicals in the whole plant, this approach leads to heavy consumption of farmlands and damage to precious natural plants. Taking crocetin—the most important active ingredient and pigment mainly distributed in the stigmas of Crocus sativus, as an example, it is estimated that 20 g of crocetin (in 1 kg of dry Crocus sativus stigma) requires 110,000–170,000 flowers and over 400 h of hand-labor (Frusciante et al. 2014). Besides, Crocus sativus grows slowly and only propagates by vegetative production. Plant cell culture is a technique which can enable plant cells to produce the full spectrum of characteristic second metabolites as mother plants under strictly controlled environmental conditions (Georgiev et al. 2018). In vitro plant production approach is independent of geographical and seasonal limitations, providing an alternative approach to increase the productivity of plant metabolites of interests. Selection and development of high-yielding cell lines as well as optimization of cell cultivation process, including optimization of culture conditions, recycling the cells via immobilization, and bioreactor redesign for scale-up cultivation, are the most commonly used strategies for improving metabolite accumulation (Fig. 1) (Bhojwani and Dantu 2013). As these traditional strategies in pigment production have been summarized in previous reviews (Murthy et al. 2014; Steingroewer et al. 2013), herein, we will mainly focus on two extension techniques for triggering the biosynthesis of secondary metabolites based on bionic innovation (elicitation) and continuous release of desired bioactive compounds (exudation).
Elicitation
The biosynthesis of secondary metabolites in plants is activated in response to pathogens (or insects) attack or various abiotic stresses (e.g., radiation, heavy metal and mineral, temperature, salinity) (Ramirez-Estrada et al. 2016). These stress signaling compounds are designated as “elicitors”, the majority of which are perceived by membrane receptors first and entail various sequential reactions (including phosphorylation or dephosphorylation of plasma membrane proteins, [Ca2+] enhancement, Cl−and K+ efflux/H + influx, NADPH oxidase activation, defense gene expression, and secondary metabolite biosynthesis) to trigger the biosynthesis of secondary metabolites (Ramirez-Estrada et al. 2016; Zhang et al. 2012). According to the studies on production of the pigments—betalain, shikonins, carotenoids and flavonoids (Georgiev et al. 2008; Malik et al. 2016; Rizzello et al. 2014; Savitha et al. 2006), biotic elicitors can be divided to four classes: (1) exogenous compounds secreted by microorganisms and insects when attacking plants (Warhade and Badere 2018); (2) exogenous compounds formed from the degradation of microbial cells by plant enzymes, such as fungal and bacterial lysates and polysaccharides of the microbial cell walls (e.g., chitin, glucans) (Bahabadi et al. 2014; Tang et al. 2019); (3) plant cell wall fragments degraded by pathogens; (4) the intracellular proteins or compounds synthesized by plants in response to pathogen attack or abiotic stress (e.g., plant hormones). On the other hand, many abiotic elicitors (like heavy metals, UV irradiation, and inorganic salts) also play important roles in activating plant secondary product biosynthesis.
Exudation
Plant pigments are usually stored in different tissues, making their isolation difficult. A considerable number of studies have been executed to export pigment compounds, which can be summarized into three methods: (1) membrane permeabilization using chemical agents (e.g., Tween 20, DMSO, isopropanol, Triton X-100, polysaccharides like chitosan, and high ionic strength) or physical approaches (e.g., ultrasonication, pulsed electric fields, and high hydrostatic pressure) (Saw et al. 2012); (2) in situ extraction (e.g., usage of n-hexadecane and liquid paraffin in liquid–liquid system) (Zare et al. 2010); (3) introduction of specific transport mechanisms, including vesicle trafficking, membrane transporters, and glutathione S-transferase (GST) (Zhao 2015). However, the specific transporters as well as the mechanisms are still poorly understood, which may be a future research focus.
Microbial cultivation
Traditional mutagenesis
Typical pigment-producing microorganisms include Phaffia rhodozyma (yeast, producing red pigments—carotenoids), Monascus sp (fungus, producing monascus red pigments), Blakeslea trispora (fungus, producing orange pigments like beta-carotene), Streptomyces cyaneus (fungus, producing black pigment—melanin), and Serratia sp (bacteria, producing red pigment—prodigiosin) (Chao et al. 2018; El-Batal et al. 2017; Kim and Ku 2018; Mussagy et al. 2021; Sun et al. 2021). Traditional random mutagenesis using ultraviolet (UV), 1-methyl-3-nitro-1-nitrosoguanidine (NTG) and ethyl methane sulfonate (EMS) have greatly improved microbial pigment production (Yolmeh and Khomeiri 2016; Yolmeh et al. 2017). Nevertheless, these methods suffer from severe drawbacks such as long searching time and low success rate.
Discovery of key impact factors and comprehensive optimization
The accumulation of pigments is generally related to cell growth and affected by nutrient factors (carbon source, nitrogen source, C/N ratio), microbiological parameters (spores age, seed age, inoculum age) and environmental conditions. In terms of environmental conditions, several essential aspects should be evaluated and optimized for the solid-state fermentation (SSF)—the processes performed on non-soluble materials. These aspects include the humidity of medium, physical and structural properties of substrates, temperature, pH and agitation (de Castro and Sato 2015). The low moisture content indicates that this cultivation process can only be carried out by a limited number of microorganisms, mainly yeasts and fungi. The other cultivation process is submerged fermentation (SmF) which based on liquid culture for most microorganisms, and is highly influenced by factors including temperature, pH, and agitation (Morales-Oyervides et al. 2020). Furthermore, it was revealed that production of some pigments is induced by certain stress conditions. For instance, Velmurugan et al. demonstrated that total darkness stimulated accumulation of pigments in Monascus purpureus, Isaria farinosa, Emericella nidulans, Fusarium verticillioides and Penicillium purpurogenum (Velmurugan et al. 2010). For photosynthetic microorganisms, light sources, light intensity, and light photoperiod were illustrated to be the key factors affecting pigment production (Cheirsilp and Torpee 2012; Kuo et al. 2012; Liu et al. 2019b; Zhou et al. 2015b).
Based on discovery of the key impact factors, comprehensive optimization is required to obtain high-yield production of desired pigments. Response surface method (RSM) can solve multivariate data from appropriately designed experiments, and help improve pigment production by optimization of culture medium (Seyedin et al. 2015), process parameters (Sehrawat et al. 2017), and extraction conditions (Zhong et al. 2019). The recently emerged artificial neural networks (ANN) is more flexible and accurate in comparison to RSM (Shafi et al. 2018), having proved effective in improving the production of red pigment in M. purpureus MTCC 369 (Singh et al. 2015).
Development and application of novel immobilization methods
Immobilization technology provides a feasible approach to improve the stability and reusability of the cells, facilitating easier downstream cell separation and continuous operation. Immobilization of bacteria and yeast has been applied for production of carotenoids (Alipour et al. 2017). For molds, SSF is deemed to be superior to liquid-state fermentation (LSF) in pigment production. Extensive studies showed that immobilization cultivation by using cell entrapment and adsorption methods can mimic the solid-state environment of SSF and effectively improve pigment production from molds (Liu et al. 2010). To eliminate nutrient mass transfer limitation from traditional immobilization, membrane-surface liquid culture (MSLC) and modified system-self-immobilization biomembrane-surface liquid culture were developed, whereby the molds could grow on the upper side of the membrane and the other side contacts with liquid medium for supplementation of nutrition (Wang et al. 2012). On the other hand, cultivation of phototrophic dinoflagellates, which can produce pigments like carotenoids and xanthophyll peridinin, is hindered by their sensitivity to hydrodynamic stress. Immobilization of microalgae in a biofilm on sheet-like surfaces could circumvent this problem (Olivieri et al. 2014). Employment of biofilm photobioreactors for cultivation of marine dinoflagellate Symbiodinium led to significantly higher biomass yield and peridinin productivity over suspension culture (Benstein et al. 2014).
Rational engineering of natural producers
Mining of natural pathways
Pre-omics
Isotope labeling (1H, 3H, 13C and 15 N) and nuclear magnetic resonance spectroscopy (NMR) analyses can give clues on the biosynthetic pathways via direct observation of isotopic shifts. In the study by Dong et al. (2021), a dual stable isotope labeling approach named DLEMMA was employed to identify and track phenylpropanoid pathway in Arabidopsis thaliana. It was found that PAP1 was a key enzyme for accumulation of cyanidin-type anthocyanins and quercetin-type flavonols. In addition to chemical analysis, a variety of molecular techniques have been employed to identify genes and gene clusters responsible for production of the target metabolites (Fig. 2). The most common strategies can be summarized as: (1) gene deletion and complementation (Bitok et al. 2017); (2) amplification of target gene based on conserved sequences of known enzymes from other organisms and other methods derived thereof (Jones et al. 2008); (3) sequencing the DNA fragments flanking both sides of the known functional enzymes by genome walking (Shapter and Waters 2014); and (4) using a known gene as the bait to reveal unknown regulators or related genes (Lu et al. 2018).
Overview of strategies for exploration of metabolic pathways in nature producers. Pre-omics analysis methods consist of chemical analysis (e.g., isotope labeling, NMR analysis) and molecular techniques (e.g., gene deletion, gene expression, DNA sequencing by genome walking); omics technologies include genomics, transcriptomics, proteomics and metabolomics. Both of them offer powerful tools to discover new genes, gene clusters, transcription factors as well as pathways for synthesis of bioactive compounds
Omics
In recent years, combined applications of “omics” techniques—via comparative systematic analysis among different species or the same species under different cultivation/induction conditions—offer powerful tools to discover new genes, gene clusters, transcription factors as well as pathways for synthesis of bioactive compounds (Fig. 2). Via transcriptome analysis of purplish-red leaf and green leaf of Paeonia qiui, several new candidate genes influencing anthocyanin accumulation were discovered, including MYB1, MYB2, bHLH1 and WD40-1 (Luo et al. 2017). Integrated transcriptomics and metabolomics in tomato revealed 38 candidate carotenoid-correlated genes and demonstrated the important role of the ethylene response factor SIERF6 in ripening and carotenoid production (Lee et al. 2012).
Omics-based information on gene distribution and expression regulation can also be used to guide the discovery of unknown genes or clusters (Medema and Osbourn 2016). As demonstrated by Shimura et al., new metabolic pathways or unknown genes can be discovered based on the identification of physically clustered groups (Shimura et al. 2007). Another typical rule is “co-expression”: specialized metabolites are synthesized in certain cell types, growth phases, or in response to environmental induction, for which the corresponding genes usually co-ordinate transcripts activated by specific transcription factors. Therefore, if the gene encoding one of the pathway enzymes has been identified, the other functional enzymes in the same pathway might be elucidated by co-expression analysis using transcriptomics and by metabolites analysis via metabolomics (Geu-Flores et al. 2012).
Genetic engineering of natural producers
The rapid development of bioinformatics, sequencing technology, and genetic engineering has enabled gene manipulation of natural producers to improve pigment production. Herein, we take the progress of Monascus azaphilone pigments (MonAzPs) as an example for illustration. MonAzPs have been used as food colorants and food preservatives in East Asian countries for more than a millennium, while the genetic background of Monascus sp. for producing red pigments remained unclear until recently. In 2013, the key gene cluster (the PKS-FAS gene cluster) involved in the biosynthesis of azaphilone pigments was identified by T-DNA random mutagenesis in M. purpureus (Balakrishnan et al. 2013), whereby MppR1 and MpPKS5 were confirmed as the key enzymes. The complete genome information of M. purpureus YY-1 was obtained by next-generation sequencing and optical mapping techniques, which together with transcriptomic analyses revealed the potential biosynthesis pathway and the regulatory mechanism of pigment production (Yang et al. 2015). On this basis, the individual steps of MonAzPs pathway of M. ruber M7 were elucidated by a systematic functional investigation (Chen et al. 2017b). These advances in elucidating genetic background of natural pigment producers give a roadmap towards improved synthesis of desired products. Downregulation of the citrinin biosynthesis, a competing metabolic branch to MonAzPs biosynthesis, improved the production of pigments by 60% (Liang et al. 2018). Disruption of the ergosterol biosynthetic pathway increased membrane permeability and led to the secretion of Monascus pigments, resulting in 2.06-fold increase in pigment production (Liu et al. 2019a). In short, upregulation of the MonAzPs biosynthetic pathway, complemented with downregulation of competing metabolic branches and improvement of product secretion, offers effective approaches for enhancing Monascus pigments in the native producers. Apart from MonAzPs, genetic engineering is being performed in natural producers of other pigments, such as carotenoids (Ganapathy et al. 2016). For further improvement, the difficulty lies in the lack of understanding on the regulation of pigment synthesis.
Efforts towards heterologous biosynthesis of pigments
Despite the great progress achieved thus far in pigment production by natural biosynthesis, the yield of target compounds remains limited due to low productivity and incomplete understanding of the genetic background of natural producers. Over the past two decades, metabolic engineering has emerged as a powerful tool to develop heterologous cell factories for producing natural pigments. Herein, the efforts towards heterologous biosynthesis of pigments, in the aspects of pathway design, pathway construction and pathway optimization are summarized.
Pathway design
The first critical step for heterologous biosynthesis of pigments is the identification of functional genes and responsible pathways. Natural synthetic pathways usually suffer from metabolic bottlenecks caused by the presence of rate-limiting enzymes and feedback inhibition. Synthetic biology provides an approach to design novel pathways via assembly of catalytic elements and pathway modules from diverse organisms so as to circumvent low-activity enzymes and avoid feedback regulation. For example, insufficient isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP) supply is a common bottleneck for accumulation of carotenoids. Introduction of a recombinant mevalonate pathway (MVA pathway) consisting of the bottom portion from Streptococcus pneumonia and the top portion from Enterococcus faecalis led to increased β-carotene production in E. coli (Yoon et al. 2009). For production of anthocyanins and flavonoids, shikimate pathway (SK) is crucial for providing the key precursor coumaric acid. In SK, the conversion of 2-dehydro-3-deoxyarabinoheptulosonate-7-phosphate (DAHP) into the important intermediate EP3P was catalyzed by only Aro1 in yeast systems. In contrast, this catalysis requires the cooperation of seven distinct enzymes—AroB, AroD, AroE, YdiB, AroL, AroK, and AroA, in E. coli. The addition of aroL from E. coli into the native SK in yeast displayed a positive effect on p-coumaric acid production (Rodriguez et al. 2015).
Pathway construction
The long biosynthetic pathway of complex natural pigments, such as carotenoids, flavonoids and quinones, raises the issue of pathway assembly in heterologous hosts. Construction of multiple plasmids using the traditional approach involving multiple cloning steps such as PCR, endonuclease digestion and ligation, is time-consuming, inefficient, and limited by the availability of restriction sites in the plasmids. In recent years, novel DNA assembly methods and multi-locus integration methods have been developed and adopted for pigment production (Fig. 3).
Overview of innovative methods for pathway assembly. A Assembly of multiple genes into one large plasmid or one genomic locus. Restriction digestion/ligation-based methods with usage of type II and type IIs restriction enzymes are employed for in vitro gene assembly. Sequence homology-based methods are adopted to join multiple DNA fragments within one step in vivo or in vitro via short overlap. B Multi-locus integration of long pathway by splitting the whole pathway into several segments and then inserting them into multiple sites within one step or by multi-round integration, utilizing multiple markers, recycling markers or adopting marker-free systems. C Combination of multi-locus integration approach and in vivo assembly method to facilitate long-pathway construction
DNA assembly
Restriction digestion/ligation-based methods using type II and type IIs restriction enzymes such as BioBrick (Shetty et al. 2008), BglBrick (Lee et al. 2011) and Golden gate (Engler et al. 2009), are the typical in vitro gene assembly method for large plasmid construction. The other type of in vitro gene assembly method is the sequence homology-based methods like Gibson (Gibson et al. 2009) and CPEC (Quan and Tian 2009), whereby multiple DNA fragments can be easily joined together within one step via short overlaps (Fig. 3A). Apart from in vitro gene assembly methods, in vivo HR-based methods like E. coli RecET/Red system (Fu et al. 2012) and B. subtilis DNA assembly (Itaya et al. 2008) have been developed and employed for pathway construction and even chromosome creation. In in vivo DNA assembler, each DNA cassette is designed with > 40 bp homologous arms. Based on simple operation plus the high efficiency of the native HR mechanism within yeast, several DNA fragments could be assembled and inserted into the designated sites in the chromosome. By employment of an in vivo DNA assembler, a xylose-utilizing zeaxanthin biosynthetic pathway consisting of eight genes (~ 19 kb) was assembled within one-step transformation in yeast (Shao et al. 2009). As compared to in vitro gene assembly, the in vivo gene assembly via employment of the intracellular HR mechanism may omit the addition of cloning enzymes which may lower cloning efficiencies.
Multi-locus integration
Although DNA assembly provides an effective approach for rapid construction of long pathways, successful rate could be affected depending on whether all bioparts are assembled in a single designated site. As an alternative strategy, multi-locus integration splits the pathway into several segments and inserts them into multiple sites via one-step manipulation or multi-round integration (Fig. 3B). The conventional way is to integrate target genes in different loci using different markers, but this strategy is limited by the low available number of selection markers. Cre-loxp-based system and URA-FOA negative selection were then developed for removal and reuse of selection marker (Aguiar et al. 2014). Inspired by these designs, we developed a marker recyclable integrative toolbox for pathway assembly in S. cerevisiae, with the “loxp-kanMX-loxp” cassette conferring resistance against the antibiotic G418 and meanwhile facilitating marker recycling (Ye et al. 2017). By combining this with the decentralized assembly strategy, a total of 11 genes encoding the carotenoid biosynthetic pathway, with length of ~ 22 kb, was integrated into the genome of yeast within 5 rounds of reiterative recombination, leading to successful heterologous production of 16.3 mg∙g−1 dcw carotenoids.
In order to improve the efficiency of HR and achieve marker-free genomic engineering in the multi-locus integration method, several CRISPR–Cas9-facilitated multiplexed integration strategies, such as delta integration CRISPR–Cas (Shi et al. 2016), “Wicket” (Hou et al. 2018), co-transformation of multiple gRNA and donor DNA cassettes (Horwitz et al. 2015), have been developed for assembly of long pathways or modulating the copy numbers of integrated genes. Taking “wicket” design as an example, a short DNA harboring two 50 bp HAs and a 23 bp CRISPR–Cas9 target sequence were designed as docks for integration of exogenous DNA fragments (Hou et al. 2018). Using this approach, multicopies of β-carotene synthetic genes crtE, crtYB and crtI were integrated into the yeast genome, without needing any selective marker.
Multi-locus integration with in vivo assembly
As mentioned previously, both DNA assembly and multi-locus integration can significantly facilitate long-pathway construction. In 2015, the Jay Keasling group developed a new method, named CasEMBLR, for multi-DNA assembly via a combination of multi-locus genomic integration method and in vivo DNA assembly with the usage of CRISPR/Cas9 system. To validate its applicability, 15 DNA parts for carotenoid biosynthesis were successfully assembled and integrated into three targeted genomic loci (URA3, HIS3 and ADE2) of yeast (Jakociunas et al. 2015).
Pathway optimization
Introduction of a heterologous pathway can endow the cell factory with the capacity to produce desired compounds, however the productivity is generally low due to the complexity of metabolic networks in all living systems. For instance, rate-limiting steps may result from undesirable biological characteristics of bio-parts (e.g., promoter, enzyme, cofactor, transporter); metabolic flux to the target compound may be limited by feedback regulation, diverse competing branches or insufficient precursor supply; moreover, overexpression of proteins may lead to heavy metabolic burden, resulting in significant biomass decrease. Therefore, pathway optimization is essential to overcome these bottlenecks for achieving high-efficiency bioproduction. Herein, pathway optimization strategies and their applications in pigment biosynthesis are illustrated, classified into three levels: engineering of rate-limiting bio-parts/factors (“Engineering of rate-limiting bio-parts/factors” Section , Fig. 4), engineering of the metabolic network (“Engineering of the metabolic network” Section, Fig. 5), and engineering of cellular systems (“Engineering of cellular systems” Section, Fig. 6).
Overview of strategies for engineering of rate-limiting bio-parts in heterologous biosynthesis. I Promoter engineering is used for fine-tuning of gene expression via screening and characterization of native promoter pool as well as construction of synthetic promoter library. II Protein engineering is employed to optimize enzymatic properties via directed evolution and rational design. III Cofactor engineering is adopted to improve the supply of reducing forces by regulation of endogenous cofactor system or introduction of heterologous cofactor regeneration systems. IV Transporter mining is applied to eliminate the transportation bottlenecks via omics analysis and literature retrieval
Overview of strategies for engineering of metabolic network in heterologous biosynthesis. A “Push–pull–block” strategy is used to direct more metabolic flux to the target compounds, in which “push” indicates enhancing the precursor supply towards target products, “pull” means strengthening the formation ability of the target products, and “block” represents downregulation of the competing pathways. B Inter- and intra-modular pathway engineering strategy is proposed to regulate metabolic balance among multiple modules as well as individual genes within each module. C Multiplex genome engineering like multiplex automated genome engineering (MAGE) is applied for large-scale programming of cell factory. In MAGE, synthetic ssDNA pools (color lines) are transformed into engineered hosts for generation of a diverse set of genomic modifications
Overview of strategies for engineering of cellular systems in heterologous biosynthesis. A Two-stage cultivation is employed to maintain normal cell growth while maximizing pigment production, in which environmental signals (like temperature, light, pH and oxygen) and intra-/inter-cellular signals (like QS system) are used as inducers to trigger the switch-on and switch-off of gene expression. B Co-culture engineering provides an alternative approach for efficient pathway engineering via dividing the complex long pathways into several pathway sections in different strains
Engineering of rate-limiting bio-parts/factors
Promoter engineering
Precise regulation of gene expression is critical to improve product formation and cell growth, as excessive gene expression will generate metabolic burden while insufficient expression may cause accumulation of metabolic intermediates. Promoter optimization is the most direct way to modulate gene expression via controlling the transcription level of specific genes. However, its application is dependent on the availability of endogenous promoters with proper feature. To enrich promoter sources, the native promoter pools in microorganisms and plants are screened via omics and characterized using reporters, and synthetic promoter libraries have been constructed by means of error-prone PCR, site-directed mutagenesis, saturation mutagenesis, and hybrid-promoter design (Fig. 4I). As these above methods have been previously reviewed (Xu et al. 2019; Zhou et al. 2017c), they shall not be described in detail here.
Protein engineering
Enzymes are the basic catalytic elements for biosynthesis of natural products. Innate enzymatic properties, including catalytic activity, substrate specificity and allosteric regulation characteristics, are often incompatible with the engineering hosts. Poor performance of enzymes may generate bottlenecks, unwanted by-products and feedback inhibition, resulting in low synthetic efficiency and productivity.
Directed evolution can mimic Darwinian evolution to achieve functional changes by altering the enzyme structure, and the process typically comprises library construction and artificial selection (Fig. 4II). Random mutations can be introduced into the protein via either error-prone PCR or DNA shuffling. In terms of pigments biosynthesis, colorimetric assays are used as the most intuitive and effective high-throughput screening method. For production of high-purity lycopene, we have created a CrtYB variant with solely phytoene synthase activity via directed evolution, which eliminated the metabolic flux loss towards β-carotene and dramatically improved lycopene yield (Xie et al. 2015a). Apart from altering substrate specificity, the catalytic activity of the pathway enzymes such as OBKT (β-carotene ketolase) and GGPPS (geranylgeranyl diphosphate synthase) was also improved by directed evolution, leading to improved biosynthesis of astaxanthin (Zhou et al. 2017b).
With the growing number of structural data and increasing availability of mechanistic information, rational design is playing a more important role in protein engineering (Fig. 4II). In this approach, engineering targets can be identified via homologous modeling followed by molecular docking or molecular dynamics simulations or via sequence conservatism analysis, followed by site-specific mutagenesis, which significantly reduces the onerous task of screening that is required for directed evolution. For instances, isopentenyl phosphate kinase (IPK) can catalyze the conversion of dimethylallyl alcohol into the fundamental building block of isoprenoids, IPP and DMAPP. Rational design based on protein coevolution analysis generated a positive IPK mutant with eightfold activity improvement, leading to 97% higher β-carotene production (Liu et al. 2016b).
Cofactor engineering
Biosynthetic pathways of pigments commonly comprise multiple redox reactions driven by the reducing forces provided by cofactors, NAD(P)H. Although native cofactor systems exist in microorganisms and plants, they are often insufficient to support heterologous biosynthesis.
Regulation of endogenous cofactor systems, via strengthening cofactor generation or restraining cofactor consumption, provides a feasible strategy to maintain cellular redox balance (Fig. 4 III). In S. cerevisiae, enhancement of NADPH supply by overexpression of POS5 (a mitochondrial NADH kinase) and ZWF1 (glucose-6-phosphate dehydrogenase involved in the pentose phosphate pathway) led to 59.9% and 81.4% increase of lycopene and β-carotene production, respectively (Zhao et al. 2015). In addition to regulation of endogenous cofactor systems, NADPH generation can also be enhanced via introduction of heterologous cofactor regeneration systems (Fig. 4 III). By introducing a heterologous Entner–Doudoroff (ED) pathway from Zymomonas mobilis into E. coli, the NADPH regeneration rate was increased by up to 25-fold, achieving 97% improvement of carotenoid production (Ng et al. 2015). Comprehensive regulation of NADH supply in Monascus purpureus by addition of exogenous cofactor compounds and disturbing nuoI (encoding NADH-quinone oxidoreductase) significantly increased Monascus pigment production (Liu et al. 2021a).
Transporter engineering
In metabolic engineering, extracellular export of natural compounds like fat-soluble compounds (e.g., carotenoids) is a big challenge. In recent years, based on transcriptomic analysis and literature mining (Fig. 4 IV), many novel transporters have been revealed for specific pigments and then applied to eliminate the transport bottlenecks.
Depending on the mechanism, transporters can be divided into the ATP-binding cassette (ABC) transporters (driven by ATP hydrolysis) and facilitator superfamily (making use of electric gradients). When microorganisms or plants are exposed to noxious environment or engineered to produce heterologous biochemicals, efflux pumps will be activated to export harmful substances for cell survival. Based on this mechanism, the transcriptomes of two engineered S. cerevisiae strains with different carotenoids production capacities were comparatively analyzed, suggesting induction of genes involved in pleiotropic drug resistance including the ABC transporter-pdr10 in the strain with high carotenoids production (Verwaal et al. 2010). To confirm its effects on secretion of carotenoids, pdr10 from S. cerevisiae was introduced into Rhodosporidium toruloides in our previous study, leading to production improvement of total carotenoids from 1.9 μg/mg to 2.9 μg/mg in a bi-phasic culture (Lee et al. 2016). In addition to omics analysis, literature retrieval has also been used for transporter mining. For example, an array of efflux pumps reportedly responsible for secondary metabolite transports were introduced in anthocyanins-producing E. coli, and an anthocyanin-associated transporter yadH was identified, overexpression of which resulted in 15% increase of anthocyanin production (Lim et al. 2015).
Engineering of the metabolic network
Push–pull–block strategy
In heterologous biosynthesis, the efficiency of cell factories is often limited by the insufficient precursor supply and strong competitive metabolic branches. To direct more metabolic flux to the target compound, “push–pull–block” strategies have been developed and employed for biosynthesis of diverse chemicals and biofuels (Li et al. 2016): “push” indicates pushing more precursor supply towards target products, “pull” means strengthening the target biosynthetic pathway, and “block” or “restrain” represents downregulation of the competing pathways (Fig. 5A).
Isoprene is the monomeric building block of carotenoids. In our previous study, a rational push–pull–block strategy was employed for enhancement of isoprene biosynthesis in S. cerevisiae via engineering of the native acetyl-CoA and MVA pathways. This strategy was decomposed into upregulation of precursor supply in the acetyl-CoA and MVA pathway (by overexpression of ACS2/ERG10 and tHMG1, respectively), increasing the isoprene branch flux (via overexpression of IDI1 and ISPS), and downregulation of the competitive pathway (through decreasing the promoter strength of ERG20). With this combined regulation strategy, a total of 782-fold improvement of isoprene production was achieved in the engineered strain (Lv et al. 2014). Similar strategies have also enhanced the synthesis of pigments like flavonoids (Lyu et al. 2019b).
Inter- and intra-modular pathway engineering
In heterologous biosynthesis of complex pigments, overexpression of a large set of pathway genes often results in severe imbalance in pathway flux. Multivariate modular metabolic engineering partitions the complicated pathway of natural compounds into multiple modules, and the carbon flux of each individual module will be maximized and balanced to enable global fine-tuning of the whole metabolic network. The metabolic levels of different modules can be adjusted via altering plasmid copy number, promoter strength, RBS variety and enzyme properties (Fig. 5B). For example, the eight-step pinocembrin biosynthetic pathway was divided into four modules and optimized via altering the copy number of plasmids, leading to more than tenfold increase in pinocembrin production (Wu et al. 2013).
In addition to inter-module engineering, intra-module engineering is also viral to reduce the accumulation of intermediates (Fig. 5B). As illustrated in our previous study (Lv et al. 2016), a combinatorial strategy of protein engineering and modular pathway engineering was applied to simultaneously improve the intra- and inter-pathway balance for biosynthesis of isoprene in E. coli. Specifically, the whole biosynthetic pathway of isoprene was partitioned into the upstream module consisting of the native MEP pathway, and the downstream module containing isoprene synthase. The intra-module engineering within the upstream module via directed coevolution of DXS/DXR/IDI resulted in 60% improvement of isoprene production. Inter-module engineering between the upstream and downstream module via promoter manipulation further increased isoprene production by 4.7-fold. In a more recent study, a multidimensional heuristic process (MHP) was developed to co-ordinate intra-module activities and inter-module balance for biosynthesis of astaxanthin. A total of 15 genes were distributed in four modules and balanced by varying promoter strength, and intra-module balance was coordinated via RBS and enzyme variants, resulting in 320 mg/L of astaxanthin (Zhang et al. 2018a).
Multiplex genome engineering
Due to the complexity of biological systems, it is often hard to meet industrial demand on titers of desired compounds by solely engineering the target pathways. To date, a series of multiplex genome engineering methods have been developed for large-scale programming of cell factories, which can be classified into three types: (1) RNA interference-based (RNAi) method, for which a large RNAi library was coupled with high-throughput screening to generate accumulated beneficial modifications (Si et al. 2015). (2) Recombinase-based method, e.g., multiplex automated genome engineering (MAGE) (Wang et al. 2009) and trackable multiplex recombineering (TRAR) (Warner et al. 2010). (3) Multiplex CRISPR-based technologies, in which numerous gRNA or Cas enzymes are expressed simultaneously (McCarty et al. 2020). Herein, we will take MAGE (Fig. 5C) as an example to briefly introduce its application in metabolic engineering of E. coli for pigment production (Wang et al. 2009). MAGE depends on single-stranded DNA (ssDNA)-based genetic modification. Directed by λ-Red ssDNA-binding protein β, the sodas are loaded to the lagging strand of the replication fork for subsequent incorporation. Transformation of synthetic oligo pools enable generation of a diverse set of genetic modifications. Targeting 24 targeting genomic sites related to the 1-deoxy-D-xylulose-5-phosphate (DXP) biosynthesis pathway, fivefold increase of lycopene production was obtained with employment of MAGE method in E. coli.
Engineering of cellular systems
Dynamic control for cell growth–metabolism balance
Static pathway engineering, e.g., constitutive overexpression of pathway genes and deletion of competing pathways, has been proven as a powerful strategy to improve the yield of valuable products. However, it often results in decreased biomass due to severe metabolic burden and accumulation of toxic intermediates.
Two-phase cultivation has been widely recognized as a powerful strategy to circumvent the trade-offs between product accumulation and cell growth. Unlike traditional inducible expression systems, the newly developed environmental signal-responsive systems use cultivation parameters (like temperature, light, pH and oxygen) as inducers to trigger the switch-on and switch-off of gene expression, eliminating the requirement of adding expensive inducers (like isopropyl-β-D-thiogalactoside and galactose) (Fig. 6A). In our previous study, a glucose-responsive dynamic control system with PGAL promoters has been developed by deletion of GAL80, which has been successfully applied for β-carotene production (Xie et al. 2014). By employment of a lycopene-indicated high-throughput screening method, a temperature-sensitive Gal4 mutant was screened out from directed evolution library and introduced into the GAL80-knockout yeast strain. With this design, target genes under PGAL can be precisely switched on by altering the culture temperature, which resulted in 44% higher biomass and 177% increased lycopene production (Zhou et al. 2018), and facilitated high-density fermentation of astaxanthin (Zhou et al. 2019).
A further extension of dynamic control is autonomous regulation systems, in which target enzymes can be activated by intracellular intermediates or intercellular signals (Fig. 6A). For example, quorum-sensing circuits offer a powerful strategy to autonomously regulate cell metabolism. At high cell densities, quorum-sensing microbes can secrete sufficient molecular signals to alter the expression of endogenous genes. LuxI-LuxR and AHL-EsaR are two of the best-studied QS systems. A bifunctional QS circuit combining both LuxR and EsaR systems was developed using 3-oxohexanoyl homoserine lactone as the signal to dynamically regulate the expression of genes under Plux and PesaR in a cell density-dependent manner, and successfully improved flavonoid titer (Dinh and Prather 2019).
Co-culture engineering
Despite the progress in engineering single strains, introduction and regulation of the whole target biosynthetic pathway often raises overwhelming metabolic burden and encounter the intrinsic limitation of monocellular environment. Recently, co-culture engineering has emerged as an alternative approach for pathway engineering via division of the complex and long biosynthetic pathways into different host strains. As compared to mono-culture engineering, co-culture systems display notable advantages: (1) reducing metabolic burden by division of labor to different strains; (2) improving flexibility for balancing pathway modules via optimizing the population ratio of engineered strains; (3) providing diversified intracellular environments for functional overexpression of a large variety of enzymes involved; (4) reducing negative cross-interference among pathway modules; (5) enabling utilization of different carbon sources (Fig. 6B).
In recent years, considerable progress has been achieved in employment of modular co-culture system for biosynthesis of complex natural colored compounds (e.g., flavonoids and curcuminoids). One typical example is the biosynthesis of anthocyanins by E. coli polycultures, in which 15 exogenous enzymes were divided into 4 modules across four E. coli strains (Jones et al. 2017). Similar E. coli co-culture systems have also been adopted for biosynthesis of other flavonoids like sakuranetin (Wang et al. 2020), naringenin (Ganesan et al. 2017), resveratrol (Hong et al. 2020), and flavan-3-ols (Jones et al. 2016). In addition, efforts have also been made to employ cross-species co-culture systems for biosynthesis of complex compounds. For example, a synergistic E. coli-S. cerevisiae culture system was developed for naringenin production from D-xylose, generating 21.16 mg/L of the target product (Zhang et al. 2017). Although co-culture engineering exhibited significant advantages over mono-culture engineering, it is not suitable for all biosynthetic systems. For instance, many pathway intermediates cannot easily traverse cellular membranes, and different specifies may not be compatible in the same environment due to differing requirements of nutrients, temperature and pH.
Major natural pigments and advances in biotechnological production
Depending on their structural characteristics, natural pigments may be classified into five major classes: tetrapyrroles, carotenoids, flavonoids, curcuminoids, and betalains. The general introduction of each class of pigments, including structure, classification, function and biosynthetic pathway, as well as the advances in biotechnological production of the typical products from each group are individually summarized in this section.
Tetrapyrroles
Structure, classification and function
Tetrapyrroles, also called ‘pigments of life’, represent a small group of complex pigments which are the most abundant in living organisms. Structurally, tetrapyrroles are composed of four pyrrole-derived rings, joined together by methine bridges to form linear bile pigments or cyclic porphyrins (Solymosi and Mysliwa-Kurdziel 2017). Cyclic tetrapyrroles may differ in the oxidation state of pyrrole rings, peripheral substitutions, and the centrally chelated metal ions. They can be further subdivided into several groups—chlorophylls, hemes, cytochrome C, vitamin B12, and coenzyme F430, all of which contribute to the wide color range of natural pigments, from red, yellow to green, blue, and purple (Velisek et al. 2007). Moreover, degradation of cyclic tetrapyrroles generates linear tetrapyrroles like bilins, which loses one bridge carbon.
The most famous representatives of these ‘pigments of life’ are chlorophyll (‘plant blood’, in charge of the green color in plants) and heme (giving red color in animal blood). In natural producers, tetrapyrroles play important roles in a series of key metabolic processes, such as photosynthesis (e.g., chlorophyll) and electron transfer (e.g., cytochrome c) in plants, as well as transportation of oxygen (e.g., heme) in animals. More importantly, they have been proven to possess good antioxidant, anticancer, antimutagenic and anticlastogenic activities, showing great potential for applications in food and pharmaceutical industries (Beata and Solymosi 2016).
Biosynthetic pathway
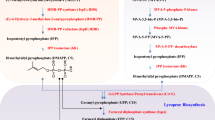
The biosynthetic pathway of tetrapyrroles can be sectioned into six stages (Kobayashi and Masuda 2016) (Fig. 7): (1) biosynthesis of the common precursor- 5-aminolevulinic acid (ALA); (2) formation of the pyrrole unit porphobilinogen (PBG); (3) formation of uroporphyrinogen III, the carbon skeleton of porphyrins; (4) decarboxylation of uroporphyrinogen III to generate coproporphyrinogen III via uroporphyrinogen decarboxylase; (5) conversion of coproporphyrinogen III to protoporphyrinogen IX under the catalysis of coproporphyrinogen oxidase; (6) generation of protoporphyrin IX, the completely conjugated ring system to show color, by losing six hydrogen atoms under the action of protoporphyrinogen oxidase.
Typical product—heme
Heme is the most famous representative of tetrapyrroles, which gives red color in animal blood and serves critical biological functions in transporting O2 and electrons, modulating gene expression, as well as regulating protein stability and cell differentiation. Due to its natural red color displayed in blood, it has a long history of use as a meat additive. Nowadays, it is widely used as an iron supplement in the healthcare industry, owing to its high bioavailability (Hoppe et al. 2013), and is drawing increasing attention in production of plant-based meat alternatives by increasing the meat-like color and flavor (Gerhard 2020; Simsa et al. 2019).
Free heme sources are traditionally isolated from biological samples, typically animal blood, with obvious issues of this production mode being low-yielding and ethical animal welfare concerns. Bioproduction of heme on a large scale is therefore greatly preferred. The biosynthetic pathway of heme starts with the formation of 5-aminolevulinate (ALA) via the C4 pathway (in humans, animals, fungi and few bacteria) and the C5 pathway (in plant and most bacteria), followed by cyclization, modification of the side chains, and incorporation of reduced iron. In 2003, an entire heme biosynthetic pathway including hemA (encoding ALA synthase of the C4 pathway) from Rhodobacter capsulatus, hemB, hemC, hemD and hemF genes from E. coli, hemE from Synechocystis, and hemY, hemH from B. subtilis was assembled in E. coli, leading to accumulation of 3.3 μmol/L of heme (Kwon et al. 2003). In another study, co-expression of ALA synthase (hemA), NADP-dependent malic enzyme (MaeB) and dicarboxylic acid transporter (DctA) in E. coli resulted in 6.4 mg/L of heme (Kwon et al. 2009), and additional overexpression of pantothenate kinase (coaA) led to 0.49 μmol/g DCW of heme (Lee et al. 2013). In a more recent study, comprehensive regulation of heme synthesis and secretion was conducted to improve heme production. Respective expression of the C4 and C5 pathways in E. coli revealed the superior capacity of the C5 pathway for producing ALA. Downstream pathways for heme biosynthesis were subsequently optimized by elimination of the limiting steps and balancing enzyme expression. In addition, three competing pathways including acetate and lactate synthesis, and heme degradation pathway, were blocked via knocking out pta (encoding phosphate acetyl transferase), ldhA (encoding lactate dehydrogenase) and yfeX (encoding a heme dechelatase). The resulting E. coli strain produced 7.88 mg/L of heme. Further overexpression of a heme exporter cmABC led to total heme production of 239.2 mg/L, among which 151.4 mg/L was secreted (Zhao et al. 2018). These studies demonstrated that microbial metabolic engineering by enhancement of the upstream C4/C5 pathway, together with downregulation with the heme degradation pathway and improvement of heme secretion, provides a highly promising option for enhancing the production of free heme.
Carotenoids
Structure, classification and function
Carotenoids are a family of lipophilic isoprenoid pigments produced by numerous bacteria (e.g., Corynebacterium michiganense, Micrococcus roseus, Brevibacterium spp., Bradyrhizobium spp., Gordonia jacobaea and Dietzia natronolimnaea), fungi (e.g., Blakeslea trispora, Phycomyces blakesleeanus, Rhodotorula spp., Xanthophyllomyces dendrorhous), and microalgae (e.g., the genus of Chlorella, Dunaliella, Coelastrella and Haematococcus). (Carlos Mata-Gomez et al. 2014; Mannazzu et al. 2015; Mussagy et al. 2019a). Recent studies have demonstrated that some multicellular organisms like aphids, adelgids, phylloxerids, and gall midges, possess the capability for de novo synthesis of carotenoids (Cobbs et al. 2013; Moran and Jarvik 2010; Novakova and Moran 2012; Zhao and Nabity 2017); in contrast, most higher animals (including humans) can only take carotenoids from their diet (Rodriguez-Concepcion et al. 2018). Structurally, carotenoids originate from the condensation of C5 isoprenoid units to generate a vast class of over 600 carotenoid structures, among which C40 carotenoids are the most abundant in nature (Kiokias et al. 2016). These pigments are mainly classified into two subgroups: (1) hydrocarbons-carotenes (e.g., lycopene, α-carotene, and β-carotene); and (2) xanthophylls, the oxygenated derivatives of carotenes (e.g., lutein, zeaxanthin, astaxanthin, canthaxanthin). They display in yellow, orange, red or even colorless, and possess excellent antioxidant, anticancer, and anti-inflammation activities. According to the reports from Allied Market Research, the global carotenoid market value was $1.5 billion in 2017, and is estimated to reach nearly $2.1 billion by 2025 (Dawande 2018). Structurally, carotenoids originate from the condensation of C5 isoprenoid units to generate a vast class of over 600 carotenoid structures, among which C40 carotenoids are the most abundant in nature.
Biosynthetic pathway
The biosynthetic process of carotenoids can be summarized as four steps, as shown in Fig. 8: (1) synthesis of the building units—isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP); (2) chain elongation by successive condensation reaction of IPP to DMAPP generating the growing polyprenyl diphosphate chain; (3)cyclization of linear isopentenyl pyrophosphate precursors to form the carotenoid carbon skeleton; (4) modification of the carotenoid carbon skeleton to generate diverse carotenoids. Among numerous carotenoids, β-Carotene and astaxanthin are the most commercially valuable products.
Schematic of carotenoids biosynthesis. Most carotenoids go through four synthetic steps consisting generation of building unit, chain elongation, cyclization, and modification. DXS, 1-deoxy-d-xylulose-5-phosphate synthase; DXR, 1-deoxy-d-xylulose-5-phosphate reductoisomerase; ISPD, 4-diphosphocytidyl-2C-methyl-D-erythritol synthase; IspE, 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase; IspF, 2C-methyl-D-erythritol-2,4-cyclodiphosphate synthase; IspG, 1-hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate synthase; IspH, 1-hydroxy-3-methyl-2-(E)-butenyl-4-diphosphate reductase; HMGS, HMG-CoA synthase; HMGR, HMG-CoA reductase; MK, mevalonate kinase; PMK, phosphomevalonate kinase; MVD1, mevalonate diphosphate decarboxylase; IPPI/IDI1, isopentenyl-diphosphate isomerase; GGPPS, GGPP synthase; PSY, phytoene synthase; PDS, phytoene desaturase; ZDS, ζ‐carotene desaturase; CRTISO, carotenoid isomerase; CrtB, phytoene synthase; CrtI, phytoene desaturase; DXP, 1-deoxy-D-xylulose-5-phosphate; MEP, 2-C-methyl-D-erythrito-l4-phosphate; CDP2ME, 4-diphosphocytidyl-2-C-methylerythritol; MEP-cPP, 2-C-methyl-D-erythritol-2,4-cyclo-diphosphate; HMBPP, (E)-4-hydroxy-3-methylbut-2-enyl-diphosphate; IPP, isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate; GGPP, geranylgeranyl diphosphate
Scrutinizing the first step, IPP and DMAPP may generated from one of two distinct pathways, the mevalonate (MVA) pathway and the 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway (Lange et al. 2000; Rohdich et al. 2003; Rohmer et al. 1993; Wölwer-Rieck et al. 2014). The MVA pathway is ubiquitous in bacteria, most fungi, and plants (in the cytoplasm), which initiates from acetyl-CoA and ends up with the production of DMAPP under the catalysis of acetoacetyl-CoA thiolase (ERG10), HMG synthase (HMGS), 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR), MVA kinase (MK) and phosphor-MVA kinase (PMK), diphospho-MVA decarboxylase (MVD1), and IPP isomerase (IDI) (Allen et al. 1967; Vranova et al. 2013). In the MVA pathway, HMGR is identified as the rate-limiting enzyme. The MEP pathway is present in most bacteria, green algae, and plants (in the chloroplast) (Rohdich et al. 2003). It starts from the condensation of glyceraldehyde-3-phosphate (G3P) and pyruvate to generate 1-deoxy-D-xylulose 5-phosphate (DXP), which is further catalyzed by 1-deoxy-D-xylulose 5-phosphate synthase (DXS), DXP reductoisomerase (DXR), 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase (ISPD), 4-(cytidine 5′-diphospho)-2-C-methyl-D-erythritol kinase (ISPE), 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase (ISPF), 4-hydroxy-3-methylbut-2-enyldiphosphate (HMBPP) synthase (ISPG), HMBPP reductase (ISPH), and IPP isomerase (IPPI), to yield IPP/DMAPP (the end product in the first step). The processes catalyzed by DXS (Lv et al. 2013a; Ramos et al. 2014; Xue and Ahring 2011), DXR (Lv et al. 2013a; Zhao et al. 2011) and IPPI (Lv et al. 2013b; Ramos et al. 2014) are identified to be the rate-limiting steps involved in the MEP pathway. In the second step, lycopene is the typical product from IPP/DMAPP, under catalysis of geranylgeranyl diphosphate synthase (GGPPS), phytoene synthase (PSY), phytoene desaturase (PDS), ZISO f-carotene isomerase (ZDS), and carotenoid isomerase CRTISO in plants, or under the catalysis of GGPP synthase (CRTE), phytoene synthase (CRTB), and phytoene desaturase/isomerase (CRTI) in non-photosynthetic bacteria. In the next step, α/β-carotene can be generated from lycopene under the catalysis of lycopene β-cyclase (LCYB) and/or lycopene ε-cyclase (LCYE). Further modification of carbon skeleton in the last step will result in other active carotenoids, e.g., lutein, zeaxanthin, and astaxanthin.
Typical product—β-carotene
β-Carotene is the most extensively studied carotenoid, abundant in leafy vegetables and is sold as a dietary supplement due to its well-known pro-vitamin A characteristic and excellent antioxidant activity. As the precursor of Vitamin A, β-carotene can help accelerate human growth, maintain a healthy immune system and good vision (Chen and Zhong 2015).
At present, the natural source-based production methods of β-carotene are either extraction from plants, silkworm excrement or algae, or cultivation of yeast and fungi. The extraction process of β-carotene from carrot, palm, seabuckthorn and other raw materials is complex. Moreover, its efficiency is limited by the long growth cycle of plants and the impact of climate and geography on their growth. Algae have proven to be more advantageous than plants in terms of productivity and production cycle. However, due to the harsh cultivation conditions and degradation issues, large-scale cultivation remains difficult. Microbial cultivation is a promising alternative with the advantage of continuous production, and has achieved stellar progress in the last decade (Table 1). Blakeslea trispora and Rhodotorula glutinis are the best-explored natural producers for β-carotene. The improvement in biosynthetic ability of these two strains have stemmed from the optimization of cultivation equipment133 and the culture medium (Chang-he et al. 2020; Tkáčová et al. 2017), random mutagenesis and addition of exogenous stimulants (Hu et al. 2013; Jing et al. 2016; Luo et al. 2021). The incomplete host genetic information and the lack of tools for genetic-level operation have thus far prevented more precise rational engineering of these strains. Researchers turned to engineering of E. coli and S. cerevisiae, the two model for prokaryotic and eukaryotic organisms, respectively. Herein we mainly focus on such studies reported in the last decade.
Via multiple strategies such as coordinating the inherent MEP pathway (Yuan et al. 2006) and β-carotene pathway (Zhao et al. 2013), introducing exogenous MVA pathway to increase the supply of precursors (Yang and Guo 2014; Yoon et al. 2009), cofactor engineering (Wu et al. 2020b), increasing membrane biosynthesis (Wu et al. 2017) and changing membrane morphology (Wu et al. 2019a) as well regulation of inner- and outer-membrane vesicle formation (Yang et al. 2021) to alleviate the burden caused by accumulation of hydrophobic macromolecule products on cell membrane, the β-carotene production of E.coli reached as high as 44.8 mg/g DCW. However, concerns such as the food safety of E. coli, the requirement of expensive inducers, and its susceptibility to phage infection, have thus far prevented the commercialization of β-carotene produced by engineered E. coli.
As a generally recognized as safe (GRAS) microorganism, S. cerevisiae has been widely used to produce carotenoids as well as other isoprenoids. In the early stage, the main research focuses were on improving the genetic stability of β-carotene synthetic genes in engineered strains and optimizing culture temperature (Shi et al. 2014; Sun et al. 2015). Overexpression of the rate-limiting enzyme in the MVA pathway and several novel genes outside the isoprenoid pathway and adjustment of the copy number of downstream pathway genes were then employed to balance the synthetic pathway and improve β-carotene production (Li et al. 2017; Yan et al. 2012). Deleting genes in bypassing pathways including lipid, amino acid and ergosterol syntheses, genes encoding transcription factors (e.g., Rox1) that represses many enzymes in the MVA and ergosterol synthesis pathways, as well as some genes involved in the stability of mRNA and protein (Ozaydin et al. 2013) also resulted in increased the accumulation of precursors and further increased final production of β-carotene. In addition, dynamic regulation of the synthetic pathway to separate β-carotene accumulation from cell growth greatly improved β-carotene production (Xie et al. 2015b). Recently, it was found that the synthesis and addition of unsaturated fatty acids also had a significant influence on β-carotene synthesis (Liu et al. 2016a; Sun et al. 2016).
The oleaginous yeast Yarrowia lipolytica has emerged as a promising microbial cell factory, due to its biochemical characteristics such as intrinsic high flux of acetyl-CoA and therefore a native high capacity to accumulate lipid-based chemicals. By strengthening and balancing the biosynthetic pathway (Gao et al. 2017; Larroude et al. 2018; Liu et al. 2021b; Qiang et al. 2020), production of up to 6.5 g/L β-carotene was achieved in Y. lipolytica.
Typical product—astaxanthin
Astaxanthin (3, 3′-dihydroxy-β,β-carotene-4,4′-dione), a red ketocarotenoid belonging to the terpene family, has received intensive attention worldwide due to its strong antioxidant activity and in particular, widespread use as feed additive for farmed salmon. The global market for astaxanthin was valued at USD 1,371.24 million in 2020 and is estimated to increase at an annual growth rate of 16.8% by 2028 (https://www.grandviewresearch.com/industry-analysis/global-astaxanthin-market). In recent years, microbial biosynthesis has emerged as a promising alternative astaxanthin production route to alga extraction and chemical synthesis. As summarized in Table 2, heterologous production of astaxanthin in microorganisms has been achieved in E. coli, S. cerevisiae, Y. lipolytica and Corynebacterium glutamicum 158.
Pathway optimization of astaxanthin could be divided into two parts: the upstream pathway section converting acetyl-CoA to β-carotene and the downstream pathway section converting β-carotene to astaxanthin. The upstream pathway has often been engineered using common strategies for terpene biosynthesis, such as improving the metabolic flux of MVA or MEP pathway (Kildegaard et al. 2017; Park et al. 2018) and downregulating competitive pathways like ergosterol synthesis (Kildegaard et al. 2017). To maximize the conversion of β-carotene to astaxanthin, the substrate preference and activity of β-carotene hydroxylase and ketolase need to be balanced. The carotenoids profile and astaxanthin contents vary among different combinations of β-carotene hydroxylase and ketolase from different organisms (Lu et al. 2017; Qi et al. 2020). To balance the activity of these enzymes, strategies including RBS site combination (Zelcbuch et al. 2013; Zhang et al. 2018b), promoter engineering and copy number adjustment (Gong et al. 2020; Kildegaard et al. 2017; Tramontin et al. 2019) were used.
Protein engineering also contributes to improvement of astaxanthin biosynthesis, including directed evolution (Zhou et al. 2017a, 2019), fusion expression (Nogueira et al. 2019) and construction of multi-enzyme complex (Qu et al. 2019).
Except for the synthetic pathway and the enzymes thereof, the properties of the chassis cell also influence astaxanthin yield. As an excellent antioxidant, high oxidative pressure is supposed to increase astaxanthin content. Using hydrogen peroxide as selective pressure, ARTP (atmospheric and room temperature plasma) mutagenesis (Jiang et al. 2020) and CRISPRi method (Lu and Liu 2019) generated mutant strains with improved astaxanthin production, uncovering gene targets related to ROS level, chronological lifespan and cell morphology.
To sum up, the typical strategies for heterologous production of astaxanthin in microorganisms include: (1) enhancing the supply of β-carotene as the precursor; (2) balancing the activities of β-carotene hydroxylase and β-carotene ketolase by screening, protein engineering or expression level regulation; (3) uncovering new engineering targets by random mutagenesis and omics analyses.
Flavonoids
Structure, classification and function
Flavonoids are a highly diverse group of plant secondary metabolites derived from the phenylpropanoid metabolisms. They are distinguished by the characteristic structural backbone of C6−C3−C6, consisting of two aryl rings linked by a heterocyclic ring. A variety of modifications across this backbone, e.g., hydroxylation, ring repositioning, acetylation, glycosylation, and methylations, result in the vast chemical diversity of flavonoids. Based on these structural modifications, flavonoids can be divided into six major categories: flavanones, flavones, isoflavones, flavonols, catechins, and anthocyanins (Dudnik et al. 2018).
Flavonoids are abundant in fruits, vegetables, grains, trees, and flowers, and possess a variety of biological functions. For example, flavonoids are responsible for the color of petals and buds as well as the aroma of flowers and fruits to attract pollinating insects. Specifically, most of flavanones, flavones, isoflavones, flavonols are light yellow or yellow-colored, whereas anthocyanin hues range from red, pink, magenta, to violet, purple, and blue. Flavonoids can also protect plants from diverse environmental stresses like UV, heat, frost, drought, flooding, etc., by modifying the flavonoid structure for survival (Panche et al. 2016). Apart from plants, many in vivo or in vitro assays also demonstrate potential health-promoting functions of flavonoids to humans. They have been recommended as a dietary functional ingredient because of their high antioxidant capacity and protective effects against many infectious and degenerative diseases, such as inflammation, cardiovascular diseases, cancers, Alzheimer’s disease (AD), and other age-related diseases (Carrizzo et al. 2013; Chirumbolo 2014; Rahman et al. 2006; Scalbert et al. 2005; Sekizawa et al. 2013; Xiao and Hogger 2015; Yamagata et al. 2015). These health-promoting bioactivities of flavonoids, together with their diverse colors and high solubility in water, broaden their usage in the field of pharmaceutics and make them excellent candidates of food colorants, functional food and beverages, and dietary supplements. The global flavonoid market is predicted to reach USD 1.06 billion by 2025. Notably, the increasing demand for anthocyanins as healthful coloring agents in the food and beverage industry is the major driving factor. As multiple anthocyanins have been approved by the European Food Safety Authority (Panel on Food and Nutrient Sources, 2013) for use in foods, their universal usage as alternatives to synthetic dyes is expected, once bioproduction capability meets demand.
Biosynthetic pathway
As shown in Fig. 9 (Lyu et al. 2019a), flavonoids are derived from L-phenylalanine (Phe) or L-tyrosine (Tyr). Both amino acids can be converted into p-coumaric acid by non-oxidative deamination with distinct enzymes. Subsequently, p-coumaric acid: CoA ligase (4CL) mediates the addition of a coenzyme A group to p-coumaric acid to generate 4-coumaroyl-CoA, which was then condensed with three molecules of malonyl-CoA to create chalcones by chalcone synthase (CHS). Finally, chalcones are isomerized to flavanones through chalcone isomerase (CHI) or by spontaneous reactions, resulting in flavanone structure, e.g., naringenin. Naringenin can undergo various modifications by a series of “decorating” enzymes, giving rise to the vast diversity of flavonoids. For instance, the biosynthesis of flavanones or isoflavonones can be achieved by introduction of flavone synthase (FNS) or isoflavone synthase (IFS), respectively, whereas additional combination of flavanone 3-hydroxylase (F3H), dihydroflavonol reductase (DFR), anthocyanidin synthase (ANS) and anthocyanidin 3-O-glucosyltransferase (3GT) will lead to production of anthocyanins from flavanones.
Schematic of flavonoids biosynthesis. PAL, phenylalanine ammonia lyase; C4H, cinnamate 4-hydroxylase; 4CL, p-coumaric acid: CoA ligase; TAL, tyrosine ammonia lyase; CHS, chalcone synthase; CHI, chalcone isomerase; FNS, flavone synthase; IFS, isoflavone synthase; F3H, flavanone 3-hydroxylase; FLS, flavonol synthase; F3′H, flavonoid 3′-hyroxylase; F3′5′H, flavonoid 3′,5′-hyroxylase; DFR, dihydroflavonol reductase; ANS, anthocyanidin synthase
Typical product—anthocyanins
Anthocyanins are water-soluble red, magenta and blue flavonoids. As multiple anthocyanins have already been approved by the European Food Safety Authority (Panel on Food and Nutrient Sources, 2013), their universal application as healthier and safer alternatives to synthetic dyes is expected imminently. Extensive genetic modification studies have been conducted to improve the productivity of anthocyanins in plants. Due to the complexity of the anthocyanin biosynthetic pathway, most strategies focused on regulation of R2R3 MYB transcription factors and MYC-like basic helix–loop–helix (bHLH) and WD40-repeat proteins. One typical example is enrichment of anthocyanins in tomato via expression of bHLH-type Delila (Del) and MYB-related factor Rosea1 (Ros1), which resulted in 2.83 mg/g (fresh weight) of anthocyanin (Butelli et al. 2008). As reviewed by Naing et al., great success has been achieved in metabolic engineering of plants (Naing and Kim 2018). However, this production approach as aforementioned, relies heavily on seasonal and environmental conditions and suffers from the difficulty of product purification as well as slow plant growth.
Microbial production provides an alternative approach to produce anthocyanin. In 2005, heterologous production of anthocyanins was first reported in E. coli (Yajun et al. 2005). A four-step metabolic pathway, consisting F3H from Malus domestica, DFR from Anthurium andraeanum, ANS from M. domestica, and 3-GT from Petunia hybrid, was constructed in E. coli, generating 5.6 μg/L of pelargonidin 3-O-glucoside and 6.0 μg/L of cyanidin 3-O-glucoside. In the following studies, systematic engineering strategies, including enhancing precursor supply (e.g., UDP-glucose), enzyme fusion, balancing gene expression level, and optimization of cultivation condition (e.g., pH), have been employed to further enhance the yield and titer of anthocyanin (Lim et al. 2015; Shrestha et al. 2019). As reported by Lim et al. (2015) and Shrestha et al. (2019), up to 350 mg/L and 439 mg/L of cyanidin 3-O-glucoside were obtained in E. coli, by using catechin as the substrate. Similar combinatorial engineering strategies also led to ~ 40 mg/L of cyanidin 3-O-glucoside in Corynebacterium glutamicum (Zha et al. 2018). More recently, de novo biosynthesis of anthocyanins has been made possible in E. coli and S. cerevisiae, via employment of a polyculture system and introduction of the whole pathway from phenylalanine to anthocyanin (Eichenberger et al. 2018; Jones et al. 2017). This approach greatly reduces cost via removing the need for addition of expensive flavonoid substrates during cultivation. Typical examples of metabolic engineering of plants and microorganisms for anthocyanins production are listed in Table 3.
Curcuminoids—curcumin
Structure, classification and function
Curcuminoids are yellow polyphenolic compounds, with a typical structural backbone C6–C7–C6, derived from turmeric (the rhizome of the herb Curcuma longa mainly distributed in tropical and sub-tropical South-East Asia). As one of the most popular medicinal herbs, Curcuma longa has been used for the treatment of jaundice and other liver ailments in China, Japan, and India for many centuries. Curcumin is identified as the main active chemical constituent while bisdemethoxycurcumin and dicinnamoylmethane are found to be the other functional curcuminoids in turmeric. The studies on pharmacological activities of curcuminoids have been well reviewed previously (Amalraj et al. 2017), displaying prospects of curcuminoids in disease treatments. In addition to medical applications, curcuminoids have a long history as food coloring and flavoring agent. The World Health Organization (WHO) stated the acceptable daily intake of curcuminoids is in the range of 0–3 mg/kg. The safety of curcuminoids, as a food additive, has also been proved by FDA in USA (Amalraj et al. 2017).
Biosynthetic pathway
Curcuminoids and flavonoids share the precursor pathway starting from phenylalanine or tyrosine to coumaroyl-CoA. For biosynthesis of curcuminoids (Fig. 10), coumaroyl-CoA will be converted to feruloyl-CoA first by the actions of p-coumaroyl shikimate transferase (CST), p-coumaroyl 5-O-shikimate 3’-hydroxylase (CS3’H), and caffeoyl-CoA O-methyltransferase (CCoAOMT). After that, curcuminoids can be produced by type III polyketide synthases, including diketide-CoA synthase (DCS) and curcuminoid synthase (CURS) (Lan et al. 2018).
Typical product—curcumin
Curcumin is the archetypal curcuminoid. It has been used as a natural coloring agent as well as food additive (to optimize food flavor) for centuries. For improved industrial commercialization, one effective strategy is to accelerate plant growth and induce plant defense systems via elicitation approaches. For example, bacterization of turmeric resulted in 13.6% increase of curcumin production (4.16 g/100 g) via promoting plant growth (Chauhan et al. 2017); elicitation by chitosan treatment improved the yield of curcumin by 100% (Sathiyabama et al. 2016). On the other hand, in vitro propagation provides another solution to resolve the problem of yield variability and low efficiency of vegetative propagation in traditional breeding. As reported by Pistelli et al., more than 260 μg/g fresh weight of curcumin was obtained from in vitro microrhizomes (Pistelli et al. 2012). In a more recent study, optimization of plant growth regulators, including sucrose, α-naphthalene acetic acid, 6-benzylaminopurine and light quality, achieved 534 μg/g of curcumin in in vitro microrhizomes of Curcuma aromatic (Wu et al. 2015).
Furthermore, in the past two decades, much efforts have also been made in the heterologous synthesis of curcumin in microorganisms (e.g., E. coli and Aspergillus oryzae) by synthetic biology (Table 4). By fine-tuning gene expression with employment of heat shock promoters (dnaK and ibpA) and optimization of RBS, 17 μM curcumin was obtained from ferulic acid in the strain expressing 4CL-DCS-CURS1 (Rodrigues et al. 2017). Optimization of cultivation conditions, including E. coli subspecies, induction parameters, culture media and carbon source concentration, led to high titers of curcumin (817.7 μM) from ferulic acid (Couto et al. 2017). In order to avoid addition of the expensive precursor ferulic acid, de novo biosynthetic pathway of curcumin from glucose or tyrosine was developed, via co-expression of TAL, 4-coumarate 3-hydroxylase (C3H), caffeic acid 3-O-methyltransferase (COMT), 4CL and CUS (or DCS and CURS1), while the yield of curcumin was quite low (0.2–0.7 mg/L) (Wang et al. 2015). In a later study, randomly modification of the 5’-untranslational region (UTR) sequences of all six enzymes involved in curcumin synthesis with design of MAGE oligomers and introduction of λ red recombination system resulted in approximately 38.2-fold improvement of curcumin production (3.8 mg/L) from glucose (Kang et al. 2018). In another study, in order to reduce cell metabolic burden caused by introduction of multiple genes, the whole curcumin biosynthetic pathway from tyrosine was divided into two modules and transferred into two separate E. coli strains, which led to 1.6-fold increase of final curcumin production (15.9 mg/L) (Rodrigues et al. 2020).
Apart from E. coli, the filamentous fungus Aspergillus oryzae has also been used for production of curcuminoids via metabolic engineering. In a recent study, overexpression of CUS in A. oryzae was reported to produce 64 μg/plate of curcumin from feruloyl-N-acetylcysteamine (feruloyl-CoA analog). Enhancement of malonyl-CoA supply, via disruption of SnfA (an inhibitor of ACC enzyme) and SCAP (involved in acetyl-CoA-consuming sterol biosynthesis pathway), led to sixfold increase of curcumin yield (404 μg/plate) (Kan et al. 2019). From these studies, it can be understood that improving the de novo production ability and relieving the metabolic burden caused by introduction of multiple genes are key approaches for achieving high-yield production of curcumin by heterologous hosts.
Betalains
Structure, classification and function
Betalains, also known as chromo-alkaloids, are water-soluble nitrogen-containing pigments. They are mainly present in plants of the order Caryophyllales and some higher fungi like Amanita and Hygrocybe genera. Betalamic acid is the common chromophore to all betalains. According to the structural differences in the betalain subgroups, betalains can be subdivided into betacyanins (red–violet) and betaxanthins (yellow–orange), whereby betacyanins are formed by conjugating betalamic acid with cyclo-dihydroxyphenylalanine (cDOPA) glucoside while betaxanthins result from the condensation of betalamic acid with amino acids or amines (Gandia-Herrero and Garcia-Carmona 2013). Typical betacyanins include betanin, isobetanin, probetanin, and neobetanin; and the main betaxanthins present in plants are vulgaxanthin, miraxanthin, portulaxanthin, and indicaxanthin.
In the plant kingdom, betalains and anthocyanins are uniquely mutually exclusive, having never been detected in the same plant despite their similar biological functions (mainly coloration). Different from anthocyanins, the color of betalains does not rely on pH (no change in the pH range of 3 to 7) and thus are good colorants for food with taste varying from sour to neutral. The main edible plants containing betalains include beets, Swiss chard, cactus fruit, and amaranth. Among them, red beetroot, the plant mostly cultivated in America and Britain, is the major commercially exploited source of betalains. Red beet pigments have been approved for usage in food processing by Europe (EU, E162) and the USA (FDA, Title 21 of Code of Federal Regulations-CFR-73.40) (Scotter 2011; Wrolstad and Culver 2012), commercialized as either liquid concentrates or spray-dried powders, containing from 0.3% to 1% of pigment, and are used as colorants for manufactured food products, such as red wine, meat, ice cream, soft drinks, sugar sweets, etc. (Akbar Hussain et al. 2018). They are also used as coloring agents in the pharmaceutical industry in drug formulations.
In recent years, betalains have also attracted increasing attention due to their pharmacological activities, including antioxidant, antimicrobial, and anticancer. Both fruit extracts rich in betalains and purified betalain compounds could act as free radical scavengers to protect proteins and NDA from oxidative damage (Escribano et al. 2017; Kumar et al. 2015; Sawicki and Wiczkowski 2018; Swarna et al. 2013). These discoveries would surely accelerate further development of betalains for application in the food and health care industries.
Biosynthetic pathway
As compared with other important pigments like carotenoids and flavonoids, the biosynthetic pathway of betalains is much less well understood. By now, two pathways have been proposed (Gandia-Herrero and Garcia-Carmona 2013; Tanaka et al. 2008), of which the widely accepted one starts from tyrosine while the other, derived from tyramine, is still under exploration. The tyrosine pathway (Fig. 11) begins with the hydroxylation of tyrosine to dihydroxyphenylalanine (DOPA) under the catalysis of tyrosinase. Subsequently, DOPA may proceed in 2 divergent metabolic directions: either converted to betalamic acid by DOPA dioxygenase and spontaneous reaction, or to cycol-DOPA via a tyrosinase-catalyzed reaction followed by a spontaneous cyclization process.
Schematic of betalains biosynthesis. Betalains can be subdivided into betacyanins (red–violet) and betaxanthins (yellow–orange), whereby betacyanins are formed by conjugating betalamic acid with cyclo-dihydroxyphenylalanine (cDOPA) glucoside while betaxanthins result from the condensation of betalamic acid with amino acids or amines. DOPA, dihydroxyphenylalanine
Typical product—betanin
Among betalains, the main red pigment from red beet—betanin, is the most frequently utilized for commercial applications owing to its robust stability, high extinction coefficient and color intensity (Esatbeyoglu et al. 2015; Imtiyaj Khan and Giridhar 2015). In the food industry, betanin is commonly obtained from beet juice or root macerates via conventional solid–liquid extraction. Recent application of microwave-assisted extraction and high-pressure CO2-assisted extraction techniques (Cardoso-Ugarte et al. 2014; Ciriminna et al. 2018) have dramatically increased the extraction efficiencies. Besides, great efforts have been made in in vitro production of betanin via plant cell and tissue culture in diverse species (Georgiev et al. 2008), e.g., Amaranthus tricolor (Biswas et al. 2013), Pereskia aculeata Miller (Lage et al. 2015), and Celosia cristata (Warhade and Badere 2018). Selection of highly productive cell lines, optimization of inoculum conditions, addition of growth precursor, elicitation and permeabilization are also major effective strategies.
In parallel to the progress made in production enhancement from native producers, research has also been directed to heterologous biosynthesis of betalains via metabolic engineering. Identification of dihydroxyphenylalanine (DOPA) dioxygenase (DOD) enabled in vitro synthesis of yellow betaxanthins in E. coli, Solanum tuberosum (potato) and Antirrhinum majus, in the culture systems fed with the substrate DOPA (Gandia-Herrero and Garcia-Carmona 2012; Harris et al. 2012). Subsequently, with the discovery of new P450 enzyme CYP76AD1 which is responsible for producing cyclo-DOPA from DOPA, heterologous production of betanidin (the precursor of betanin) in yeast was achieved by introduction of DODA1 from Beta vulgaris and CYP76AD1 from beet (Hatlestad et al. 2012). In a recent study, CYP76AD1 is elucidated to catalyze both DOPA formation and subsequent conversion to cyclo-DOPA; and another P450 enzyme CYP76AD6 uniquely possesses tyrosine hydroxylase activity (Polturak et al. 2016). Based on discovery of enzymes in charge of the first committed step, de novo production of betalains has been successfully achieved in engineered tobacco, tomato, potato, eggplant, and ornamental petunia by expression of DODA1, CYP76AD1 and cycl0-DOPA-5-O-glucosyltransferase (cDOPA5GT) (Polturak et al. 2016, 2017). Similar operations in yeast led to 17 mg/L of betanin (Grewal et al. 2018) and two thirds of pigments were found to be secreted into the media, which would significantly reduce the efforts of pigment extraction as compared to plants. In a more recent study, expression of de-regulated arogenate dehydrogenase (ADH) resulted in sevenfold increase in betalain yield (855 mg/kg·FW) in planta (Timoneda et al. 2018). To date, these studies mainly focused on pathway mining and assembly, and more pathway optimization strategies are required to boost heterologous production of betalain.
Challenges and future prospects
As compared with chemical synthesis, natural pigments produced using either natural producers or engineered organisms are currently not cost competitive, due to the generally low contents and high production cost. For example, the chemical synthesis of β-carotene was realized by using β-ionone as the precursor as early as 1960s, by Roche (since 1954) and BASF (since 1960). As compared with natural β-carotene derived from plants and cultivation of yeast, fungi, or algae, the price of chemical synthesis is two times cheaper. Nevertheless, according to the Global Market Size Analysis by GRAND VIEW RESEARCH, synthetic β-carotene sources only accounted for a very small share in the global industry due to the growing health awareness and various disadvantages regarding the usage of synthetic products (https://www.grandviewresearch.com/industry-analysis/beta-carotene-market). By contrast, natural β-carotene kept witnessing steady growth trend in these years, especially for algae route, which accounted for over 35% of total revenue in 2015. Therefore, utilization of natural pigments is the future trend in the relevant fields; however, how to reduce the production cost is one of the biggest issues to accelerate large-scale production and commercialization.
One main cost source for pigment biosynthesis is the cultivation expense. Specifically, strong light is required for cultivation of pigment-producing algae and the cell density is often limited. For plant extraction, there are additional issues of farmland occupation and climate dependence. Although microbial cultivation largely circumvents these issues, it currently suffers from the main bottleneck of high production costs especially when expensive glucose or sucrose is commonly used as the feedstock. To lower the cost of natural pigments, approaches should be developed for effective utilization of significantly cheaper agrowastes as the feedstocks, including proper pretreatment of biomass wastes and detoxification of the hydrolysates, as well as strain engineering towards efficient co-utilization of sugars and enhanced inhibitor tolerances.
Another obstacle for commercialization of natural pigments is the low productivity of natural producers. As mentioned before, great advances have been achieved by optimization of plant extraction, in vitro tissue culture, microbial cultivation, as well as heterologous synthesis by metabolic engineering to improve the productivity of bio-producers; whereas each approach still faces specific bottlenecks: (1) extraction from natural pigment-producing plants suffers from long cultivation cycle; (2) in vitro tissue culture encounters difficulties in scaling up; (3) pigment production by isolated microbial strains still encounters a glass ceiling because the regulatory mechanisms of all the different pigment biosynthesis are yet to be fully understood and microbial classis are often resistant to gene manipulations; moreover, some pigment-producing microorganisms may have biosafety issues; (4) using microorganisms with well-annotated genetic backgrounds (E. coli, S. cerevisiae, B. subtilis, Y. lipolytica, etc.) and sufficient manipulation tools as hosts to construct cell factories is therefore an emergent focus for biotechnological production of natural pigments. As compared with other high-valued compounds, the color characteristic of natural pigments endows heterologous engineering with huge advantages on protein engineering, directed-evolution, as well as sensor-based dynamic control, upon the natural high-throughput screening methods using pigments as direct indicators. Despite the significant advances made in biosynthesis of pigments via metabolic engineering, a number of challenges remain further down the road towards industrialization.
Challenge 1: Tricky enzyme expression issue. The yields of pigments produced by cultivation are generally low due to the incompatibility among the pathway enzymes and between the enzymes and the intracellular environment of the host. Dividing up a long biosynthetic pathway into multiple modules and matching them with compatible hosts after individual optimizations may thus serve as a more ideal approach for engineering complex pathways.
Challenge 2: Conflict between product accumulation and cell growth, caused by the accumulation of toxic intermediates and poor metabolic balance during static metabolic engineering. In order to allay this trade-off phenomenon, many strategies on dynamic regulation via two-stage cultivation and continuous control have been proposed. However, in many cases, the sensors and actuators for target compounds are unclear and the exploration is time-consuming. Besides, two-stage cultivation strategies will require sufficiently detailed mathematical models and improved genetic circuits (Venayak et al. 2015). These challenges offer the opportunity to motivate the integration of artificial intelligence, system biology and metabolic engineering.
Challenge 3: Lack of color diversity. Considering the rich pool of unculturable microorganisms, metagenomics may be conducted for mining of synthetic pathways towards novel pigments. Design of artificial synthetic pathways for generating brand-new pigments based on the understanding of the synthetic mechanism and chemical nature of existing pigments is another future direction.
Challenge 4: Storage of lipophilic pigments. Distinct with above three points, this challenge only occurs with heterologous production of lipophilic compounds, but not for hydrophilic products. The natural lipophilic nature of the pigments leads to their accumulation in cell membranes, which however exerts burden on the chassis cells. The generally higher production of carotenoids in oleaginous yeast and their storage in lipid droplets in natural algal producers implies storage of such products in natural or artificial subcellular compartments may be a promising solution.
Apart from above four challenges, safety is another major issue with the fermented pigments using genetically modified organisms for food application, with concerns about endotoxin and introduction of selective markers. The regulation (EC) No. 1829/2003 of the European Parliament and the Council made clear definition with the genetically modified food and feed sources, which are recognized to be the food products using genetically modified organisms (GMO) as the material, containing or produced from GMO. In EU, the commercialization of all GMOs and derived products like color additives must undergo risk assessment and regulatory approval by European Food Safety Authority (EFSA), according to the regulation (EC) No. 1829/2003. Detailed guidance for the risk assessment of genetically modified microorganism and their products as well as the presence at low level of genetically modified plant materials, are adopted by the EFSA GMO Panel in 2011 and 2017 (Naegeli et al. 2017; Organisms 2011). Based on the assessment of the taxonomic identity, safety concerns and the body of knowledge, many microorganisms belonging to filamentous fungi, bacteriophages, Streptomycetes, Oomycetes, Enterococcus faecium and E. coli were excluded in list of the qualified presumption of safety (QPS) of EFSA. This does not mean that all these strains are not allowed to be used the hosts to produce food products. As long as the safety of the strain is proven, it may still be authorized by the EU. Actually, to date, a number of natural products produced by genetic engineered E. coli has been authorized (Bampidis et al. 2020, 2019, 2021). If the genetic modification process does not introduce resistance genes, the possibility of passing the EFSA assessment will also be greatly improved. In future, more GRAS chassis systems should be explored with seamless cloning techniques for safe bioproduction of pigments.
Conclusions
In summary, great strides have been devoted to the in vitro production of natural pigments via plant cell/tissue culture or optimization of microbial cultivation approaches. More remarkably, synthetic biology together with omics technologies have shed lights on the potential of development of heterologous systems for pigment production, by pathway exploration, pathway construction, and pathway optimization. Based on the studies in the last decade, omics technologies contributed significantly to mining of metabolic pathways and regulation mechanisms of pigment production; and substantial progress have been achieved by metabolic engineering strategies at the levels of parts, network and systems. With the advent and continuing rapid progress of novel biotechnologies, the prospect of large-scale commercial production of diverse natural pigments is likely to be realized in the near future.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
References
Aguiar TQ, Dinis C, Domingues L (2014) Cre-loxP-based system for removal and reuse of selection markers in Ashbya gossypii targeted engineering (vol 68, pg 1, 2014). Fungal Genet Biol 70:11–11
Ahmadkelayeh S, Hawboldt K (2020) Extraction of lipids and astaxanthin from crustacean by-products: a review on supercritical CO2 extraction. Trends Food Sci Technol 103:94–108
Akbar Hussain E, Sadiq Z, Zia-Ul-Haq M (2018) Betalains: biomolecular aspects. Springer, Cham
Akdemir H, Silva A, Zha J, Zagorevski DV, Koffas MAG (2019) Production of pyranoanthocyanins using Escherichia coli co-cultures. Metab Eng 55:290–298
Alipour S, Habibi A, Taavoni S, Varmira K (2017) Beta-carotene production from soap stock by loofa-immobilized Rhodotorula rubra in an airlift photobioreactor. Process Biochem 54:9–19
Allen CM, Alworth W, Macrae A, Bloch K (1967) A long chain terpenyl pyrophosphate synthetase from Micrococcus Lysodeikticus. J Bio Chem 242(8):1895–2000
Amalraj A, Pius A, Gopi S, Gopi S (2017) Biological activities of curcuminoids, other biomolecules from turmeric and their derivatives—a review. J Tradit Complement Med 7(2):205–233
Astorg P (1997) Food carotenoids and cancer prevention—an overview of current research. Trends Food Sci Technol 8:406–413
Bahabadi S, Sharifi M, Murata J, Satake H (2014) The effect of chitosan and chitin oligomers on gene expression and lignans production in Linum album cell cultures. J Med Plants 13:46–53
Balakrishnan B, Karki S, Chiu SH, Kim HJ, Suh JW, Nam B, Yoon YM, Chen CC, Kwon HJ (2013) Genetic localization and in vivo characterization of a Monascus azaphilone pigment biosynthetic gene cluster. Appl Microbiol Biotechnol 97(14):6337–6345
Bampidis V, Azimonti G, Bastos MD, Christensen H, Dusemund B, Kouba M, Durjava MK, Lopez-Alonso M, Puente SL, Marcon F, Mayo B, Pechova A, Petkova M, Sanz Y, Villa RE, Woutersen R, Costa L, Cubadda F, Dierick N, Flachowsky G, Glandorf B, Herman L, Mantovani A, Saarela M, Wallace RJ, Anguita M, Tarres-Call J, Ramos F, Us EPAPS (2019) Safety and efficacy of L-leucine produced by fermentation with Escherichia coli NITE BP-02351 for all animal species. Efsa J. https://doi.org/10.2903/j.efsa.2019.5689
Bampidis V, Azimonti G, Bastos MD, Christensen H, Dusemund B, Kouba M, Durjava MK, Lopez-Alonso M, Puente SL, Marcon F, Mayo B, Pechova A, Petkova M, Ramos F, Sanz Y, Villa RE, Woutersen R, Glandorf B, Herman L, Maradona MP, Saarela M, Anguita M, Brozzi R, Galobart J, Gregoretti L, Innocenti M, Lopez-Galvez G, Sofianidis K, Pettenati E, Vettori MV, Us EPAPS (2020) Safety and efficacy of L-cysteine hydrochloride monohydrate produced by fermentation using Escherichia coli KCCM 80180 and Escherichia coli KCCM 80181 as a flavouring additive for all animal species. Efsa J. https://doi.org/10.2903/j.efsa.2020.6003
Bampidis V, Azimonti G, Bastos MD, Christensen H, Dusemund B, Kouba M, Lopez-Alonso M, Puente SL, Marcon F, Mayo B, Pechova A, Petkova M, Ramos F, Sanz Y, Villa RE, Woutersen R, Herman L, Anguita M, Galobart J, Pettenati E, Tarres-Call J, Us EPAPS (2021) Safety and efficacy of a feed additive consisting of l-histidine monohydrochloride monohydrate produced using Escherichia coli NITE SD 00268 for all animal species (Kyowa Hakko Europe GmbH). Efsa J. https://doi.org/10.2903/j.efsa.2021.6622
Beata M-K, Solymosi K (2016) Chlorophylls and their derivatives used in food industry and medicine. Mini Rev Med Chem. https://doi.org/10.2174/1389557516666161004161411
Begum H, Yusoff FM, Banerjee S, Khatoon H, Shariff M (2016) Availability and utilization of pigments from microalgae. Crit Rev Food Sci Nutr 56(13):2209–2222
Benstein RM, Cebi Z, Podola B, Melkonian M (2014) Immobilized growth of the peridinin-producing marine dinoflagellate Symbiodinium in a simple biofilm photobioreactor. Mar Biotechnol 16(6):621–628
Bhojwani SS, Dantu PK (2013) Production of industrial phytochemicals plant tissue culture: an introductory text. Springer, Delhi, pp 275–286
Biswas M, Das SS, Dey S (2013) Establishment of a stable Amaranthus tricolor callus line for production of food colorant. Food Sci Biotechnol 22(1):1–8
Bitok JK, Lemetre C, Ternei MA, Brady SF (2017) Identification of biosynthetic gene clusters from metagenomic libraries using PPTase complementation in a Streptomyces host. Fems Microbio Lett. https://doi.org/10.1093/femsle/fnx155
Butelli E, Titta L, Giorgio M, Mock H-P, Matros A, Peterek S, Schijlen EGWM, Hall RD, Bovy AG, Luo J, Martin C (2008) Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat Biotechnol 26(11):1301–1308
Cardoso-Ugarte GA, Sosa-Morales ME, Ballard T, Liceaga A, Martin-Gonzalez MFS (2014) Microwave-assisted extraction of betalains from red beet (Beta vulgaris). Lwt Food Sci Technol 59(1):276–282
Carlos Mata-Gomez L, Cesar Montanez J, Mendez-Zavala A, Noe Aguilar C (2014) Biotechnological production of carotenoids by yeasts: an overview. Micro Cell Fact. https://doi.org/10.1186/1475-2859-13-12
Carrizzo A, Forte M, Damato A, Trimarco V, Salzano F, Bartolo M, Maciag A, Puca AA, Vecchione C (2013) Antioxidant effects of resveratrol in cardiovascular, cerebral and metabolic diseases. Food Chem Toxicol 61:215–226
Chandler DR (2001) Mauve: how one man invented a color that changed the world. Relig Humanism 35(3–4):77–78
Chang J-J, Thia C, Lin H-Y, Liu H-L, Ho F-J, Wu J-T, Shih M-C, Li W-H, Huang C-C (2015) Integrating an algal β-carotene hydroxylase gene into a designed carotenoid-biosynthesis pathway increases carotenoid production in yeast. Bioresour Technol 184:2–8
Chang-he D, Meng Y, Hong-li L (2020) Study on the optimization of beta-carotene production by fermentation of Blakeslea trispora. Food Res Dev 41(16):188–194
Chao F, Hao H, Yan L, Wen-zhong W (2018) Critical control points of industrial carotenoid production by Blakeslea trispora. Food Ferment Ind 5:284–290
Chauhan AK, Maheshwari DK, Dheeman S, Bajpai VK (2017) Termitarium-inhabiting Bacillus spp. enhanced plant growth and bioactive component in turmeric Curcuma longa L.). Curr Microbiol 74(2):184–192
Cheirsilp B, Torpee S (2012) Enhanced growth and lipid production of microalgae under mixotrophic culture condition: effect of light intensity, glucose concentration and fed-batch cultivation. Bioresour Technol 110:510–516
Chen H, Zhong Q (2015) Thermal and UV stability of beta-carotene dissolved in peppermint oil microemulsified by sunflower lecithin and Tween 20 blend. Food Chem 174:630–636. https://doi.org/10.1016/j.foodchem.2014.11.116
Chen L, Zhang YH, Wang S, Zhang Y, Huang T, Cai YD (2017a) Prediction and analysis of essential genes using the enrichments of gene ontology and KEGG pathways. PLoS ONE 12(9):e0184129
Chen W, Chen R, Liu Q, He Y, He K, Ding X, Kang L, Guo X, Xie N, Zhou Y, Lu Y, Cox RJ, Molnar I, Li M, Shao Y, Chen F (2017b) Orange, red, yellow: biosynthesis of azaphilone pigments in Monascus fungi. Chem Sci 8(7):4917–4925
Chirumbolo S (2014) Dietary assumption of plant polyphenols and prevention of allergy. Curr Pharm Des 20(6):811–839
Chou Y-L, Ko C-Y, Yen C-C, Chen L-FO, Shaw J-F (2019) Multiple promoters driving the expression of astaxanthin biosynthesis genes can enhance free-form astaxanthin production. J Microbiol Methods 160:20–28
Ciriminna R, Fidalgo A, Danzi C, Timpanaro G, Ilharco LM, Pagliaro M (2018) Betanin: a bioeconomy insight into a valued betacyanin. Acs Sustain Chem Eng 6(3):2860–2865
Cobbs C, Heath J, Stireman JO, Abbot P (2013) Carotenoids in unexpected places: gall midges, lateral gene transfer, and carotenoid biosynthesis in animals. Mol Phylogenet Evol 68(2):221–228
Couto MR, Rodrigue JL, Rodrigues LR (2017) Optimization of fermentation conditions for the production of curcumin by engineered Escherichia coli. J R Soc Interface. https://doi.org/10.1098/rsif.2017.0470
Dawande R (2018) Carotenoids market by product (astaxanthin, capsanthin, lutein, beta-carotene, lycopene, and others), source (natural and synthetic), and application (animal feed, human food, dietary supplement, and others) - global opportunity analysis and industry forecast, 2018–2025. Nutraceutical/Wellness food:266
de Castro RJS, Sato HH (2015) Enzyme production by solid state fermentation: general aspects and an analysis of the physicochemical characteristics of substrates for agro-industrial wastes valorization. Waste Biomass Valorization 6(6):1085–1093
Dinh CV, Prather KLJ (2019) Development of an autonomous and bifunctional quorum-sensing circuit for metabolic flux control in engineered Escherichia coli. Proc Natl Acad Sci USA 116(51):25562–25568
Dong YH, Feldberg L, Rogachev I, Aharoni A (2021) Characterization of the production of anthocyanin pigment 1 Arabidopsis dominant mutant using dlemma dual isotope labeling approach. Phytochemistry. https://doi.org/10.1016/j.phytochem.2021.112740
Dudnik A, Gaspar P, Neves AR, Forster J (2018) Engineering of microbial cell factories for the production of plant polyphenols with health-beneficial properties. Curr Pharm Des 24(19):2208–2225
Eichenberger M, Hansson A, Fischer D, Durr L, Naesby M (2018) De novo biosynthesis of anthocyanins in Saccharomyces cerevisiae. FEMS Yeast Res. https://doi.org/10.1093/femsyr/foy046
El-Batal AI, El-Sayyad GS, El-Ghamery A, Gobara M (2017) Response surface methodology optimization of melanin production by Streptomyces cyaneus and synthesis of copper oxide nanoparticles using gamma radiation. J Cluster Sci 28(3):1083–1112
Engler C, Gruetzner R, Kandzia R, Marillonnet S (2009) Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS ONE. https://doi.org/10.1371/journal.pone.0005553
Esatbeyoglu T, Wagner AE, Schini-Kerth VB, Rimbach G (2015) Betanin-A food colorant with biological activity. Mol Nutr Food Res 59(1):36–47. https://doi.org/10.1002/mnfr.201400484
Escribano J, Cabanes J, Jiménez-Atiénzar M, Ibañez-Tremolada M, Gómez-Pando LR, García-Carmona F, Gandía-Herrero F (2017) Characterization of betalains, saponins and antioxidant power in differently colored quinoa (Chenopodium quinoa) varieties. Food Chem 234:285–294
Fangyu D, Feng L, Wenming S, Jianlin C, Bin W, Bingfang H (2018) Efficient synthesis of crocins from crocetin by a microbial glycosyltransferase from Bacillus subtilis 168. J Agric Food Chem 66(44):11701–11708
Fathi Z, Tramontin LRR, Ebrahimipour G, Borodina I, Darvishi F (2021) Metabolic engineering of Saccharomyces cerevisiae for production of beta-carotene from hydrophobic substrates. FEMS Yeast Res. https://doi.org/10.1093/femsyr/foaa068
Frusciante S, Diretto G, Bruno M, Ferrante P, Pietrella M, Prado-Cabrero A, Rubio-Moraga A, Beyer P, Gomez-Gomez L, Al-Babili S, Giuliano G (2014) Novel carotenoid cleavage dioxygenase catalyzes the first dedicated step in saffron crocin biosynthesis. Proc Natl Acad Sci USA 111(33):12246–12251
Fu J, Bian X, Hu S, Wang H, Huang F, Seibert PM, Plaza A, Xia L, Mueller R, Stewart AF, Zhang Y (2012) Full-length RecE enhances linear-linear homologous recombination and facilitates direct cloning for bioprospecting. Nat Biotechnol 30(5):440
Ganapathy A, Jayavel S, Natesan S (2016) Draft genome sequence of carotenoid producing yellow pigmented Planococcus maritimus MKU009. J Genomics 4:23–25
Gandia-Herrero F, Garcia-Carmona F (2012) Characterization of recombinant Beta vulgaris 4,5-DOPA-extradiol-dioxygenase active in the biosynthesis of betalains. Planta 236(1):91–100
Gandia-Herrero F, Garcia-Carmona F (2013) Biosynthesis of betalains: yellow and violet plant pigments. Trends Plant Sci 18(6):334–343
Ganesan V, Li Z, Wang X, Zhang H (2017) Heterologous biosynthesis of natural product naringenin by co-culture engineering. Syn Syst Biotechno 2(3):236–242
Gao S, Tong Y, Zhu L, Ge M, Zhang Y, Chen D, Jiang Y, Yang S (2017) Iterative integration of multiple-copy pathway genes in Yarrowia lipolytica for heterologous beta-carotene production. Metab Eng 41:192–201
Georgiev V, Ilieva M, Bley T, Pavlov A (2008) Betalain production in plant in vitro systems. Acta Physiol Plant 30(5):581–593
Georgiev V, Slavov A, Vasileva I, Pavlov A (2018) Plant cell culture as emerging technology for production of active cosmetic ingredients. Eng Life Sci 18(11):779–798
Gerhard GS (2020) Heme as a taste molecule. Curr Diab Rep. https://doi.org/10.1007/s13668-020-00320-6
Geu-Flores F, Sherden NH, Courdavault V, Burlat V, Glenn WS, Wu C, Nims E, Cui Y, O’Connor SE (2012) An alternative route to cyclic terpenes by reductive cyclization in iridoid biosynthesis. Nature 492(7427):138–142
Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA III, Smith HO (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6(5):343-U41
Gong Z, Wang H, Tang J, Bi C, Li Q, Zhang X (2020) Coordinated expression of astaxanthin biosynthesis genes for improved astaxanthin production in Escherichia coli. J Agr Food Chem 68(50):14917–14927
Grewal PS, Modavi C, Russ ZN, Harris NC, Dueber JE (2018) Bioproduction of a betalain color palette in Saccharomyces cerevisiae. Metab Eng 45:180–188
Harris NN, Javellana J, Davies KM, Lewis DH, Jameson PE, Deroles SC, Calcott KE, Gould KS, Schwinn KE (2012) Betalain production is possible in anthocyanin-producing plant species given the presence of DOPA-dioxygenase and L-DOPA. BMC Plant Biol. https://doi.org/10.1186/1471-2229-12-34
Hatlestad GJ, Sunnadeniya RM, Akhavan NA, Gonzalez A, Goldman IL, McGrath JM, Lloyd AM (2012) The beet R locus encodes a new cytochrome P450 required for red betalain production. Nat Genet 44(7):816-U130
Heider SAE, Peters-Wendisch P, Wendisch VF, Beekwilder J, Brautaset T (2014) Metabolic engineering for the microbial production of carotenoids and related products with a focus on the rare C50 carotenoids. Appl Microbiol Biotechol 98(10):4355–4368
Henke NA, Wendisch VF (2019) Improved astaxanthin production with corynebacterium glutamicum by application of a membrane fusion protein. Mar Drugs. https://doi.org/10.3390/md17110621
Henke N, Heider S, Peters-Wendisch P, Wendisch V (2016) Production of the marine carotenoid astaxanthin by metabolically engineered Corynebacterium glutamicum. Mar Drugs. https://doi.org/10.3390/md14070124
Hong J, Im DK, Oh MK (2020) Investigating E. coli coculture for resveratrol production with c-13 metabolic flux analysis. J Agr Food Chem 68(11):3466–3473
Hoppe M, Brun B, Larsson MP, Moraeus L, Hulthen L (2013) Heme iron-based dietary intervention for improvement of iron status in young women. Nutrition 29(1):89–95
Horwitz AA, Walter JM, Schubert MG, Kung SH, Hawkins K, Platt DM, Hernday AD, Mahatdejkul-Meadows T, Szeto W, Chandran SS, Newman JD (2015) Efficient multiplexed integration of synergistic alleles and metabolic pathways in yeasts via CRISPR-Cas. Cell Syst 1(1):88–96
Hou S, Qin Q, Dai J (2018) Wicket: a versatile tool for the integration and optimization of exogenous pathways in Saccharomyces cerevisiae. Acs Synth Biol 7(3):782–788
Hu X, Ma X, Tang P, Yuan Q (2013) Improved beta-carotene production by oxidative stress in Blakeslea trispora induced by liquid paraffin. Biotechnol Lett 35(4):559–63
Hu JJ, Nagarajan D, Zhang QG, Chang JS, Lee DJ (2018) Heterotrophic cultivation of microalgae for pigment production: a review. Biotechnol Adv 36(1):54–67. https://doi.org/10.1016/j.biotechadv.2017.09.009
Imtiyaj Khan M, Giridhar P (2015) Plant betalains: chemistry and biochemistry. Phytochemistry 117:267–295. https://doi.org/10.1016/j.phytochem.2015.06.008
Itaya M, Fujita K, Kuroki A, Tsuge K (2008) Bottom-up genome assembly using the Bacillus subtilis genome vector. Nat Methods 5(1):41–43
Jakociunas T, Rajkumar AS, Zhang J, Arsovska D, Rodriguez A, Jendresen CB, Skjodt ML, Nielsen AT, Borodina I, Jensen MK, Keasling JD (2015) CasEMBLR: cas9-facilitated multiloci genomic integration of in vivo assembled dna parts in Saccharomyces cerevisiae. Acs Synth Biol 4(11):1226–1234
Jeong TH, Cho YS, Choi S-S, Kim G-D, Lim HK (2018) Enhanced production of astaxanthin by metabolically engineered non-mevalonate pathway in Escherichia coli. Korean J Microbiol Biotechnol 46(2):114–119
Jiang G, Yang Z, Wang Y, Yao M, Chen Y, Xiao W, Yuan Y (2020) Enhanced astaxanthin production in yeast via combined mutagenesis and evolution. Biochem Eng J. https://doi.org/10.1016/j.bej.2020.107519
Jin J, Wang Y, Yao M, Gu X, Li B, Liu H, Ding M, Xiao W, Yuan Y (2018) Astaxanthin overproduction in yeast by strain engineering and new gene target uncovering. Biotechnol Biofuels 11:230
Jing K, He S, Chen T, Lu Y, Ng IS (2016) Enhancing beta-carotene biosynthesis and gene transcriptional regulation in Blakeslea trispora with sodium acetate. Biochem Eng J 114:10–17
Jones CG, Keeling CI, Ghisalberti EL, Barbour EL, Plummer JA, Bohlmann J (2008) Isolation of cDNAs and functional characterisation of two multi-product terpene synthase enzymes from sandalwood, Santalum Album L. Arch. Biochem Biophys 477(1):121–30
Jones JA, Vernacchio VR, Sinkoe AL, Collins SM, Ibrahim MHA, Lachance DM, Hahn J, Koffas MAG (2016) Experimental and computational optimization of an Escherichia coli co-culture for the efficient production of flavonoids. Metab Eng 35:55–63
Jones JA, Vernacchio VR, Collins SM, Shirke AN, Xiu Y, Englaender JA, Cress BF, McCutcheon CC, Linhardt RJ, Gross RA, Koffas MAG (2017) Complete biosynthesis of anthocyanins using E. coli polycultures. Mbio. https://doi.org/10.1128/mBio.00621-17
Kan E, Katsuyama Y, Maruyama J-i, Tamano K, Koyama Y, Ohnishi Y (2019) Production of the plant polyketide curcumin in Aspergillus oryzae: strengthening malonyl-CoA supply for yield improvement. Biosci Biotech Bioch 83(7):1372–1381
Kang S-Y, Heo KT, Hong Y-S (2018) Optimization of artificial curcumin biosynthesis in E-coli by randomized 5′-UTR sequences to control the multienzyme pathway. Acs Synth Biol 7(9):2054–2062
Katsuyama Y, Matsuzawa M, Funa N, Horinouch S (2008) Production of curcuminoids by Escherichia coli carrying an artificial biosynthesis pathway. Microbiology 154:2620–2628
Kildegaard KR, Adiego-Pérez B, Doménech Belda D, Khangura JK, Holkenbrink C, Borodina I (2017) Engineering of Yarrowia lipolytica for production of astaxanthin. Synth Syst Biotechnol 2(4):287–294
Kim D, Ku S (2018) Beneficial effects of monascus sp kccm 10093 pigments and derivatives: a mini review. Molecules. https://doi.org/10.3390/molecules23010098
Kiokias S, Proestos C, Varzakas T (2016) A review of the structure, biosynthesis, absorption of carotenoids-analysis and properties of their common natural extracts. Curr Res Nutr Food Sci 4:25–37
Kobayashi K, Masuda T (2016) Transcriptional regulation of tetrapyrrole biosynthesis in Arabidopsis thaliana. Front Plant Sci. https://doi.org/10.3389/fpls.2016.01811
Krinsky NI, Landrum JT, Bone RA (2003) Biologic mechanisms of the protective role of lutein and zeaxanthin in the eye. Annu Rev Nutr 23:171–201
Kumar SS, Manoj P, Giridhar P, Shrivastava R, Bharadwaj M (2015) Fruit extracts of Basella rubra that are rich in bioactives and betalains exhibit antioxidant activity and cytotoxicity against human cervical carcinoma cells. J Funct Foods 15:509–515
Kuo FS, Chien YH, Chen CJ (2012) Effects of light sources on growth and carotenoid content of photosynthetic bacteria Rhodopseudomonas palustris. Bioresour Technol 113:315–8
Kwon SJ, de Boer AL, Petri R, Schmidt-Dannert C (2003) High-level production of porphyrins in metabolically engineered Escherichia coli: systematic extension of a pathway assembled from overexpressed genes involved in heme biosynthesis. Appl Environ Microb 69(8):4875–4883
Kwon O-H, Kim S, Hahm D-H, Lee SY, Kim P (2009) Potential application of the recombinant Escherichia coli-synthesized heme as a bioavailable iron source. J Microbiol Biotechnol 19(6):604–609
Lagashetti AC, Dufossé L, Singh SK, Singh PN (2019) Fungal pigments and their prospects in different industries. Microorganisms 7(12):604
Lage DdA, Tirado MdS, Vanicore SR, de Carvalho Sabino KC, Albarello N (2015) Production of betalains from callus and cell suspension cultures of Pereskia aculeata miller, an unconventional leafy vegetable. Plant Cell Tiss Org 122(2):341–350
Lan TTP, Huy ND, Luong NN, Nghi NV, Tan TH, Quan LV, Loc NH (2018) Identification and characterization of genes in the curcuminoid pathway of Curcuma zedoaria roscoe. Curr Pharm Biotechnol 19(10):839–846
Lange BM, Rujan T, Martin W, Croteau R (2000) Isoprenoid biosynthesis: the evolution of two ancient and distinct pathways across genomes. Proc Natl Acad Sci USA 97(24):13172–13177
Larroude M, Celinska E, Back A, Thomas S, Nicaud JM, Ledesma-Amaro R (2018) A synthetic biology approach to transform Yarrowia lipolytica into a competitive biotechnological producer of beta-carotene. Biotechnol Bioeng 115(2):464–472
Lee TS, Krupa RA, Zhang F, Hajimorad M, Holtz WJ, Prasad N, Lee SK, Keasling JD (2011) BglBrick vectors and datasheets: a synthetic biology platform for gene expression. J Biol Eng. https://doi.org/10.1186/1754-1611-5-12
Lee JM, Joung J-G, McQuinn R, Chung M-Y, Fei Z, Tieman D, Klee H, Giovannoni J (2012) Combined transcriptome, genetic diversity and metabolite profiling in tomato fruit reveals that the ethylene response factor SlERF6 plays an important role in ripening and carotenoid accumulation. Plant J 70(2):191–204
Lee MJ, Kim H-J, Lee J-Y, Kwon AS, Jun SY, Kang SH, Kim P (2013) Effect of gene amplifications in porphyrin pathway on heme biosynthesis in a recombinant Escherichia coli. J Microbiol Biotechnol 23(5):668–673
Lee JJL, Chen L, Cao B, Chen WN (2016) Engineering Rhodosporidium toruloides with a membrane transporter facilitates production and separation of carotenoids and lipids in a bi-phasic culture. Appl Microbiol Biotechnol 100(2):869–877
Lemuth K, Steuer K, Albermann C (2011) Engineering of a plasmid-free Escherichia coli strain for improved in vivo biosynthesis of astaxanthin. Microb Cell Fact 10(1):29
Levisson M, Patinios C, Hein S, de Groot PA, Daran J-M, Hall RD, Martens S, Beekwilder J (2018) Engineering de novo anthocyanin production in Saccharomyces cerevisiae. Microb Cell Fact. https://doi.org/10.1186/s12934-018-0951-6
Li S, Huang JC (2018) Assessment of expression cassettes and culture media for different Escherichia coli strains to produce astaxanthin. Nat Prod Bioprospect 8(5):397–403
Li Q-l, Tian J (2017) Safety evaluation and solutions of food synthetic pigments. Food Ind 38:268–271
Li M, Schneider K, Kristensen M, Borodina I, Nielsen J (2016) Engineering yeast for high-level production of stilbenoid antioxidants. Sci Rep. https://doi.org/10.1038/srep36827
Li J, Shen J, Sun Z, Li J, Li C, Li X, Zhang Y (2017) Discovery of several novel targets that enhance beta-carotene production in Saccharomyces cerevisiae. Front Microbiol 8:1116
Liang B, Du XJ, Li P, Sun CC, Wang S (2018) Investigation of citrinin and pigment biosynthesis mechanisms in Monascus purpureus by transcriptomic analysis. Front Microbiol 9:1374
Lim CG, Wong L, Bhan N, Dvora H, Xu P, Venkiteswaran S, Koffas MAG (2015) Development of a recombinant Escherichia coli strain for overproduction of the plant pigment anthocyanin. Appl Environ Microb 81(18):6276–6284
Lin YJ, Chang JJ, Lin HY, Thia C, Kao YY, Huang CC, Li WH (2017) Metabolic engineering a yeast to produce astaxanthin. Bioresour Technol 245(Pt A):899–905
Liu J, Ren Y, Yao S (2010) Repeated-batch cultivation of encapsulated Monascus purpureus by polyelectrolyte complex for natural pigment production. Chin J Chem Eng 18(6):1013–1017
Liu P, Sun L, Sun Y, Shang F, Yan G (2016a) Decreased fluidity of cell membranes causes a metal ion deficiency in recombinant Saccharomyces cerevisiae producing carotenoids. J Ind Microbiol Biotechnol 43(4):525–35
Liu Y, Yan Z, Lu X, Xiao D, Jiang H (2016b) Improving the catalytic activity of isopentenyl phosphate kinase through protein coevolution analysis. Sci Rep. https://doi.org/10.1038/srep24117
Liu J, Chai X, Guo T, Wu J, Yang P, Luo Y, Zhao H, Zhao W, Nkechi O, Dong J, Bai J, Lin Q (2019a) Disruption of the ergosterol biosynthetic pathway results in increased membrane permeability, causing overproduction and secretion of extracellular monascus pigments in submerged fermentation. J Agric Food Chem. https://doi.org/10.1021/acs.jafc.9b05872
Liu S, Daigger GT, Kang J, Zhang G (2019b) Effects of light intensity and photoperiod on pigments production and corresponding key gene expression of Rhodopseudomonas palustris in a photobioreactor system. Bioresour Technol 294:122172
Liu T, Dong C, Qi M, Zhang B, Huang L, Xu Z, Lian J (2020) Construction of a stable and temperature-responsive yeast cell factory for crocetin biosynthesis using CRISPR-Cas9. Front Bioeng Biotech 8:653
Liu J, Wu J, Cai X, Zhang S, Liang Y, Lin Q (2021a) Regulation of secondary metabolite biosynthesis in Monascus purpureus via cofactor metabolic engineering strategies. Food Microbiol 95:103689
Liu L, Qu YL, Dong GR, Wang J, Hu CY, Meng YH (2021b) Elevated β-carotene production using codon-adapted CarrA&B and metabolic balance in engineered Yarrowia lipolytica. Front Microbiol. https://doi.org/10.3389/fmicb.2021.627150
Lu Q, Liu JZ (2019) Enhanced astaxanthin production in Escherichia coli via morphology and oxidative stress engineering. J Agric Food Chem 67(42):11703–11709
Lu Q, Bu Y-F, Liu J-Z (2017) Metabolic engineering of Escherichia coli for producing astaxanthin as the predominant carotenoid. Mar Drugs. https://doi.org/10.3390/md15100296
Lu SW, Zhang Y, Zhu K, Yang W, Jl Ye, Chai L, Xu Q, Deng X (2018) The citrus transcription factor CsMADS6 modulates carotenoid metabolism by directly regulating carotenogenic genes. Plant Physiol 176(4):2657–2676
Luo JR, Duan JJ, Huo D, Shi QQ, Niu LX, Zhang YL (2017) Transcriptomic analysis reveals transcription factors related to leaf anthocyanin biosynthesis in Paeonia qiui. Molecules. https://doi.org/10.3390/molecules22122186
Luo W, Wang Y, Yang P, Qu Y, Yu X (2021) Multilevel regulation of carotenoid synthesis by light and active oxygen in Blakeslea trispora. J Agr Food Chem 69(37):10974–10988
Lv X, Xu H, Yu H (2013a) Significantly enhanced production of isoprene by ordered coexpression of genes dxs, dxr, and idi in Escherichia coli. Appl Microbiol Biotechnol 97(6):2357–2365
Lv X, Xie W, Lu W, Guo F, Gu J, Yu H, Ye L (2014) Enhanced isoprene biosynthesis in Saccharomyces cerevisiae by engineering of the native acetyl-CoA and mevalonic acid pathways with a push-pull-restrain strategy. J Biotechnol 186:128–136
Lv X, Gu J, Wang F, Xie W, Liu M, Ye L, Yu H (2016) Combinatorial pathway optimization in Escherichia coli by directed co-evolution of rate-limiting enzymes and modular pathway engineering. Biotechnol Bioeng 113(12):2661–2669
Lyu XM, Lee J, Chen WN (2019a) Potential natural food preservatives and their sustainable production in yeast: terpenoids and polyphenols. J Agr Food Chem 67(16):4397–4417
Lyu XM, Zhao GL, Ng KR, Mark R, Chen WN (2019b) Metabolic engineering of Saccharomyces cerevisiae for de novo production of kaempferol. J Agr Food Chem 67(19):5596–5606
Ma T, Zhou Y, Li X, Zhu F, Cheng Y, Liu Y, Deng Z, Liu T (2016) Genome mining of astaxanthin biosynthetic genes from Sphingomonas sp. ATCC 55669 for heterologous overproduction in Escherichia coli. Biotechnol J 11(2):228–237
Markets Ma (2017) Lutein market by form (powder and crystalline, oil suspension, beadlet, emulsion), source (natural, synthetic), application (food, beverages, dietary supplements, animal feed), production process, and region—global forecast to 2022. markets and markets
Malik S, Bhushan S, Sharma M, Ahuja PS (2016) Biotechnological approaches to the production of shikonins: a critical review with recent updates. Crit Rev Biotechnol 36(2):327–40
Manivasagan P, Bharathiraja S, Santha Moorthy M, Mondal S, Seo H, Dae Lee K, Oh J (2018) Marine natural pigments as potential sources for therapeutic applications. Crit Rev Biotechnol 38(5):745–761
Mannazzu I, Landolfo S, da Silva TL, Buzzini P (2015) Red yeasts and carotenoid production: outlining a future for non-conventional yeasts of biotechnological interest. World J Microb Biot 31(11):1665–1673
McCarty NS, Graham AE, Studena L, Ledesma-Amaro R (2020) Multiplexed CRISPR technologies for gene editing and transcriptional regulation. Nat Commun 11(1):1281–1281
Medema MH, Osbourn A (2016) Computational genomic identification and functional reconstitution of plant natural product biosynthetic pathways. Nat Prod Rep 33(8):951–62
Miura Y, Kondo K, Saito T, Shimada H, Fraser PD, Misawa N (1998) Production of the carotenoid lycopene, beta-carotene, and astaxanthin in the food yeast Candida utilis. Appl Environ Microbiol 64(4):1226–1229
Morales-Oyervides L, Ruiz-Sanchez JP, Oliveira JC, Sousa-Gallagher MJ, Mendez-Zavala A, Giuffrida D, Dufosse L, Montanez J (2020) Biotechnological approaches for the production of natural colorants by talaromyces/penicillium: a review. Biotechnol Adv. https://doi.org/10.1016/j.biotechadv.2020.107601
Moran NA, Jarvik T (2010) Lateral transfer of genes from fungi underlies carotenoid production in aphids. Science 328(5978):624–627. https://doi.org/10.1126/science.1187113
Mota GCP, Moraes LBSd, Oliveira CYB, Oliveira DWS, Abreu JLd, Dantas DMM, Gálvez AO (2021) Astaxanthin from Haematococcus pluvialis: processes, applications, and market. Prep Biochem Biotechnol. https://doi.org/10.1080/10826068.2021.1966802
Murthy HN, Lee EJ, Paek KY (2014) Production of secondary metabolites from cell and organ cultures: strategies and approaches for biomass improvement and metabolite accumulation. Plant Cell Tissue Organ 118(1):1–16
Mussagy CU, Winterburn J, Santos-Ebinuma VC, Brandao Pereira JF (2019a) Production and extraction of carotenoids produced by microorganisms. Appl Microbiol Biot 103(3):1095–1114
Mussagy CU, Winterburn J, Santos-Ebinuma VC, Pereira JFB (2019b) Production and extraction of carotenoids produced by microorganisms. Appl Microbiol Biot 103(3):1095–1114
Mussagy CU, Pereira JFB, Dufosse L, Raghavan V, Santos-Ebinuma VC, Pessoa A (2021) Advances and trends in biotechnological production of natural astaxanthin by Phaffia rhodozyma yeast. Crit Rev Food Sci Nutr. https://doi.org/10.1080/10408398.2021.1968788
Naegeli H, Birch AN, Casacuberta J, De Schrijver A, Gralak MA, Guerche P, Jones H, Manachini B, Messean A, Nielsen EE, Nogue F, Robaglia C, Rostoks N, Sweet J, Tebbe C, Visioli F, Wal J-M, Devos Y, Dumont AF, Lanzoni A, Paoletti C, Paraskevopoulos K, Waigmann E, Modified EPG (2017) Guidance for the risk assessment of the presence at low level of genetically modified plant material in imported food and feed under regulation (EC) No 1829/2003. Efsa J. https://doi.org/10.2903/j.efsa.2017.5048
Naing AH, Kim CK (2018) Roles of R2R3-MYB transcription factors in transcriptional regulation of anthocyanin biosynthesis in horticultural plants. Plant Mol Biol 98(1–2):1–18
Nam HK, Choi JG, Lee JH, Kim SW, Oh DK (2013) Increase in the production of beta-carotene in recombinant Escherichia coli cultured in a chemically defined medium supplemented with amino acids. Biotechnol Lett 35(2):265–71
Nanou K, Roukas T, Papadakis E (2012) Improved production of carotenes from synthetic medium by Blakeslea trispora in a bubble column reactor. Biochem Eng J 67:203–207
Navab M, Dwyer KM, Hassan K, Sun P, Shircore A, Hama-Levy S, Hough G, Wang X, Drake T, Merz NB, Fogelman AM (2001) Oxygenated carotenoid lutein and progression of early atherosclerosis: the Los Angeles atherosclerosis study. Circulation 103:2922–2927
Ng CY, Farasat I, Maranas CD, Salis HM (2015) Rational design of a synthetic Entner-Doudoroff pathway for improved and controllable NADPH regeneration. Metab Eng 29:86–96
Nogueira M, Enfissi EMA, Welsch R, Beyer P, Zurbriggen MD, Fraser PD (2019) Construction of a fusion enzyme for astaxanthin formation and its characterisation in microbial and plant hosts: a new tool for engineering ketocarotenoids. Metab Eng 52:243–252
Novakova E, Moran NA (2012) Diversification of genes for carotenoid biosynthesis in aphids following an ancient transfer from a fungus. Mol Biol Evol 29(1):313–323
Olivieri G, Salatino P, Marzocchella A (2014) Advances in photobioreactors for intensive microalgal production: configurations, operating strategies and applications. J Chem Technol Biot 89(2):178–195
Organisms EPoGM (2011) Guidance on the risk assessment of genetically modified microorganisms and their products intended for food and feed use. EFSA J 9(6):2193
Osbourn AE, O’Maille PE, Rosser SJ, Lindsey K (2012) Synthetic biology. New Phytol 196(3):671–677
Ozaydin B, Burd H, Lee TS, Keasling JD (2013) Carotenoid-based phenotypic screen of the yeast deletion collection reveals new genes with roles in isoprenoid production. Metab Eng 15:174–83
Panche AN, Diwan AD, Chandra SR (2016) Flavonoids: an overview. J Nutr Sci 5:e47
Panel on Food A, Nutrient Sources added to F (2013) Scientific opinion on the re-evaluation of anthocyanins E 163 as a food additive. EFS2. https://doi.org/10.2903/j.efsa.2013.3145
Park SY, Binkley RM, Kim WJ, Lee MH, Lee SY (2018) Metabolic engineering of Escherichia coli for high-level astaxanthin production with high productivity. Metab Eng 49:105–115
Pistelli L, Bertoli A, Gelli F, Bedini L, Ruffoni B, Pistelli L (2012) Production of curcuminoids in different in vitro organs of Curcuma longa. Nat Prod Commun 7(8):1037–1042
Polturak G, Breitel D, Grossman N, Sarrion-Perdigones A, Weithorn E, Pliner M, Orzaez D, Granell A, Rogachev I, Aharoni A (2016) Elucidation of the first committed step in betalain biosynthesis enables the heterologous engineering of betalain pigments in plants. New Phytol 210(1):269–283
Polturak G, Grossman N, Vela-Corcia D, Dong Y, Nudel A, Pliner M, Levy M, Rogachev I, Aharoni A (2017) Engineered gray mold resistance, antioxidant capacity, and pigmentation in betalain-producing crops and ornamentals. Proc Natl Acad Sci USA 114(34):9062–9067
Qi D-D, Jin J, Liu D, Jia B, Yuan Y-J (2020) In vitro and in vivo recombination of heterologous modules for improving biosynthesis of astaxanthin in yeast. Microb Cell Fact 19(1):103
Qiang S, Wang J, Xiong XC, Qu YL, Liu L, Hu CY, Meng YH (2020) Promoting the synthesis of precursor substances by overexpressing hexokinase (Hxk) and hydroxymethylglutaryl-coa synthase (Erg13) to elevate β-carotene production in engineered Yarrowia Lipolytica. Front Microbiol. https://doi.org/10.3389/fmicb.2020.01346
Qu JL, Cao S, Wei QX, Zhang HW, Wang R, Kang W, Ma T, Zhang L, Liu TG, Au SWN, Sun F, Xia J (2019) Synthetic multienzyme complexes, catalytic nanomachineries for cascade biosynthesis in vivo. ACS Nano 13(9):9895–9906
Quan J, Tian J (2009) Circular polymerase extension cloning of complex gene libraries and pathways. PLoS ONE. https://doi.org/10.1371/journal.pone.0006441
Rahman I, Biswas SK, Kirkham PA (2006) Regulation of inflammation and redox signaling by dietary polyphenols. Biochem Pharmacol 72(11):1439–52
Ramirez-Estrada K, Vidal-Limon H, Hidalgo D, Moyano E, Golenioswki M, Cusido RM, Palazon J (2016) Elicitation, an effective strategy for the biotechnological production of bioactive high-added value compounds in plant cell factories. Molecules. https://doi.org/10.3390/molecules21020182
Ramos KR, Valdehuesa KN, Liu H, Nisola GM, Lee WK, Chung WJ (2014) Combining De Ley-Doudoroff and methylerythritol phosphate pathways for enhanced isoprene biosynthesis from D-galactose. Bioprocess Biosyst Eng 37(12):2505–13
Rapp G (2009) Pigments and colorants archaeomineralogy. Springer, Berlin, pp 201–221
Rizzello F, De Paolis A, Durante M, Blando F, Mita G, Caretto S (2014) Enhanced production of bioactive isoprenoid compounds from cell suspension cultures of Artemisia annua L. using beta-cyclodextrins. Int J Mol Sci 15(10):19092–105
Rodrigues JL, Araujo RG, Prather KLJ, Kluskens LD, Rodrigues LR (2015) Production of curcuminoids from tyrosine by a metabolically engineered Escherichia coli using caffeic acid as an intermediate. Biotechnol J 10(4):599-U315
Rodrigues JL, Couto MR, Araujo RG, Prather KLJ, Kluskens L, Rodrigues LR (2017) Hydroxycinnamic acids and curcumin production in engineered Escherichia coli using heat shock promoters. Biochem Eng J 125:41–49
Rodrigues JL, Gomes D, Rodrigues LR (2020) A combinatorial approach to optimize the production of curcuminoids from tyrosine in Escherichia coli. Front Bioeng Biotech. https://doi.org/10.3389/fbioe.2020.00059
Rodriguez A, Kildegaard KR, Li MJ, Borodina I, Nielsen J (2015) Establishment of a yeast platform strain for production of p-coumaric acid through metabolic engineering of aromatic amino acid biosynthesis. Metab Eng 31:181–188
Rodriguez-Amaya DB (2016) Natural food pigments and colorants. Curr Opin Food Sci 7:20–26
Rodriguez-Concepcion M, Avalos J, Bonet ML, Boronat A, Gomez-Gomez L, Hornero-Mendez D, Limon MC, Melendez-Martinez AJ, Olmedilla-Alonso B, Palou A, Ribot J, Rodrigo MJ, Zacarias L, Zhu CF (2018) A global perspective on carotenoids: metabolism, biotechnology, and benefits for nutrition and health. Prog Lipid Res 70:62–93
Rohdich F, Hecht S, Bacher A, Eisenreich W (2003) Deoxyxylulose phosphate pathway of isoprenoid biosynthesis. Discovery and function of ispDEFGH genes and their cognate enzymes. Pure Appl Chem 75(2–3):393–405
Rohmer M, Knani M, Simonin P, Sutter B, Sahm H (1993) Isoprenoid biosynthesis in bacteria—a novel pathway for the early steps leading to isopentenyl diphosphate. Biochem J 295:517–524
Saini RK, Keum YS (2019) Microbial platforms to produce commercially vital carotenoids at industrial scale: an updated review of critical issues. J Ind Microbiol Biot 46(5):657–674
Saini DK, Chakdar H, Pabbi S, Shukla P (2020) Enhancing production of microalgal biopigments through metabolic and genetic engineering. Crit Rev Food Sci 60(3):391–405
Sathiyabama M, Bernstein N, Anusuya S (2016) Chitosan elicitation for increased curcumin production and stimulation of defence response in turmeric (Curcuma longa L.). Ind Crop Prod 89:87–94
Savitha BC, Thimmaraju R, Bhagyalakshmi N, Ravishankar GA (2006) Different biotic and abiotic elicitors influence betalain production in hairy root cultures of Beta vulgaris in shake-flask and bioreactor. Process Biochem 41(1):50–60
Saw NMMT, Riedel H, Cai ZZ, Kutuk O, Smetanska I (2012) Stimulation of anthocyanin synthesis in grape (Vitis vinifera) cell cultures by pulsed electric fields and ethephon. Plant Cell Tissue Organ 108(1):47–54
Sawicki T, Wiczkowski W (2018) The effects of boiling and fermentation on betalain profiles and antioxidant capacities of red beetroot products. Food Chem 259:292–303
Scaife MA, Ma CA, Ninlayarn T, Wright PC, Armenta RE (2012) Comparative analysis of β-carotene hydroxylase genes for astaxanthin biosynthesis. J Nat Prod 75(6):1117–1124
Scalbert A, Manach C, Morand C, Remesy C, Jimenez L (2005) Dietary polyphenols and the prevention of diseases. Crit Rev Food Sci Nutr 45(4):287–306
Scotter MJ (2011) Methods for the determination of European Union-permitted added natural colours in foods: a review. Food Addit Contam Part A 28(5):527–596
Seddon JM, Ajani UA, Sperduto RD, Hiller R, Blair N, Burton TC, Farber MD, Gragoudas ES, Haller J, Miller DT, Yannuzzi LA, Willett W (1994) Dietary carotenoids, Vitamins A, C, and E, and advanced age-related macular degeneration. JAMA 272(18):1413–1420
Sehrawat R, Panesar PS, Swer TL, Kumar A (2017) Response surface methodology (RSM) mediated interaction of media concentration and process parameters for the pigment production by Monascus purpureus MTCC 369 under solid state fermentation. Pigment Resin Technol 46(1):14–20
Sekizawa H, Ikuta K, Mizuta K, Takechi S, Suzutani T (2013) Relationship between polyphenol content and anti-influenza viral effects of berries. J Sci Food Agr 93(9):2239–2241
Seyedin A, Yazdian F, Hatamian Zarmi A, Rasekh B, Mir-Derikvand M (2015) Natural pigment production by monascus purpureus: bioreactor yield improvement through statistical analysis. Appl Food Biotechnol 2(2):23–30
Shafi J, Sun Z, Ji M, Gu Z, Ahmad W (2018) ANN and RSM based modelling for optimization of cell dry mass of Bacillus sp. strain B67 and its antifungal activity against Botrytis cinerea. Biotechnol Biotechnol Equip 32(1):58–68
Shao Z, Zhao H, Zhao H (2009) DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res. https://doi.org/10.1093/nar/gkn991
Shapter FM, Waters DLE (2014) Genome Walking. In: Henry RJ, Furtado A (eds) Cereal Genomics: methods and protocols. Methods in Molecular Biology, vol 1099. Humana Press Inc, 999 Riverview Dr, Ste 208, Totowa, Nj 07512–1165 USA, pp 133–146
Shetty RP, Endy D, Knight TF Jr (2008) Engineering BioBrick vectors from BioBrick parts. J Biol Eng 2:5
Shi F, Zhan W, Li Y, Wang X (2014) Temperature influences beta-carotene production in recombinant Saccharomyces cerevisiae expressing carotenogenic genes from Phaffia rhodozyma. World J Microbiol Biotechnol 30(1):125–33
Shi S, Liang Y, Zhang MM, Ang EL, Zhao H (2016) A highly efficient single-step, markerless strategy for multi-copy chromosomal integration of large biochemical pathways in Saccharomyces cerevisiae. Metab Eng 33:19–27
Shimura K, Okada A, Okada K, Jikumaru Y, Ko K-W, Toyomasu T, Sassa T, Hasegawa M, Kodama O, Shibuya N, Koga J, Nojiri H, Yamane H (2007) Identification of a biosynthetic gene cluster in rice for momilactones. J Biol Chem 282(47):34013–34018
Shrestha B, Pandey RP, Darsandhari S, Parajuli P, Sohng JK (2019) Combinatorial approach for improved cyanidin 3-O-glucoside production in Escherichia coli. Microb Cell Fact. https://doi.org/10.1186/s12934-019-1056-6
Si T, Luo Y, Bao Z, Zhao H (2015) RNAi-Assisted genome evolution in Saccharomyces cerevisiae for complex phenotype engineering. ACS Synth Biol 4(3):283–291
Sigurdson GT, Tang PP, Giusti MM (2017) Natural colorants: food colorants from natural sources. Annu Rev Food Sci T 8(8):261–280
Simsa R, Yuen J, Stout A, Rubio N, Fogelstrand P, Kaplan DL (2019) Extracellular heme proteins influence bovine myosatellite cell proliferation and the color of cell-based meat. Foods. https://doi.org/10.3390/foods8100521
Singh N, Goel G, Singh N, Kumar Pathak B, Kaushik D (2015) Modeling the red pigment production by Monascus purpureus MTCC 369 by Artificial Neural Network using rice water based medium. Food Biosci 11:17–22
Solymosi K, Mysliwa-Kurdziel B (2017) Chlorophylls and their derivatives used in food industry and medicine. Mini Rev Med Chem 17(13):1194–1222
Steingroewer J, Bley T, Georgiev V, Ivanov I, Lenk F, Marchev A, Pavlov A (2013) Bioprocessing of differentiated plant in vitro systems. Eng Life Sci 13(1):26–38
Sun Z, Liu J, Zeng X, Huangfu J, Jiang Y, Wang M, Chen F (2011) Protective actions of microalgae against endogenous and exogenous advanced glycation endproducts (AGEs) in human retinal pigment epithelial cells. Food Funct 2(5):251–8
Sun L, Shang F, Duan C-q, Yan G-l (2015) Reduction of fatty acid flux at low temperature led to enhancement of β-carotene biosynthesis in recombinant Saccharomyces cerevisiae. Korean J Chem Eng 32(7):1354–1360
Sun Y, Sun L, Shang F, Yan G (2016) Enhanced production of β-carotene in recombinant Saccharomyces cerevisiae by inverse metabolic engineering with supplementation of unsaturated fatty acids. Process Biochem 51(5):568–577
Sun L, Atkinson CA, Lee Y-G, Jin Y-S (2020) High-level beta-carotene production from xylose by engineered Saccharomyces cerevisiae without overexpression of a truncated HMG1 (tHMG1). Biotechnol Bioeng 117(11):3522–3532
Sun D, Zhou XG, Liu C, Zhu JR, Ru YR, Liu WJ, Liu JW (2021) Fnr negatively regulates prodigiosin synthesis in Serratia sp. ATCC 39006 during aerobic fermentation. Front Microbiol. https://doi.org/10.3389/fmicb.2021.734854
Swarna J, Lokeswari TS, Smita M, Ravindhran R (2013) Characterisation and determination of in vitro antioxidant potential of betalains from Talinum triangulare (Jacq.) Willd. Food Chem 141(4):4382–4390
Tanaka Y, Sasaki N, Ohmiya A (2008) Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J 54(4):733–49
Tang HB, Ye ZW, Liu C, Guo LQ, Lin JF, Wan H, Yun F, Kang LZ (2019) Increasing of the contain of carotenoids in caterpillar mushroom, cordyceps militaris (Ascomycetes) by using the fungal elicitors cultivation. Int J Med Mushrooms 21(12):1181–1191
Timoneda A, Sheehan H, Feng T, Lopez-Nieves S, Maeda HA, Brockington S (2018) Redirecting primary metabolism to boost production of tyrosine-derived specialised metabolites in planta. Sci Reo. https://doi.org/10.1038/s41598-018-33742-y
Tkáčová J, Čaplová J, Klempová T, Čertík M (2017) Correlation between lipid and carotenoid synthesis in torularhodin-producing Rhodotorula glutinis. Ann Microbiol 67(8):541–551
Tramontin LRR, Kildegaard KR, Sudarsan S, Borodina I (2019) Enhancement of astaxanthin biosynthesis in oleaginous yeast Yarrowia lipolytica via microalgal pathway. Microorganisms. https://doi.org/10.3390/microorganisms7100472
Ukibe K, Hashida K, Yoshida N, Takagi H (2009) Metabolic engineering of Saccharomyces cerevisiae for astaxanthin production and oxidative stress tolerance. Appl Environ Microbiol 75(22):7205–7211
Velisek J, Davidek J, Cejpek K (2007) Biosynthesis of food constituents: natural pigments. Part 1—a review. Czech J Food Sci 25(6):291–315
Velmurugan P, Lee YH, Venil CK, Lakshmanaperumalsamy P, Chae JC, Oh BT (2010) Effect of light on growth, intracellular and extracellular pigment production by five pigment-producing filamentous fungi in synthetic medium. J Biosci Bioeng 109(4):346–350
Venayak N, Anesiadis N, Cluett WR, Mahadevan R (2015) Engineering metabolism through dynamic control. Curr Opin Biotechnol 34:142–152
Venil CK, Zakaria ZA, Ahmad WA (2013) Bacterial pigments and their applications. Process Biochem 48(7):1065–1079
Verwaal R, Jiang Y, Wang J, Daran J-M, Sandmann G, van den Berg JA, van Ooyen AJJ (2010) Heterologous carotenoid production in Saccharomyces cerevisiae induces the pleiotropic drug resistance stress response. Yeast 27(12):983–998
Vranova E, Coman D, Gruissem W (2013) Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu Rev Plant Biol 64(64):665–700
Wan X, Zhou X-R, Moncalian G, Su L, Chen W-C, Zhu H-Z, Chen D, Gong Y-M, Huang F-H, Deng Q-C (2021) Reprogramming microorganisms for the biosynthesis of astaxanthin via metabolic engineering. Prog Lipid Res 81:101083
Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, Forest CR, Church GM (2009) Programming cells by multiplex genome engineering and accelerated evolution. Nature 460(7257):894-U133
Wang H, Li P, Liu Y, Ren Z, Wang G (2012) Overproduction of a potential red pigment by a specific self-immobilization biomembrane-surface liquid culture of Penicillium novae-zeelandiae. Bioprocess Biosyst Eng 35(8):1407–1416
Wang S, Zhang S, Xiao A, Rasmussen M, Skidmore C, Zhan J (2015) Metabolic engineering of Escherichia coli for the biosynthesis of various phenylpropanoid derivatives. Metab Eng 29:153–159
Wang X, Li Z, Policarpio L, Koffas MAG, Zhang H (2020) De novo biosynthesis of complex natural product sakuranetin using modular co-culture engineering. Appl Microbiol Biot 104(11):4849–4861
Wang L, Liu Z, Jiang H, Mao X (2021a) Biotechnology advances in β-carotene production by microorganisms. Trends Food Sci Tech 111:322–332
Wang Y, Wang Y, Chen X, Gao N, Wu Y, Zhang H (2021b) Protoplast fusion between Blakeslea trispora 14,271 (+) and 14,272 (−) enhanced the yield of lycopene and β-carotene. World J Microb Biotechnl 37(4):58
Warhade MI, Badere RS (2018) Fusarium oxysporum cell elicitor enhances betalain content in the cell suspension culture of Celosia cristata. Physiol Mol Biol Plants 24(2):285–293
Warner JR, Reeder PJ, Karimpour-Fard A, Woodruff LBA, Gill RT (2010) Rapid profiling of a microbial genome using mixtures of barcoded oligonucleotides. Nat Biotechnol 28(8):856-U138
Wölwer-Rieck U, May B, Lankes C, Wüst M (2014) Methylerythritol and mevalonate pathway contributions to biosynthesis of mono-, sesqui-, and diterpenes in glandular trichomes and leaves of Stevia rebaudiana bertoni. J Agr Food Chem 62(11):2428–2435
Wrolstad RE, Culver CA (2012) Alternatives to those artificial FD&C food colorants. Annu Rev Food Sci T 3(1):59–77
Wu J, Du G, Zhou J, Chen J (2013) Metabolic engineering of Escherichia coli for (2S)-pinocembrin production from glucose by a modular metabolic strategy. Metab Eng 16:48–55
Wu K, Zhang X, Sun S, Wang X (2015) Factors affecting the accumulation of curcumin in microrhizomes of Curcuma aromatica salisb. Biomed Res Int. https://doi.org/10.1155/2015/740794
Wu T, Ye L, Zhao D, Li S, Li Q, Zhang B, Bi C, Zhang X (2017) Membrane engineering—a novel strategy to enhance the production and accumulation of beta-carotene in Escherichia coli. Metab Eng 43(Pt A):85–91
Wu T, Li S, Ye L, Zhao D, Fan F, Li Q, Zhang B, Bi C, Zhang X (2019a) Engineering an artificial membrane vesicle trafficking system (AMVTS) for the excretion of beta-carotene in Escherichia coli. ACS Synth Biol 8(5):1037–1046
Wu Y, Yan P, Liu X, Wang Z, Tang Y-J, Chen T, Zhao X (2019b) Combinatorial expression of different β-carotene hydroxylases and ketolases in Escherichia coli for increased astaxanthin production. J Ind Microbiol Biotechnol 46(11):1505–1516
Wu J, Chen W, Zhang Y, Zhang X, Jin J-M, Tang S-Y (2020a) Metabolic engineering for improved curcumin biosynthesis in Escherichia coli. J Agr Food Chem 68(39):10772–10779
Wu Y, Yan P, Li Y, Liu X, Wang Z, Chen T, Zhao X (2020b) Enhancing β-carotene production in Escherichia coli by perturbing central carbon metabolism and improving the NADPH supply. Front Bioeng Biotechnol. https://doi.org/10.3389/fbioe.2020.00585
Xiao JB, Hogger P (2015) Dietary polyphenols and type 2 diabetes: current insights and future perspectives. Curr Med Chem 22(1):23–38
Xie W, Liu M, Lv X, Lu W, Gu J, Yu H (2014) Construction of a controllable beta-carotene biosynthetic pathway by decentralized assembly strategy in Saccharomyces cerevisiae. Biotechnol Bioeng 111(1):125–33
Xie W, Lv X, Ye L, Zhou P, Yu H (2015a) Construction of lycopene-overproducing Saccharomyces cerevisiae by combining directed evolution and metabolic engineering. Metab Eng 30:69–78
Xie W, Ye L, Lv X, Xu H, Yu H (2015b) Sequential control of biosynthetic pathways for balanced utilization of metabolic intermediates in Saccharomyces cerevisiae. Metab Eng 28:8–18
Xu N, Wei L, Liu J (2019) Recent advances in the applications of promoter engineering for the optimization of metabolite biosynthesis. World Microbiol Biotechnol. https://doi.org/10.1007/s11274-019-2606-0
Xue J, Ahring BK (2011) Enhancing isoprene production by genetic modification of the 1-deoxy-d-xylulose-5-phosphate pathway in Bacillus subtilis. Appl Environ Microbiol 77(7):2399–2405
Yajun Y, Chemler J, Lixuan H, Martens S, Koffas MAG (2005) Metabolic engineering of anthocyanin biosynthesis in Escherichia coli. Appl Environ Microbiol 71(7):3617–3623
Yamagata K, Tagami M, Yamori Y (2015) Dietary polyphenols regulate endothelial function and prevent cardiovascular disease. Nutrition 31(1):28–37
Yan Y, Li Z, Koffas MAG (2008) High-yield anthocyanin biosynthesis in engineered Escherichia coli. Biotechnol Bioeng 100(1):126–140. https://doi.org/10.1002/bit.21721
Yan GL, Wen KR, Duan CQ (2012) Enhancement of beta-carotene production by over-expression of HMG-CoA reductase coupled with addition of ergosterol biosynthesis inhibitors in recombinant Saccharomyces cerevisiae. Curr Microbiol 64(2):159–63
Yang JM, Guo LZ (2014) Biosynthesis of beta-carotene in engineered E. coli using the MEP and MVA pathways. Microb Cell Factories. https://doi.org/10.1186/s12934-014-0160-x
Yang Y, Liu B, Du X, Li P, Liang B, Cheng X, Du L, Huang D, Wang L, Wang S (2015) Complete genome sequence and transcriptomics analyses reveal pigment biosynthesis and regulatory mechanisms in an industrial strain, Monascus purpureus YY-1. Sci Rep 5:8331
Yang D, Park SY, Lee SY (2021) Production of rainbow colorants by metabolically engineered Escherichia coli. Adv Sci 8(13):2100743
Ye L, Lv X, Yu H (2017) Assembly of biosynthetic pathways in Saccharomyces cerevisiae using a marker recyclable integrative plasmid toolbox. Front Chem Sci Eng 11(1):126–132
Ye L, Zhu X, Wu T, Wang W, Zhao D, Bi C, Zhang X (2018) Optimizing the localization of astaxanthin enzymes for improved productivity. Biotechnol Biofuels. https://doi.org/10.1186/s13068-018-1270-1
Yen H-W, Palanisamy G, Su G-C (2019) The influences of supplemental vegetable oils on the growth and β-carotene accumulation of oleaginous Yeast-Rhodotorula glutinis. Biotechnol Bioprocess Eng 24(3):522–528
Yolmeh M, Khomeiri M (2016) Using physical and chemical mutagens for enhanced carotenoid production from Rhodotorula glutinis (PTCC 5256). Biocatal Agric Biotechnol 8:158–166
Yolmeh M, Khomeiri M, Ghorbani M, Ghaemi E, Ramezanpour SS (2017) High efficiency pigment production from Micrococcus roseus (PTCC 1411) under ultraviolet irradiation. Biocatal Agric Biotechnol 9:156–161
Yoon SH, Lee SH, Das A, Ryu HK, Jang HJ, Kim JY, Oh DK, Keasling JD, Kim SW (2009) Combinatorial expression of bacterial whole mevalonate pathway for the production of beta-carotene in E. coli. J Biotechnol 140(3–4):218–26
Yuan LZ, Rouviere PE, Larossa RA, Suh W (2006) Chromosomal promoter replacement of the isoprenoid pathway for enhancing carotenoid production in E. coli. Metab Eng 8(1):79–90
Zare K, Nazemiyeh H, Movafeghi A, Khosrowshahli M, Motallebi-Azar A, Dadpour M, Omidi Y (2010) Bioprocess engineering of Echium italicum L.: induction of shikonin and alkannin derivatives by two-liquid-phase suspension cultures. Plant Cell Tissue Organ 100(2):157–164
Zelcbuch L, Antonovsky N, Bar-Even A, Levin-Karp A, Barenholz U, Dayagi M, Liebermeister W, Flamholz A, Noor E, Amram S, Brandis A, Bareia T, Yofe I, Jubran H, Milo R (2013) Spanning high-dimensional expression space using ribosome-binding site combinatorics. Nucleic Acids Res 41(9):e98
Zha J, Zang Y, Mattozzi M, Plassmeier J, Gupta M, Wu X, Clarkson S, Koffas MAG (2018) Metabolic engineering of Corynebacterium glutamicum for anthocyanin production. Microb Cell Fact. https://doi.org/10.1186/s12934-018-0990-z
Zhang B, Zheng LP, Wang JW (2012) Nitric oxide elicitation for secondary metabolite production in cultured plant cells. Appl Microbiol Biotechnol 93(2):455–466
Zhang C, Wang Z, Zhao J, Li Q, Huang C, Zhu L, Lu D (2016a) Neuroprotective effect of lutein on NMDA-Induced retinal ganglion cell injury in rat retina. Cell Mol Neurobiol 36(4):531–40
Zhang Y, Navarro E, Canovas-Marquez JT, Almagro L, Chen H, Chen YQ, Zhang H, Torres-Martinez S, Chen W, Garre V (2016b) A new regulatory mechanism controlling carotenogenesis in the fungus Mucor circinelloides as a target to generate beta-carotene over-producing strains by genetic engineering. Microb Cell Fact 15:99
Zhang W, Liu H, Li X, Liu D, Dong X-T, Li F-F, Wang E-X, Wang E-X, Li B-Z, Yuan Y-J (2017) Production of naringenin from D-xylose with co-culture of E-coli and S-cerevisiae. Eng Life Sci 17(9):1021–1029
Zhang C, Seow VY, Chen X, Too H-P (2018a) Multidimensional heuristic process for high-yield production of astaxanthin and fragrance molecules in Escherichia coli. Nat Commun. https://doi.org/10.1038/s41467-018-04211-x
Zhang C, Seow VY, Chen X, Too H-P (2018b) Multidimensional heuristic process for high-yield production of astaxanthin and fragrance molecules in Escherichia coli. Nat Commun 9:1858
Zhao J (2015) Flavonoid transport mechanisms: how to go, and with whom. Trends Plant Sci 20(9):576–85
Zhao CY, Nabity PD (2017) Phylloxerids share ancestral carotenoid biosynthesis genes of fungal origin with aphids and adelgids. PLoS ONE. https://doi.org/10.1371/journal.pone.0185484
Zhao YR, Yang JM, Qin B, Li YH, Sun YZ, Su SZ, Xian M (2011) Biosynthesis of isoprene in Escherichia coli via methylerythritol phosphate (MEP) pathway. Appl Microbiol Biotechnol 90(6):1915–1922
Zhao J, Li Q, Sun T, Zhu X, Xu H, Tang J, Zhang X, Ma Y (2013) Engineering central metabolic modules of Escherichia coli for improving beta-carotene production. Metab Eng 17:42–50
Zhao X, Shi F, Zhan W (2015) Overexpression of ZWF1 and POS5 improves carotenoid biosynthesis in recombinant Saccharomyces cerevisiae. Lett Appl Microbiol 61(4):354–360
Zhao XR, Choi KR, Lee SY (2018) Metabolic engineering of Escherichia coli for secretory production of free haem. Nat Catal 1(9):720–728
Zhong M, Huang S, Wang H, Huang Y, Xu J, Zhang L (2019) Optimization of ultrasonic-assisted extraction of pigment from Dioscorea cirrhosa by response surface methodology and evaluation of its stability. Rsc Adv 9(3):1576–1585
Zhou P, Ye L, Xie W, Lv X, Yu H (2015a) Highly efficient biosynthesis of astaxanthin in Saccharomyces cerevisiae by integration and tuning of algal crtZ and bkt. Appl Microbiol Biotechnol 99(20):8419–28
Zhou Q, Zhang P, Zhang G (2015b) Biomass and pigments production in photosynthetic bacteria wastewater treatment: effects of light sources. Bioresour Technol 179:505–509
Zhou P, Xie W, Li A, Wang F, Yao Z, Bian Q, Zhu Y, Yu H, Ye L (2017a) Alleviation of metabolic bottleneck by combinatorial engineering enhanced astaxanthin synthesis in Saccharomyces cerevisiae. Enzyme Microb Technol 100:28–36
Zhou P, Xie W, Li A, Wang F, Yao Z, Bian Q, Zhu Y, Yu H, Ye L (2017b) Alleviation of metabolic bottleneck by combinatorial engineering enhanced astaxanthin synthesis in Saccharomyces cerevisiae. Enzyme Microb Tech 100:28–36
Zhou S, Du G, Kang Z, Li J, Chen J, Li H, Zhou J (2017c) The application of powerful promoters to enhance gene expression in industrial microorganisms. World J Microb Biotechnol. https://doi.org/10.1007/s11274-016-2184-3
Zhou P, Xie W, Yao Z, Zhu Y, Ye L, Yu H (2018) Development of a temperature-responsive yeast cell factory using engineered Gal4 as a protein switch. Biotechnol Bioeng 115(5):1321–1330
Zhou P, Li M, Shen B, Yao Z, Bian Q, Ye L, Yu H (2019) Directed coevolution of beta-carotene ketolase and hydroxylase and its application in temperature-regulated biosynthesis of astaxanthin. J Agric Food Chem 67(4):1072–1080
Acknowledgements
Not applicable.
Funding
This work was funded by the National Natural Science Foundation of China (22108097), Natural Science Foundation of Jiangsu Province (BK20200616), National Key Research and Development Program of China (Grant No. 2018YFA0901800), China Postdoctoral Science Foundation (2020M671339), and Zhejiang Provincial Natural Science Foundation of China (Grant No. LZ20B060002).
Author information
Authors and Affiliations
Contributions
XL and YL: writing—original draft preparation. RY and LY: conceptualization, methodology. HY and WNC: supervision. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that there are no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lyu, X., Lyu, Y., Yu, H. et al. Biotechnological advances for improving natural pigment production: a state-of-the-art review. Bioresour. Bioprocess. 9, 8 (2022). https://doi.org/10.1186/s40643-022-00497-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40643-022-00497-4