Abstract
The production of safe and high-quality silage has always been the main concern. This experiment aimed to investigate the impact of waste dried soybean curd residue (SR) and Lactobacillus plantarum CCZZ1 on the fermentation quality and microbial community of total mixed ration (TMR) silage based on Napier grass (Pennisetum purpureum). Napier grass was made into TMR, and SR at 3%, 6% or 9% on dry material basis was included, which replaced the equivalent amount of corn meal, then they were inoculated without or with Lactobacillus plantarum CCZZ1 (105 cfu g−1; LP). The research results showed that incorporating SR even at 3% resulted in significant reduction in ammonia nitrogen content (87.3 g kg−1 total nitrogen vs. 109.7 g kg−1 total nitrogen), increased lactic acid content (34.4 g kg−1 DM vs. 25.5 g kg−1 DM), and higher relative abundance of Lactobacillus (94.5% vs. 32.2%). Additionally, it led to decreased relative abundances of pathogenic microorganisms such as Escherichia coli (< 0.1% vs. 9.68%), Staphylococcus epidermidis (< 0.1% vs. 9.46%), and Streptococcus pneumoniae (< 0.1% vs. 8.53%) during the ensiling process. When SR was used together with LP inoculation, they were further improved. These findings suggest that the inclusion of SR, even at a 3% level without LP inoculation, can effectively improve the fermentation quality and microbial profile of TMR silage based on fresh Napier grass. This offers a promising technical approach to utilizing SR and producing safe and high-quality TMR silage based on fresh grass.
Graphical Abstract

Highlights
-
1.
Ensiling is an efficacious store way for fresh grass-based TMR in tropical and subtropical regions.
-
2.
SR inclusion offers a promising approach to produce safe and high-quality TMR silage.
-
3.
SR greatly increases the relative abundance of desirable Lactobacillus in TMR silage.
-
4.
SR greatly decreases harmful microorganisms like E. coli, S. epidermidis, and S. pneumoniae during ensiling.
Similar content being viewed by others
Introduction
Total mixed ration (TMR) is a feed mixture containing raw materials such as concentrate, by-products, forage, minerals and vitamins according to ruminant animal's nutrient requirements [1]. Therefore, TMR has the advantage of balanced nutrition. However, TMR is prone to aerobic deterioration after being exposed to the air [2]. This feature is not conducive to the circulation of TMR as a commodity. In addition, the aerobic deterioration of TMR will not only result in the loss of nutrients, but also produce harmful metabolites that will cause damage to animal health [3]. Ensiling could preserve the nutrients of TMR for a longer period and facilitate long-distance transportation [3]. Compared to TMR, ensiled TMR could improve aerobic stability, nutrient digestibility and reduce methane emissions [4, 5]. On the other hand, the lactic acid content of fermented TMR increased, which improved the palatability of TMR [6]. Simultaneously, TMR silage does not cause environmental pollution from the effluent of unwilted grass silage, due to its low moisture content.
With the rapid economic and social development in China, the competition between human food and animal feed has become more pronounced. As a result, the intensification of ruminant production necessitates the development of innovative feeding strategies. Additionally, the production of hay is more challenging in certain regions due to climate factors [7]. In such circumstances, using grass-based TMR silage could be a viable solution. Napier grass (Pennisetum purpureum) is an important tropical grass [8]. High yields, wide adaptation and good palatability of Napier grass promote it to be widespread use in silage production for ruminant animals [9]. Nevertheless, the utilization of Napier grass as silage is constrained by its low dry matter (DM) content and high buffer capacity [10], which often lead to unfavorable fermentation and effluent production during ensiling, posing detrimental effects on animal production as well as the environment [11]. Consequently, the use of inoculants or absorbents is common practice to enhance fermentation quality and reduce effluent production of high moisture grass silage. Homofermentative lactic acid bacteria (LAB) can quickly reduce pH, reduce nutritional losses and improve the fermentation quality during ensiling [12]. Soybean curd residue is a by-product of the tofu production, with an annual output in China of about 2.8 million tons [13]. The crude protein (CP) content of soybean curd residue is as high as 15% of DM, and the non-fractionable protein content in CP is as high as 61.75%, which can be used as a good source of protein in dairy cow feed [14]. Additionally, in China, dried soybean curd residue (SR) is cheap and easy to obtain and might be a kind of promising and efficient absorbent for high-quality silage production due to its high moisture absorption [15]. However, the studies on SR addition in high moisture grass-based TMR silage are rare. Therefore, it is indispensable to better understand the effect of SR and LAB addition on the ensiling characteristics and microbial community of TMR silage based on fresh Napier grass. In this study, we assumed that the high moisture fresh Napier grass might affect the overall fermentation quality of TMR silage during ensiling process, and adding SR or LAB might be able to counteract these negative effects. The objective of this study was to assess the fermentation quality and microbial community of TMR silage based on fresh Napier grass with or without the incorporation of SR and LAB. The findings aim to offer novel technical approaches to mitigate the adverse impacts of fresh Napier grass TMR silage production and utilization, particularly on the health of humans and animals.
Materials and methods
Silage making and sample dealing
Napier grass was planted on an experimental field at South China Agricultural University. The experimental field locates at 23°14′N and 113°38′E, the annual average temperature was 23.1 ℃, the annual average rainfall was 1623.6–1899.8 mm. Napier grass was planted on June 12, 2021, artificially harvested at a height of 1.5 m on October 12, 2021, immediately transported to the laboratory, and chopped into Sects. 1–2 cm thick (DM, 296.6 g kg−1 FM). The SR was purchased from an agricultural market in Tianhe District, Guangzhou, China. As shown in Table 1, the control TMR (CK) in this experiment were formulated with chopped Napier grass (1–2 cm), corn meal, soybean meal, vitamin-mineral supplement (Ainong feed, Jiangsu, China), and salt in a ratio of 45.0:50.0:3.5:1:0.5, respectively, on a basis of DM. Napier grass was made into TMR, and SR at 0%, 3%, 6% or 9% on dry material basis was included, which replaced the equivalent amount of corn meal, and they were inoculated with Lactobacillus plantarum CCZZ1 (105 cfu g−1 FM; LP) or without. Approximately 200 g well-mixed TMR was filled into a plastic film bag (200 × 300 mm), vacuumed and sealed with a vacuum sealer. Each treatment was made in triplicates. In total, 48 bags were stored in a room at the temperature 25 ℃ to 28 ℃, and all the bags were opened at 60 d of ensiling for the chemical and microbial analyses.
Chemical and microbial quantity analyses
DM content was determined using a forced draft oven at 70 ℃ for 48 h. The CP content was analyzed according to the method of Association of Official Analytical Chemists [16]. The neutral detergent fiber (NDF) and acid detergent fiber (ADF) contents were determined according to the method of van Soest et al. [17]. The water-soluble carbohydrate (WSC) content was analyzed using the anthrone method [18]. For pH, ammonia nitrogen (NH3-N) and organic acid determination, 20 g of silage samples were mixed with 180 mL of distilled water, stored at 4 ℃ for 18 h, and then filtered [19]. The pH of this filtrate was measured by a glass electrode pH meter (PHS‐3C, CSDIHO Co., Ltd, Shanghai, China). The content of NH3-N was determined by the method of Broderick and Kang [20]. The content of organic acid was analyzed using HPLC (Shodex RS Pak KC-811, Showa Denko K.K., Kawasaki, Japan; detector: RID10A, Shimadzu Co., Ltd, Kyoto, Japan; eluent: 0.1% H3PO4, 1.0 mL min−1; temperature: 40 ℃). Fermentation quality was comprehensively assessed by V-score, which was determined from the proportion ammonia-n in the total nitrogen and volatile fatty acid contents in the silage [21]. Fermentation quality of silage was divided into 3 grades according to V-score points: excellent (V-score ≥ 80), acceptable (60 ≤ V-score < 80), and poor (V-score < 60).
A total of 10 g of each sample was dissolved in 90 mL of sterile saline solution and shaken for 30 min and serially diluted from 10–1 to 10–7 for microbial analysis. The number of LAB were enumerated by plate count on MRS medium agar (Huankai Biological Co., Ltd, Guangzhou, China) under anaerobic conditions at 37 ℃ for 2 days. Aerobic bacteria were counted on nutrient agar (Huankai Biological Co., Ltd) after 2 days of incubation at 37 ℃ under aerobic conditions. Yeasts and molds were enumerated by plate count on potato dextrose agar (Huankai Biological Co., Ltd), acidified to a pH of 3.5 with a sterilized tartaric acid solution, following 3 days of incubation at 30 ℃ under aerobic conditions [19].
Bacterial diversity analysis
Total DNA in silage samples was extracted using the TGuide S96 Magnetic Soil/Stool DNA Kit (Tiangen Biotech (Beijing) Co., Ltd.) according to manufacturer instructions. The DNA concentration of the samples was measured with the Qubit dsDNA HS Assay Kit and Qubit 4.0 Fluorometer (Invitrogen, Thermo Fisher Scientific, Oregon, USA). The V3–V4 regions of 16S rDNA were amplified using primers 338F (ACTCCTACGGGAGGCAGCAG) and 806R (GGACTACHVGGGTATCTAAT), then PCR products were sequenced and the raw sequences were analyzed according to the method of Tian et al. [22]. The functional genes of the bacterial communities were predicted using PICRUSt [23].
Statistical analysis
All data in this study were analyzed using the general linear model procedure of IBM SPSS.19 software for Windows (IBM Corp, New York, USA), and the means of different samples were compared (significance at P < 0.05) using Duncan’s multiple range method. The DNA sequencing data were analyzed on a free online platform of BMKCloud (http://www.biocloud.net).
Results
TMR characteristics and fermentation quality of TMR silages
The DM and WSC contents of Napier grass TMRs were 478.0–482.7 g kg−1 FM (fresh matter) and 189.2 – 192.6 g kg−1 DM, respectively (Table 1). The epiphytic LAB number was less than 4 log10cfu g−1 in all TMRs. There were no significant changes of chemical composition and microbial population sizes in TMRs when corn meal was replaced with the equivalent amount of SR at 3%, 6% or 9% fresh material basis (Table 1, P > 0.05).
Both Napier grass TMR silages with 3% SR and LP exhibited good fermentation quality, as evidenced by lower pH values, higher lactic acid concentration and the lower concentration of acetic acid, butyric acid and NH3-N compared to CK (Table 2). The NH3-N content of all SR and LP treatments was below the criterion value (100 g kg−1 TN) for well-preserved silage. Furthermore, the combined treatment of SR and LP significantly decreased NH3-N content compared to SR or LP alone (P < 0.05). However, the NH3-N content of CK exceed 100 g kg−1 TN (Table 2). The V-scores of TMR silages containing LP or SR surpassed 80, indicating an excellent grade, and were significantly higher than the CK (P < 0.05), while the CK TMR silage achieved only an acceptable grade (62, Table 2).
Microbial counts of TMR silages
Compared with the CK, the numbers of LAB and aerobic bacteria were significantly reduced in all SR and SRLP treatments (Table 3; P < 0.01). Moreover, the yeast counts in all SR treatments were below 2 log10 cfu g−1 FM (Table 3; P < 0.01). Additionally, the mold counts in all TMR silages were below 2.00 log10 cfu g−1 FM.
Bacterial community of TMR silages
Bacterial diversity of TMR silage is shown in Table 4. The coverage of all samples was approximately 1.00, and the observed species, OTUs, Chao 1, Shannon, and Simpson indices of LP, SR and SRLP treatments decreased greatly compared with those of CK (Table 4).
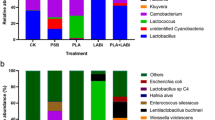
At the phylum level, Firmicutes was dominant in all treatments, and the order of proportion was as follows: SR3LP (98.1%) > SR3 (94.9%) > SR6LP (93.6%) > SR6 (91.3%) > CKLP (88.9%) > CK (59.4%) (Fig. 1A). Followed by Proteobacteria, and the order of proportion was as follows: CK (26.7%) > CKLP (11.0%) > SR6 (7.75%) > SR6LP (3.53%) > SR3 (3.32%) > SR3LP (0.95%) (Fig. 1A). At the genus level, the most abundant genus in CK was Lactobacillus (32.2%), followed by Streptococcus (11.8%), Massilia (11.3%), Escherichia (9.68%), Staphylococcus (9.46%), and uncultured_bacterium_o_Chloroplast (4.58%); differently, in CKLP, the most abundant genus was Lactobacillus (76.1%), followed by Enterobacter (10.46%), Pediococcus (9.78%). Excitingly, when replacing a part of corn meal with SR, the most abundant genus in all treatments was Lactobacillus (> 90%), and the order of proportion was as follows: SR3LP (97.3%) > SR3 (94.4%) > SR6LP (92.1%) > SR6 (90.9%) (Fig. 1B), and the heat map analysis could prove the occurrence of the core bacterial community (Fig. 2), which suggested that the addition of SR combined with or without LP effectively increased the abundance of Lactobacillus of Napier grass TMR silage. Therefore, the addition of SR effectively promoted the growth and reproduction of LAB to further promote lactic acid fermentation and improved fermentation quality. At the species level, Lactobacillus mainly consisted of L. plantarum and L. fermentum. The abundance of L. plantarum in the different treatments was 23.7% (CK), 67.3% (CKLP), 89.7% (SR6LP), 89.8% (SR3), 90.4% (SR6) and 90.4% (SR3LP). The abundance of L. fermentum was 8.3% (CK), 4.43% (SR3), 3.82% (SR3LP), 1.33% (SR6LP), 0.24% (SR6) and 0.05% (CKLP). Staphylococcus, Massilia, Escherichia and Streptococcus in CK were mainly composed of S. epidermidis (9.46%) M. eurypsychrophila (8.52%) E. coli (9.68%) and S. pneumoniae (8.53%), respectively. Enterobacter in CKLP was mainly composed of E. cloacae (10.46%), and Pediococcus was mainly composed of P. pentosaceus (9.78%) (Fig. 1C).
Relative abundance of bacterial community on phylum (A), genus (B), and species (C) level of different Napier grass total mixed ration silage. LP Lactobacillus plantarum CCZZ1; DM dry matter, SR dried soybean curd residue, SR3 and SR6 were SR levels at 3% and 6% of total mixed ration DM, respectively
Heatmap analysis of top 30 genera according to relative abundance. The X-axis represents sample identifier and the Y-axis represents the genus. Block with red color indicates the high abundance of the genus, the blue block indicates the low abundance of the genus. The hierarchical clustering tree appears on the upside of the block while the phylogenetic tree appears on the left side of the block. LP Lactobacillus plantarum CCZZ1; DM dry matter, SR dried soybean curd residue, SR3 and SR6 were SR levels at 3% and 6% of total mixed ration DM, respectively
The 16S rRNA gene-predicted function analyses are shown in Fig. 3A. In the SR and SRLP-treated silages, the abundances of phospho-transferase-system (PTS), fructose-and-mannose-metabolism, glycolysis/gluconeogenesis, alanine-aspartate-and-glutamate-metabolism, pentose-phosphate-pathway, lysine-biosynthesis, and starch-and-sucrose-metabolism were higher than those in CK silage (Fig. 3A), and these metabolisms pathways were obvious negatively correlated with glycine-serine-and-threonine metabolism and oxidative-phosphorylation (Fig. 3B).
A Heatmap of 16S rRNA gene-predicted functional profiles obtained with PICRUSt in different Napier grass total mixed ration silage, Colors reflect relative abundance from low (blue) to high (red); B heatmap of Pearson correlation analysis of different microbial function pathways. LP Lactobacillus plantarum CCZZ1; DM dry matter, SR dried soybean curd residue, SR3 and SR6 were SR levels at 3% and 6% of total mixed ration DM, respectively
Discussion
TMR characteristics, fermentation quality and microbial counts of TMR silages
It is well known that the silage with suitable contents of DM (> 300 g kg−1 FM) and WSC (> 60 g kg−1 DM), as well as LAB count (> 5 log10 cfu g−1 FM) can be well fermented [24]. In the present study, the DM and WSC contents of all TMR satisfied the prerequisite for successful fermentation, and the chemical composition of all the treatments did not exhibit significant changes, irrespective of SR replacement levels (Table 1, P > 0.05). However, the LAB counts of all TMR were less than 4 log10cfu g−1 FM, which was undesirable (Table 1). Oliveira et al. [25] found that LAB counts beyond 5 log10 cfu g−1 FM could effectively improve silage fermentation quality while LAB counts < 4 log10 cfu g−1 FM might increase DM loss and NH3-N content. The addition of L. plantarum at ensiling could promote homofermentation of TMR silage [26], which resulted in more lactic acid in all LP silages than that in the CK. Similarly, Chen et al. [27] found that inoculation with L. plantarum led to an efficient homofermentation of TMR silage based on oat and common vetch. Notably, lactic acid fermentation in the present study was promoted by either SR (even at 3%) or LP, and SRLP TMR silages were of significant higher lactic acid content and V-scores, and lower pH value and NH3-N content (far less than 100 g kg−1 TN) than CK (P < 0.05). The SR treatments significantly increased the lactic acid content (P < 0.05), and there were no significant differences among the SR replacement levels (Table 2, P > 0.05). These results suggested that SR had the same effect on promoting lactic acid fermentation as LP. All the above results indicate that SR might be a promising silage additive for improving the fermentation quality of Napier grass TMR silage.
In this study, the fermentation quality of all the SR treatments was better than that of CK (Table 2), and the microbial counts of all the SR treatments were significant less than those of CK, especially aerobic microorganisms as well as yeasts (Table 3, P < 0.01). The counts of aerobic bacteria and yeasts (P < 0.05) were not significant different among the SR replacement levels (Table 3, P > 0.05). This might be due to a low pH environment for the SR treatments, which inhibited undesirable fermentation of harmful microorganisms [24, 28]. It suggested that the addition of SR (even at 3%) combined with or without LP could effectively improve the fermentation quality and reduce the counts of aerobic bacteria and yeast in Napier grass TMR silage.
At present, there are few researches on SR as a silage additive. Li et al. [29] found that most of the soybean isoflavones were retained in soybean curd residue during the process of tofu making. Santos et al. [30] also found that soybean curd residue was a rich source of isoflavones. It is well known that isoflavones have strong antioxidant and antibacterial capacity, and microbial fermentation promotes the transformation of isoflavones from glucosides to aglycones in soy products or by-products, resulting in the improvement of physiological activities of isoflavones [30, 31]. This might be one of reasons for the significant decrease in the count of aerobic bacteria in all SR treatments, compared to CK (Table 3). Some experiments found that many aerobic bacteria have certain protein degradation ability during ensiling, resulting in protein degradation and NH3-N production [24]. Aerobic bacteria count in all SR treatments was significantly lower than that of CK, which might reduce the NH3-N production (Tables 2 and 3). The fresh grasses have high moisture content and aerobic bacteria count after harvest, in which the preparation of silage is relatively difficult [24, 32]. Moreover, the LAB counts of all TMR materials in the present study were less than 4.00 log10 cfu g−1 FM (Table 1). These reasons might result in the poor fermentation of CK (Table 3).
Bacterial community of TMR silages
The coverage of all samples was approximately 1.00, indicating that the sequencing data were large enough to reflect the microbial diversity information in all samples [33]. According to the reduction in the observed species, Shannon, Simpson indices and Chao 1, the bacterial diversity of all LP, SR and SRLP silage samples decreased greatly (Table 4). This is similar to the study in Wang et al. [34], who reported that the addition of LAB significantly reduced the observed species, Shannon, Simpson and Chao during ensiling.
In present study, the relative abundance of Proteobacteria was significantly reduced in all SR and SRLP silages during ensiling compared to CK and CKLP, which should be noted that the relative abundance of Firmicutes in SR3 silage was much higher than CKLP, and Proteobacteria in SR3 was much lower than in CKLP (Fig. 1A). It is well known that the bacteria involved in lactic acid fermentation belonging to Firmicutes rather than Proteobacteria [35]. Proteobacteria is the dominant phylum in Napier grass [36]. Therefore, the higher relative abundance of Firmicutes in SR and SRLP silage might also contribute to the higher lactic content and lower pH value of the TMR silage (Fig. 1A).
L. plantarum and L. fermentum are widely used as silage inoculants [37, 38]. L. plantarum can potentially promote the production of lactic acid, and it grows vigorously, rapidly reducing pH, and grows well on low-moisture substrates [39], thereby inhibiting the growth and reproduction of harmful microorganisms to ensure the fermentation quality of Napier grass TMR silage. This might be the main reason that the lactic acid content in SR and SRLP silages was higher and the ammonia-N content was lower than that of CK (Table 2). M. eurypsychrophila and S. pneumoniae are Gram-positive species, which can survive in an anaerobic environment and utilize glucose [40, 41], and might compete with LAB for nutrients, but the roles of these species in silage production have rarely been reported. Moreover, E. coli and S. pneumoniae are common pathogenic bacteria that are prone to negatively affect the health of humans and animals [41, 42]. E. coli may be transmitted to forage crops and their preserved products through shedding and fertilization of cow manure [43], but it cannot survive in well-preserved silage [44]. S. epidermidis is usually a Gram-positive species that is harmless in its natural environment [45]. However, it can cause virulent infections once it invades the human body. Furthermore, it is necessary to be concerned about the presence of pathogenic microorganisms during silage production due to certain potential risks to human and animal health [46]. Interestingly, although the relative abundance of these species was high in CK silage, they were almost non-existent in CKLP, SR and SRLP silages. Therefore, this could provide an additional potential control point for the prevention of the negative effects caused by S. pneumoniae, E. coli and S. epidermidis, thereby reducing the impact on the environment and animal health. Enterobacter may also compete with the LAB for nutrients and produce ammonia-N [39]. The species of P. pentosaceus could promote the fermentation process in the early ensiling stage [47], and significantly increased the lactic acid content [48]. The relative abundance of E. cloacae and P. pentosaceus was high in CKLP silage, but they were almost non-existent in SR and SRLP silages. It is well known that the abundance of dominant bacteria can decrease the diversity of the microbial community [49]. These might be due to fact that the high abundance of Lactobacillus (> 90%) in SR and SRLP silages inhibited the growth of Enterobacter. Consequently, the SR could improve TMR anaerobic fermentation quality by stimulating desirable Lactobacillus species and inhibiting undesirable microbes.
In addition, the results at the level of microbial metabolism can also prove the above statement. The abundances of the phospho-transferase-system (PTS), fructose-and-mannose-metabolism, glycolysis/gluconeogenesis, alanine-aspartate-and-glutamate-metabolism, pentose-phosphate-pathway, lysine-biosynthesis, and starch-and-sucrose-metabolism in SR and SRLP-treated silages were higher than those in CK silage (Fig. 3A). Similarly, Liu et al. [22] found that LAB inoculation could increase the abundances of these metabolism pathways in barley silage. In addition, these metabolisms were obviously negatively correlated with glycine-serine-and-threonine metabolism and oxidative-phosphorylation (Fig. 3B). Moreover, the abundances of glycine-serine-and-threonine-metabolism and oxidative-phosphorylation were reduced in SR and SRLP-treated silages (Fig. 3A). The reduced metabolisms of nitrogen, glycine, serine, and threonine could decrease the production of ammonia-n [50] since the decarboxylation of amino acid could also result in the accumulation of biogenic amines [27]. Therefore, these changes in the metabolism pathways of carbohydrate and amino acid might be the reason why LAB inoculation and SR addition significantly increased lactic acid content and decreased NH3-N content in Napier grass TMR silage (Table 2, P < 0.05).
Conclusions
Replacing a part of corn meal with dried soybean curd residue (even at 3%) can effectively improve the fermentation quality and microbial community of Napier grass-based TMR silage, indicated by reduced butyric acid and NH3-N content, increased lactic acid content and the relative abundance of Lactobacillus, decreased the relative abundances of pathogenic microorganisms like Escherichia coli, Staphylococcus epidermidis, and Streptococcus pneumoniae during ensiling. When dried soybean curd residue was used together with LP inoculation, they were further improved. The findings of this study would provide a new technical approach for the effective use of waste dried soybean curd residue and preparation of fresh grass-based TMR silage in tropical and subtropical regions.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Abbreviations
- ADF:
-
Acid detergent fiber
- cfu:
-
Colony forming units
- CP:
-
Crude protein
- DM:
-
Dry matter
- FM:
-
Fresh matter
- log:
-
Denary logarithm of the numbers
- LAB:
-
Lactic acid bacteria
- LP:
-
Lactobacillus plantarum CCZZ1
- NS:
-
Not significant
- NDF:
-
Neutral detergent fiber
- NH3-N:
-
Ammonia nitrogen
- ND:
-
No detected
- SR:
-
Dried soybean curd residue
- SR3:
-
The SR level at 3% TMR DM
- SR6:
-
The SR level at 6% TMR DM
- SR9:
-
The SR level at 9% TMR DM
- SEM:
-
Standard error of the means
- Sig:
-
Significance
- TMR:
-
Total mixed ration
- TN:
-
Total nitrogen
- WSC:
-
Water-soluble carbohydrates
References
Coppock CE, Bath DL, Harris B. From feeding to feeding systems. J Dairy Sci. 1981;64(6):1230–49. https://doi.org/10.3168/jds.S0022-0302(81)82698-7.
Wang HL, Ning TT, Hao W, Zheng ML, Xu CC. Dynamics associated with prolonged ensiling and aerobic deterioration of total mixed ration silage containing whole crop corn. Asian-Austral J. 2016;29(1):62–72. https://doi.org/10.5713/ajas.15.0319.
Wang SR, Zhang GJ, Zhao J, Dong ZH, Li JF, Shao T. Fermentation quality, aerobic stability and in vitro gas production kinetics and digestibility in total mixed ration silage treated with lactic acid bacteria inoculants and antimicrobial additives. Ital J Anim Sci. 2023;22(1):429–40. https://doi.org/10.1080/1828051X.2023.2206422.
Tian PJ, Niu DZ, Zuo SS, Jiang D, Li RR, Xu CC. Vitamin A and E in the total mixed ration as influenced by ensiling and the type of herbage. Sci Total Environ. 2020;746:141239. https://doi.org/10.1016/j.scitotenv.2020.141239.
Wang SR, Zhao J, Yu CQ, Li JF, Tao XX, Chen SF, Shao T. Nutritional evaluation of wet brewers’ grains as substitute for common vetch in ensiled total mixed ration. Ital J Anim Sci. 2020;19:1015–25. https://doi.org/10.1080/1828051X.2020.1810141.
Miyaji M, Matsuyama H, Nonaka K. Effect of ensiling process of total mixed ration on fermentation profile, nutrient loss and in situ ruminal degradation characteristics of diet. Anim Sci J. 2017;88(1):134–9. https://doi.org/10.1111/asj.12610.
McGechan MB. Operational research study of forage conservation systems for cool, humid upland climates. part 2: comparison of hay and silage systems. J Agric Eng Res. 1990;46:129–45. https://doi.org/10.1016/S0021-8634(05)80144-1.
Du Z, Yamasaki S, Oya T, Nguluve D, Tinga B, Macome F, et al. Ensiling characteristics of total mixed ration prepared with local feed resources in Mozambique and their effects on nutrition value and milk production in Jersey dairy cattle. Anim Sci J. 2020;91(1):e13370. https://doi.org/10.1111/asj.13370.
Lu F, Erik FK, Veronica M, Alastair JW, Henrik BM. Ensiling of tall fescue for biogas production: effect of storage time, additives and mechanical pretreatment. Energy Sustain Dev. 2018;47:143–8. https://doi.org/10.1016/j.esd.2018.10.001.
Zanine AD, Santos EM, Dorea JRR, Dantas PAD, da Silva TC, Pereira OG. Evaluation of elephant grass silage with the addition of cassava scrapings. Rev Bras Zootecn. 2010;39(12):2611–6. https://doi.org/10.1590/S1516-35982010001200008.
Duniere L, Sindou J, Chaucheyras-Durand F, Chevallier I, Thevenot-Sergentet D. Silage processing and strategies to prevent persistence of undesirable microorganisms. Anim Feed Sci Technol. 2013;182(1–4):1–15. https://doi.org/10.1016/j.anifeedsci.
Muck RE, Nadeau EMG, McAllister TA, Contreras-Govea FE, Santos MC, Kung L. Silage review: recent advances and future uses of silage additives. J Dairy Sci. 2018;101(5):3980–4000. https://doi.org/10.3168/jds.2017-13839.
Li B, Qiao MY, Lu F. Composition, nutrition, and utilization of Okara (soybean residue). Food Rev Int. 2012;28(3):231–52. https://doi.org/10.1080/87559129.2011.595023.
Peter WC, Kuen-Jaw C, Kwen-Sheng K, Jenn-Chung H, Bi Y. Studies on the protein degradabilities of feedstuffs in Taiwan. Anim Feed Sci Technol. 1995;55(3):215–26. https://doi.org/10.1016/0377-8401(95)00798-R.
Yin X, Wu JY, Tian J, Wang XY, Zhang JG. Dried soybean curd residue: a promising absorbent for cleaner production of high-quality silage. J Clean Prod. 2021;324:129300. https://doi.org/10.1016/j.jclepro.2021.129300.
Baur FJ, Ensminger LG. The association of official analytical chemists (AOAC). J Am Oil Chem Soc. 1977;54:171–2. https://doi.org/10.1007/BF02670789.
van Soest PJ, Robertson JB, Lewis BA. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J Dairy Sci. 1991;74(10):3583–97. https://doi.org/10.3168/jds.S0022-0302(91)78551-2.
Murphy RP. A method for the extraction of plant samples and the determination of total soluble carbohydrates. J Sci Food Agric. 1958;9:714–7. https://doi.org/10.1002/jsfa.2740091104.
Liu QH, Yang FY, Zhang JG, Shao T. Characteristics of Lactobacillus parafarraginis ZH1 and its role in improving the aerobic stability of silages. J Appl Microbiol. 2014;117(2):405–16. https://doi.org/10.1111/jam.12530.
Broderick GA, Kang JH. Automated simultaneous determination of ammonia and total amino acids in ruminal fluid and in vitro media. J Dairy Sci. 1980;63(1):64–75. https://doi.org/10.3168/jds.S0022-0302(80)82888-8.
Small SM, Regan PF. Guide book for forage evaluation (In Japanese). Guidelines for project evaluation. Tokyo: Japan Grassland Agriculture and Forage Seed Association; 2001, p. 93–96.
Tian J, Huang LY, Tian R, Wu JY, Tang RX. Fermentation quality and bacterial community of delayed filling stylo silage in response to inoculating lactic acid bacteria strains and inoculating time. Chem Biol Technol Agric. 2023;10(1):44. https://doi.org/10.1186/s40538-023-00423-6.
Liu BY, Huan HL, Gu HR, Xu NX, Shen Q, Ding CL. Dynamics of a microbial community during ensiling and upon aerobic exposure in lactic acid bacteria inoculation-treated and untreated barley silages. Bioresour Technol. 2019;273:212–9. https://doi.org/10.1016/j.biortech.2018.10.041.
Mcdonald P, Henderson AR, Heron S. The biochemistry of silage. 2nd ed. Chalcombe publications: Marlow; 1991.
Oliveira AS, Weinberg ZG, Ogunade IM, Cervantes AAP, Arriola KG, Jiang Y, et al. Meta-analysis of effects of inoculation with homofermentative and facultative heterofermentative lactic acid bacteria on silage fermentation, aerobic stability, and the performance of dairy cows. J Dairy Sci. 2017;100(6):4587–603. https://doi.org/10.3168/jds.2016-11815.
Adesogan AT, Salawu MB. The effect of different additives on the fermentation quality, aerobic stability and in vitro digestibility of pea/wheat bi-crop silages containing contrasting pea to wheat ratios. Grass Forage Sci. 2002;57(1):25–32. https://doi.org/10.1046/j.1365-2494.2002.00298.x.
Chen L, Yuan XJ, Li JF, Dong ZH, Wang SR, Guo G, et al. Effects of applying lactic acid bacteria and propionic acid on fermentation quality, aerobic stability and in vitro gas production of forage-based total mixed ration silage in Tibet. Anim Prod Sci. 2019;59(2):376–83. https://doi.org/10.1071/AN16062.
Daiane CDS, Josemar GDOF, Jhessika DSS, Milena FDS, Marcio DSV, Marco APDS, et al. Okara flour: its physicochemical, microscopical and functional properties. Food Sci Nutr. 2019;49(6):1252–64. https://doi.org/10.1108/NFS-11-2018-0317.
Li S, Zhu D, Li K, Yang Y, Lei Z, Zhang Z. Soybean curd residue: composition, utilization, and related limiting factors. Isrn Ind Eng. 2013;2013:1–8. https://doi.org/10.1155/2013/423590.
Chung WY, Kim SK, Son JY. Isoflavones contents and physiological activities of soybeans fermented with Aspergillus oryzae or Bacillus natto. J Korean Soc Food Sci Nutr. 2008;37(2):141–7. https://doi.org/10.3746/jkfn.2008.37.2.141.
Baiano A, Terracone C, Gambacorta G, La Notte E. Evaluation of isoflavone content and antioxidant activity of soy-wheat pasta. Int J Food Sci. 2009;44(7):1304–13. https://doi.org/10.1111/j.1365-2621.2009.01959.x.
Ferraretto LF, Taysom K, Taysom DM, Shaver RD, Hoffman PC. Relationships between dry matter content, ensiling, ammonia-nitrogen, and ruminal in vitro starch digestibility in high-moisture corn samples. J Dairy Sci. 2014;97(5):3221–7. https://doi.org/10.3168/jds.2013-7680.
McGarvey JA, Franco RB, Palumbo JD, Hnasko R, Stanker L, Mitloehner FM. Bacterial population dynamics during the ensiling of Medicago sativa (alfalfa) and subsequent exposure to air. J Appl Microbiol. 2013;114(6):1661–70. https://doi.org/10.1111/jam.12179.
Wang C, He LW, Xing YQ, Zhou W, Yang FY, Chen XY, et al. Fermentation quality and microbial community of alfalfa and stylo silage mixed with Moringa oleifera leaves. Bioresour Technol. 2019;284:240–7. https://doi.org/10.1016/j.biortech.2019.03.129.
Pang HL, Qin GY, Tan ZF, Li ZW, Wang YP, Cai YM. Natural populations of lactic acid bacteria associated with silage fermentation as determined by phenotype, 16S ribosomal RNA and recA gene analysis. Syst Appl Microbiol. 2011;34(3):235–41. https://doi.org/10.1016/j.syapm.2010.10.003.
Yuan XJ, Li JF, Dong ZH, Shao T. The reconstitution mechanism of napier grass microbiota during the ensiling of alfalfa and their contributions to fermentation quality of silage. Bioresour Technol. 2020;297:122391. https://doi.org/10.1016/j.biortech.2019.122391.
Adesogan AT, Salawu MB, Ross AB, Davies DR, Brooks AE. Effect of Lactobacillus buchneri, Lactobacillus fermentum, Leuconostoc mesenteroides inoculants, or a chemical additive on the fermentation, aerobic stability, and nutritive value of crimped wheat grains. J Dairy Sci. 2003;86(5):1789–96. https://doi.org/10.3168/jds.S0022-0302(03)73764-3.
Santos AO, Ávila CLS, Pinto JC. Fermentative profile and bacterial diversity of corn silages inoculated with new tropical lactic acid bacteria. J Appl Microbiol. 2015;120:266–79. https://doi.org/10.1111/jam.12980.
Pahlow G, Muck RE, Driehuis F, Elferink SJWH, Spoelstra SF. Microbiology of ensiling. In: Buxton DR, Muck RE, Harrison JH, editors. Silage Science and Technology. Madison: American Society of Agronomy, Inc., Crop Science Society of America. Silage Science and Technology; 2003. p. 31–93.
Shen L, Liu YQ, Gu ZQ, Xu BQ, Wang NL, Jiao NZ, et al. Massilia eurypsychrophila sp nov a facultatively psychrophilic bacteria isolated from ice core. Int J Syst Evol Microbiol. 2015;65:2124–9. https://doi.org/10.1099/ijs.0.000229.
Lepak AJ, Andes DR. In vivo Pharmacodynamic target assessment of delafloxacin against Staphylococcus aureus, Streptococcus pneumoniae, and Klebsiella pneumoniae in a Murine Lung Infection Model. Antimicrob Agents Chemother. 2016;60(8):4764–9. https://doi.org/10.1128/AAC.00647-16.
Wilkinson JM. Silage and health. In: Silage production in relation to animal performance, animal health, meat and milk quality. Proceedings of the 12th international silage conference, July 5–7, ed T. Natural toxins. Sweden: Swedish University of Agricultural Sciences; 1999. p. 67–8.
Russell JB, Diez-Gonzalez F, Jarvis GN. Potential effect of cattle diets on the transmission of pathogenic Escherichia coli to humans. Microbes Infect. 2000;2(1):45–53. https://doi.org/10.1016/S1286-4579(00)00286-0.
Heron SJE, Wilkinson JF, Duffus CM. Enterobacteria associated with grass silage. J Appl Bacteriol. 1993;75:13–7. https://doi.org/10.1111/j.1365-2672.1993.tb03401.x.
Otto M. Staphylococcus epidermidis—the “accidental” pathogen. Nat Rev Microbiol. 2009;7(8):555–67. https://doi.org/10.1038/nrmicro2182.
Queiroz OCM, Ogunade IM, Weinberg Z, Adesogan AT. Silage review: foodborne pathogens in silage and their mitigation by silage additives. J Dairy Sci. 2018;101(5):4132–42. https://doi.org/10.3168/jds.2017-13901.
Contreras-Govea FE, Muck RE, Broderick GA, Weimer PJ. Lactobacillus plantarum effects on silage fermentation and in vitro microbial yield. Anim Feed Sci Technol. 2013;179(1–4):61–8. https://doi.org/10.1016/j.anifeedsci.2012.11.008.
Filya I, Muck RE, Contreras-Govea FE. Inoculant effects on alfalfa silage: fermentation products and nutritive value. J Dairy Sci. 2007;90(11):5108–14. https://doi.org/10.3168/jds.2006-877.
Allen B, Kon M, Bar-Yam Y. A new phylogenetic diversity measure generalizing the shannon index and its application to phyllostomid bats. Am Nat. 2009;174(2):236–43. https://doi.org/10.1086/600101.
Wang Y, He LW, Xing YQ, Zhou W, Pian RQ, Yang FY, et al. Bacterial diversity and fermentation quality of Moringa oleifera leaves silage prepared with lactic acid bacteria inoculants and stored at different temperatures. Bioresour Technol. 2019;284:349–58. https://doi.org/10.1016/j.biortech.2019.03.139.
Acknowledgements
Not applicable.
Funding
This work was supported by the National Key R&D Program of China (2022YFE0111000) and the National Natural Science Foundation of China (31971764).
Author information
Authors and Affiliations
Contributions
XY: investigation, data curation, writing—original draft. YF: writing—original draft. RT: formal analysis, writing—review and editing. RXT: validation. JT: software, validation. JZ: methodology, conceptualization, project administration, writing—review and editing. All authors read and approved the final manuscript. All authors contributed to the article and approved the submitted version.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors listed have read the complete manuscript and have approved submission of the paper.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Yin, X., Fan, Y., Tian, R. et al. Improving the quality and reducing harmful microbes of total mixed ration silage with dried soybean curd residue. Chem. Biol. Technol. Agric. 10, 86 (2023). https://doi.org/10.1186/s40538-023-00461-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40538-023-00461-0