Abstract
Background
Sago palm (Metroxylon sagu) is a versatile crop and has been hailed as the next viable commodity in Sarawak, Malaysia. Sago palm can thrive in the harsh swampy peat, from low flooded areas to uplands and in acidic to neutral soils. Sago palm has been neglected and unfortunately, very little is known about the sago palm proteome. This study aimed to determine the best protein extraction method of Metroxylon sagu for the two-dimensional gel electrophoresis (2-DE) and its comparative analysis.
Results
To perform good proteome research, the most critical step is to establish a method that gives the best quality of extracted total proteins. Five different protein extraction protocols: polyethylene glycol (PEG) fractionation method, phenol extraction method, TCA–acetone method, the combination of phenol and TCA–acetone extraction method and imidazole method were compared to develop an optimized protein extraction method for two-dimensional gel electrophoresis analysis of Metroxylon sagu. The PEG fractionation method was found to give the most reproducible gels with the highest number of spots and highest protein concentration followed by phenol extraction method. The lowest number of spots was observed in the imidazole method. The PEG fractionation method provides improved resolution and reproducibility of 2-DE and reduces the time required to analyze samples. Partitioning Rubisco by polyethylene glycol (PEG) fractionation provides clearer detection of low abundance protein. Hence, the results from this study propose PEG fractionation as the effective protein extraction method for 2-DE proteomic studies of Metroxylon sagu.
Conclusion
In this study, the PEG fractionation method is considered as the best extraction method for 2-DE proteomic studies of Metroxylon sagu in terms of yield, gel quality, spot numbers, and quantities of proteins.

Similar content being viewed by others
Background
The sago palm (Metroxylon sagu Rottb.) is a versatile plant that can tolerate many biotic and abiotic stresses, during its growth stages. It belongs to family Arecaceae, and among fourteen starch producing genera, the genus Metroxylon is the most productive among all of them. Sago palm is found growing in zone 10 of the equator at the Malay Peninsula from Southern Thailand to east and west of Malaysia, Brunei, Indonesia and its surroundings, including Papua New Guinea and some southern region of the Philippines [10]. Papua New Guinea or the Moluccas is believed to be the centre of diversity of sago palm [22].
Sago palm is a starch producing plant and can accumulate high carbohydrate content in the trunk. It is socio-economically important for sustainable agriculture and considered as one approach for rural development in various areas in Sarawak, Malaysia. The plant is highly adaptable to environmental factors than any other crop and can be grown in low land swampy areas with high acidic content [10]. Although this species is economically important for the country, there is a lack of scientific study related to this plant, including proteomics study. Plant proteomics has become an important field in molecular biology to investigate the evolution and growth of a plant. Very early plant proteomics was confined to the relationship between plant population only [5, 28, 29]. In recent years, proteomics is used to study the distances between different species of Brassicaceae family [19], oriental and American ginseng [18]. Proteomics is the most relevant technology to further investigation of highly complex and dynamic biological systems as it offers an accurate analysis of cellular state or system changes during growth, development, and response to environmental factors [7]. Although sago palm has been neglected and was relatively given less attention compared to other crops, scientists have marked the crops as crop par excellence for sustainable agriculture [26]. Researchers work actively on the secondary metabolites of the sago palm, but there is a limited study on the proteomic part.
Extraction of proteins is the most crucial and essential step in the proteomic study because plants contain phenolic compounds, polysaccharides, and other secondary metabolites which could interfere in the protein separation and quantitation. Cellular lysis of tough plant cell walls is complicated as they retain complex assemblances of polysaccharides [12] which interfere with downstream analysis.
Selecting an efficient extraction method for a specific sample is very important to obtain high quality and quantity proteins for 2-DE to check the differential expression of proteins as well as descriptive and comparative proteomic analysis in plants. The reproducibility and high resolution of 2-DE for separating a complex mixture of proteins make it the most suitable method. However, the quality and the separation of proteins resulting from 2-DE analysis depend on the sample preparation and protein extraction method, because this subsequently affects the quality of isoelectric focusing (IEF) during the first-dimension separation [23]. Commonly used phenol extraction and TCA–acetone methods remain popular despite the availability of some new techniques [1]. Each method has some disadvantages for leaf protein analysis lower protein solubility, co-extraction of nucleic acids, protein hydrolysis by TCA [8, 14]. The first soybean leaf proteome map was constructed using the TCA–acetone extraction method [34]. The detection of the low abundance proteins was restricted because of the prevalence of ribulose-1,5-bisphosphate carboxylase oxygenase (Rubisco). Quantitative evaluation of kiwi fruit ripening in response to exogenous ethylene has been studied using the phenol extraction method [25]. Various soybean tissues, including from leaf has been extracted using the phenol extraction method [3]. This protocol, however, requires long focusing time and many volt-hours (75,000 Vh). In addition, many low abundance proteins in the leaves of the soybean were masked by the presence of Rubisco. None of these methods can detect the low abundance proteins in leaf tissue, because of the presence of Rubisco, which masks 50% of the soluble proteins [2]. Since no extraction method is universal for all kinds of samples which can capture the entire proteome, several protein extraction methods were used in this study to extract the protein from the leaf sample of Metroxylon sagu.
Materials and methods
Protein extraction methods
Five different protein extraction methods were compared, i.e., TCA–acetone method, phenol extraction method, combined phenol and TCA–acetone method, imidazole method, and PEG fractionation method to determine the most suitable method for extraction of protein from Metroxylon sagu for 2-DE analysis. Sago leaf samples used in this study were grounded finely in prechilled motor pestle in the presence of liquid nitrogen for all the five methods.
TCA–acetone method
Total proteins were extracted using the TCA–acetone method [12] with some modifications. A total of 1 g finely ground leaf sample was suspended in 10 ml of 20% (w/v) TCA–acetone with 0.2% DTT and precipitated overnight at − 20 °C. Pellet was obtained by centrifugation the mixture at 10,000×g for 20 min at 4 °C. Collected pellet was rinsed with 10 ml of 80% cold acetone (v/v),with 0.2% DDT and incubated for 60 min at − 20 °C. The pellet was collected by spinning the mixture at 10,000×g for 20 min at 4 °C, and washed twice with 80% ice-cold acetone, and air-dried and kept at − 20 °C for further use.
Phenol extraction method
Proteins were extracted according to Wang et al. [33] with some modifications. A total of 1 g of finely grounded leaf was suspended in 0.8 ml phenol (Tris-saturated pH 8.0) and 0.8 ml of SDS buffer consisting of 0.1 M Tris–HCl (pH 8.0), 2% SDS, 30% sucrose, 5% (v/v) β-mercaptoethanol, and 1 mM phenylmethylsulfonyl (PMSF). The mixture was vortexed 5 min and centrifuged at 10,000×g for 20 min at 4 °C. The aqueous phase was collected and re-extracted with an equal volume of SDS buffer by centrifugation at 10,000×g for 10 min at 4 °C. The new aqueous layer containing proteins was precipitated with five volumes of 0.1 M ammonium acetate in methanol at − 20 °C overnight. Precipitated proteins were recovered by centrifugation at 10,000×g for 15 min at 4 °C. The recovered proteins were washed and rinsed with 100% methanol containing 0.1% ammonium acetate and 80% acetone, respectively, air-dried, and stored at − 20 °C for further use.
Combined phenol and TCA–acetone method
Proteins were extracted by combining TCA–acetone and phenol extraction method with ammonium acetate in methanol precipitation [33]. One gram of finely ground leaf sample was incubated in 10 ml of 20% (w/v) TCA–acetone for 1–2 h at − 20 °C. The pellet was obtained by centrifugation at 10,000×g for 20 min at 4 °C, and the collected pellet was rinsed with 10 ml of 80% cold acetone(v/v). SDS buffer and tris-saturated phenol (pH 8.0) in the ratio of 1:1 was added to it. The mixture was vortexed for 5 min, followed by centrifugation at 10,000×g for 15 min at 4 °C. The clear phenolic phase collected was mixed with four volumes of methanol containing 0.1 M ammonium acetate and incubated at − 20 °C overnight. Precipitated proteins were recovered by centrifugation at 10,000×g for 20 min at 4 °C, rinsed twice with 80% cold acetone, dried, and stored at − 20 °C.
Imidazole method
This method is based on Nakamura et al. [21] with some modifications. One gram of ground leaf sample was incubated in 8 ml of extraction buffer consisting of 50 mM imidazole–HCl (pH 7.4), 8 mM MgCl2, 12% glycerol, 2.5% (v/v) β-mercaptoethanol, and 1 mM PMSF. The sample was vortexed for 5 min, followed by centrifugation at 9000×g for 15 min at 4 °C. The supernatant was collected using Mira cloth, and proteins were precipitated using an equal volume of 20% TCA–acetone 1 h at − 20 °C. Precipitated proteins were recovered by centrifuging the mixture at 9000×g for 15 min at 4 °C, washed thrice with 100% acetone, air-dried, and stored until further use.
PEG fractionation method
This method is based on Alam et al. [2] with some modifications. One gram of finely grounded leaf sample was suspended in 10 ml of Mg/Triton-X extraction buffer consisting of 0.5 M Tris–HCl (pH 8.3), 2% Triton-X, 20 mM MgCl2, 2% β-mercaptoethanol, and 1 mM PMSF. After vortexing for 2 min, the sample was centrifuged at 10,000×g for 15 min at 4 °C. The supernatant was collected, and the proteins were subjected to PEG fractionation, by adjusting the final concentration of 15% (w/v) of the sample by adding 50% stock solution (w/v) of polyethylene glycol (PEG), then was incubated on ice for 30 min. The supernatant collected after centrifugation was precipitated with four volumes of cold 100% acetone at − 20 °C for 3 h, followed by centrifugation at 10,000×g for 15 min at 4 °C. The recovered pellet was dissolved in 10 ml of Mg/Triton-X extraction buffer. The pellet was vortexed for 2 min and mixed with an equal volume of Tris-saturated phenol (pH 8.0). Sucrose (0.7 M) was added, mixed well, and the mixture was centrifuged at 10,000×g for 15 min at 4 °C. The upper phase was collected carefully, and proteins were precipitated by adding four volumes of 100% methanol containing 0.1 M ammonium acetate at − 20 °C overnight. The pellet was recovered by centrifugation the mixture at 10,000×g 15 min at 4 °C followed by washing thrice with 100% methanol containing 0.1 M ammonium acetate, and stored in 80% acetone at − 20 °C until use.
Protein quantification using Bradford assay
Prior to further analyses, the dried protein pellets were solubilized for 1 h in 150 µl of protein lysis buffer. The Bradford assay was carried out to determine the concentration of solubilized protein using Bradford reagent [4]. The standard protein graph of bovine serum albumin (BSA) was prepared in seven dilutions of 100 ug/ml of BSA. Samples with unknown protein concentration were prepared with different dilution factors. Absorbance was measured at 595 nm after 5 min of adding 1 ml of Bradford reagent. The total protein concentration was determined in triplicates [15].
SDS-PAGE
The quality of the proteins obtained from different protocols was evaluated according to the molecular weight using one-dimensional SDS polyacrylamide gel electrophoresis (SDS-PAGE) [16]. A 20 µl of protein sample (20 mg/mL) was mixed with 5 µl of 5 × sample loading dye (1:4 ratio), and proteins were denatured by heating at 95 °C for 5 min. The denatured proteins were centrifuged for 5 min at 16,000×g and run through 4% stacking gel followed by 12% resolving gel at 120 V for 1 h. The gels were stained with Coomassie Brilliant Blue (CBB) G-250.
2-Dimensional polyacrylamide gel electrophoresis
The extracted proteins were separated according to their isoelectric point and molecular weight by subjected to 2-DE. Dry immobilized pH gradient (IPG) strips (7 cm strip, pH 3–9 nonlinear) (Bio-Rad) were rehydrated with 125 µl of protein solution containing 250 ug/ml of proteins in an IPG re-swelling tray with 2 ml of mineral oil for 14 h. Isoelectric focusing (IEF) was performed using the PROTEAN i12 IEF system according to the following parameters: 250 V for 20 min, 4000 V for 2 h, 4000 V for 10,000V Hours. Prior to electrophoresis in the second dimension, the strips were incubated twice for 10 min each with gentle shaking in equilibration buffer (0.5 M Tris–HCl, pH 6.8; 6 M Urea; 30% glycerol, 2% SDS), first with 2% DTT, then with 2.5% iodoacetamide. The second-dimension separation of proteins was performed on 12% SDS polyacrylamide gels according to the method of Laemmli [16]. The strips were sealed with 0.5% overlay agarose with bromophenol blue before running on 100 V for 2 h. Following SDS-PAGE, the gels were fixed using deionized water for 10 min and stained for 12–14 h with Coomassie Brilliant Blue (CBB) G-250 stain with gentle shaking. Images of the stained gels were captured using a Bio-Rad gel doc.
Results
Determination of protein concentration from the five extraction methods
Evaluation of protein yield using different extraction methods was done based on the amount of protein extracted from 1 g of frozen leaf sample. The higher protein concentration of 8.9 µg/µl and 4.6 µg/µl were obtained from PEG fractionation and phenol extraction method, respectively (Table 1), followed by combined phenol and TCA–acetone method 3.9 µg/µl and TCA–acetone method 2.5 µg/µl. Imidazole method has given the lowest protein yield of 1.6 µg/µl.
Evaluation of proteins from different extraction methods using SDS-PAGE
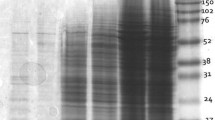
The extracted proteins were separated using one-dimensional SDS-PAGE and resolved between 10 and 245 kDa (Fig. 1). The highest number of bands was resolved using PEG as shown on lane M1 with the partitioning of the Rubisco, followed by phenol extraction methods where Rubisco is present in abundance (Fig. 1; lane M2). The combined phenol and TCA–acetone as well as the TCA–acetone methods resolved almost the same number of bands (Fig. 1; lane M4 and M5, respectively). However, most of the bands were common in all the four protocols except the imidazole method, which gives the lowest number of bands, as shown on lane M3.
Comparison of protein using 2-DE
The extracted proteins were subjected to 2-DE separation. Since the imidazole method resolves the lowest number of bands on the SDS-PAGE gel, this method was not subjected to 2-DE analysis. After staining of the gels, images of protein spots were shown in Fig. 2. The protein extracted using all protocols resulted in higher protein spots abundance on the acidic side. The PEG fractionation method, which showed the highest protein concentration and highest bands in SDS-PAGE, showed the highest number of spots at 750 (Fig. 2a), followed by phenol extraction and combined methods with 525 (Fig. 2b) and 231 spots, respectively (Fig. 2c). The lowest number of spots was shown by TCA–acetone method with 110 spots (Fig. 2d).
Discussion
The most crucial step to obtain high-quality protein is sample preparation and protein extraction. Plant tissues are rich in interfering compounds, such as polyphenols, organic compounds, nucleic acids, which are present in abundance in green tissues and can strongly hinder extraction and separation on 2-DE [20]. The purpose of this study was to compare five different protein extraction methods for high-resolution 2-DE of Metroxylon sagu. The results showed that PEG fractionation method gives the best result in comparison with the other extraction methods, in terms of reproducibility, yield, high protein definition in SDS-PAGE, and good compatibility to IEF (Fig. 1).
There are various studies about the TCA–acetone precipitation as the best extraction method over phenol precipitation for protein purification in Brassica sp. rice [24], and date palm [9]. In this study, the TCA–acetone extraction method resulted in the lowest concentration and fewer spots detected in 2-DE separation. The TCA–acetone method has a limited application to young tissues only and has lower protein solubility and co-extraction of nucleic acids [14]. On the other hand, there are various findings of the phenol extraction method gives interference-free high quality and quantity protein from diverse plants species potato [6], apple and banana [31], Cenchrus polystachion [27]. The phenol extraction method gives a better 2-DE map with more resolved spots in maize [32]. In a comparative analysis of four Aloe species, the phenol extraction method was shown to be suitable for 2-DE and MALDI-TOF–MS [11]. In our study, the phenol extraction method and combined phenol and TCA–acetone method give a high protein yield than the TCA–acetone method alone. The phenol extraction method resulted in higher concentration and protein resolution than the combination method. However, this extraction method is not suitable for detecting low abundance proteins in the leaf sample due to the presence of Rubisco, which at approximately 50% of the soluble proteins, mask them. PEG fractionation method gives best result in terms of reproducibility, yield, high protein definition with the partitioning of Rubisco in SDS-PAGE and good compatibility to IEF (Fig. 1). Better results in comparison with other methods both on resolution and number of the spots with no vertical and horizontal streaking were shown also at 2-DE analysis (Fig. 2). The partitioning of the Rubisco by PEG method resolved many abundance proteins from leaves of rice [13, 17] and sunflower [30]. PEG without interfering with the immunogenic properties of proteins can mask the surface of the protein by covalent bonding, and is nontoxic alcohol [2]. Thus, the PEG fractionation method also may provide a more detailed proteome of Metroxylon sagu, where Rubisco is prevalent. In the PEG fractionation method, the interfering substances present in many low abundance proteins may cause poor electrophoretic separation on the IPG strip during IEF, as seen in sunflower leaf proteome [30]. To overcome this problem, the TCA–acetone precipitate was re-extracted with phenol, which efficiently removes interfering substances, which results in optimal electric conductivity, less time during IEF. The prefractionation of protein samples using PEG before 2-DE can assist proteomic studies in general, because of the detection of low abundance proteins. The combination of PEG fractionation and phenol extraction methods were successfully applied for extracting proteins from leaf tissues of soybean, Miscanthus, Chinese cabbage, peanut, and tea [2]. This method can be applied to the leaf tissues of varieties of species; those contain high levels of secondary metabolites and high starch content.
Conclusion
In the present study, five distinctive protein extraction protocols, i.e. TCA–acetone method, phenol extraction method, the combination of phenol and TCA–acetone method, imidazole method, and PEG fractionation method were evaluated and compared for the whole proteome analysis of Metroxylon sagu. Overall, the best results in terms of yield, gel quality, spot numbers, and quantities were obtained using the PEG fractionation method. The prefractionation of protein samples using PEG before 2-DE can assist proteomic studies in general because low abundance proteins can be detected using the PEG fractionation method. The use of phenol in this method efficiently removes interfering substances, which results in optimal electric conductivity, less time during IEF. Smooth IEF eliminates the risk of damaging IPG strips and samples. The partition of Rubisco to the pellet permits the detection of a low-abundance regulatory protein. PEG fractionation method can be applied to the leaf tissues of varieties of species; those contain high levels of secondary metabolites and high starch content.
Availability of data and materials
The dataset supporting the conclusions of this study and the information about the materials used is included within the article.
Abbreviations
- 2-DE:
-
Two-dimensional gel electrophoresis
- BSA:
-
Bovine serum albumin
- IEF:
-
Isoelectric focusing
- IPG:
-
Immobilized pH gradient
- PAGE:
-
Polyacrylamide gel electrophoresis
- PMSF:
-
Phenylmethylsulfonyl fluoride
- SDS:
-
Sodium dodecyl sulfate
- PEG:
-
Polyethylene glycol
- TCA:
-
Trichloroacetic acid
References
Agrawal GK, Rakwal R. Rice proteomics: a move toward expanded proteome coverage to comparative and functional proteomics uncovers the mysteries of rice and plant biology. Proteomics. 2011;11:1630–49.
Alam I, Sharmin SA, Kim KH, Kim YG, Lee JJ, Lee BH. An improved plant leaf protein extraction method for high resolution two-dimensional polyacrylamide gel electrophoresis and comparative proteomics. Biotech Histochem. 2013;88:61–75.
Annamraju D, Oehrle N, Emerich D. Plant protein isolation and stabilization for enhanced resolution of two-dimensional polyacrylamide gel electrophoresis. Anal Biochem. 2008;379:192–5.
Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54.
Burstin J, de Vienne D, Dubreuil P, Damerval C. Molecular markers and protein quantities as genetic descriptors in maize. I. Genetic diversity among 21 inbred lines. Theor Appl Genet. 1994;89:943–50.
Carpentier SC, Witters E, Laukens K, Deckers P, Swennen R, Panis B. Preparation of protein extracts from recalcitrant plant tissues: An evaluation of different methods for two-dimensional gel electrophoresis analysis. Proteomics. 2005;5:2497–507.
Chen S, Harmon AC. Advances in plant proteomics. Proteomics. 2006;6:5504–16.
Chen T, Qi J-M, Xu J-T, Chen P-P, Tao A-F, Chen F-C, Chen W. Optimization of two-dimensional gel electrophoresis for kenaf leaf proteins. Agric Sci China. 2011;10:1842–50.
Devouge V, Rogniaux H, Nési N, Tessier D, Guéguen J, Larré C. Differential proteomic analysis of four near-isogenic Brassica napus varieties bred for their erucic acid and glucosinolate contents. J Proteom Res. 2007;6:1342–53.
Ehara H, Toyoda Y, Johnson Dennis V. Sago palm: multiple contributions to food security and sustainable livelihoods. Cham: Springer; 2018.
Fan JJ, Li CH, Hu G, Tan CN, Yang FQ, Chen H, Xia ZN. Comparative analysis of soluble proteins in four medicinal aloe species by two-dimensional electrophoresis and MALDI-TOF-MS. J AOAC Int. 2019;102:748–60.
Isaacson T, Damasceno CMB, Saravanan RS, He Y, Catalá C, Saladié M, Rose JKC. Sample extraction techniques for enhanced proteomic analysis of plant tissues. Nat Protoc. 2006;1:769–74.
Kim ST, Cho KS, Jang YS, Kang KY. Two-dimensional electrophoretic analysis of rice proteins by polyethylene glycol fractionation for protein arrays. Electrophoresis. 2001;22:2103–9.
Komatsu S, Ahsan N. Soybean proteomics and its application to functional analysis. J Proteomics. 2009;72:325–36.
Kruger NJ. The Bradford method for protein quantitation. Methods Mol Biol. 1994;32:9–15.
Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680.
Lee D-G, Ahsan N, Lee S-H, Kang KY, Bahk JD, Lee I-J, Lee B-H. A proteomic approach in analyzing heat-responsive proteins in rice leaves. Proteomics. 2007;7:3369–83.
Lum JH-K, Fung K-L, Cheung P-Y, Wong M-S, Lee C-H, Kwok FS-L, Leung MC-P, Hui P-K, Lo SC-L. Proteome of Oriental ginseng Panax ginseng C. A. Meyer and the potential to use it as an identification tool. Proteomics. 2002;2:1123–30.
Marques K, Sarazin B, Chane-Favre L, Zivy M, Thiellemen H. Comparative proteomics to establish genetic relationships in the Brassicaceae family. Proteomics. 2001;1:1457–62.
Matsui T-A, Sekiguchi M, Hashimoto A, Tomita U, Nishikawa T, Wada K. Functional comparison of d-serine and glycine in rodents: the effect on cloned NMDA receptors and the extracellular concentration. J Neurochem. 1995;65:454–8.
Nakamura K, Greenwood A, Binder L, Bigio EH, Denial S, Nicholson L, Zhou XZ, Lu KP. Proline isomer-specific antibodies reveal the early pathogenic tau conformation in Alzheimer's disease. Cell. 2012;149:232–44.
Rauwerdink JB. An essay on Metroxylon, the Sago Palm. Princ J Inte Palm Soc. 1986;30:165–80.
Shaw MM, Riederer BM. Sample preparation for two-dimensional gel electrophoresis. Proteomics. 2003;3:1408–17.
Shen S, Jing Y, Kuang T. Proteomics approach to identify wound-response related proteins from rice leaf sheath. Proteomics. 2003;3:527–35.
Shin M, Muneer S, Kim Y-H, Lee J-J, Bae D-W, Kwack Y-B, Kumarihami PC, H., Kim, J.-G. Proteomic analysis reveals dynamic regulation of fruit ripening in response to exogenous ethylene in kiwifruit cultivars. Environment and Biotechnology: Horticulture; 2020.
Singhal RS, Kennedy JF, Gopalakrishnan SM, Kaczmarek A, Knill CJ, Akmar PF. Industrial production, processing, and utilization of sago palm-derived products. Carbohyd Polym. 2008;72:1–20.
Somayajula D, Desai N. Optimization of protein extraction and proteomic studies in Cenchrus polystachion (L.) Schult. Heliyon. 2019;5:e02968.
Thiellement H, Bahrman N, Damerval C, Plomion C, Rossignol M, Santoni V, de Vienne D, Zivy M. Proteomics for genetic and physiological studies in plants. Electrophoresis. 1999;20:2013–26.
Vienne DD, Burstin J, Gerber S, Leonardi A, Guilloux ML, Murigneux A, Beckert M, Bahrman N, Damerval C, Zivy M. Two-dimensional electrophoresis of proteins as a source of monogenic and codominant markers for population genetics and mapping the expressed genome. Heredity. 1996;76:166–77.
Walliwalagedara C, van Keulen H, Cutright T, Wei R. Comparison of sample preparation methods for the resolution of metal-regulated proteins in helianthus annuus by 2-dimensional gel-electrophoresis. Open Proteomics Journal. 2010;3:20–5.
Wang EA, Hong CS, Shavat S, Kessell ER, Sanders R, Kushel, M.B. Wang et al. Respond. Am J Public Health. 2013;103:e6–e7.
Wang N, Wu X, Ku L, Chen Y, Wang W. Evaluation of three protein-extraction methods for proteome analysis of maize leaf midrib, a compound tissue rich in Sclerenchyma cells. Front Plant Sci. 2016;7:856–856.
Wang W, Vignani R, Scali M, Cresti M. A universal and rapid protocol for protein extraction from recalcitrant plant tissues for proteomic analysis. Electrophoresis. 2006;27:2782–6.
Xu C, Garrett WM, Sullivan J, Caperna TJ, Natarajan S. Separation and identification of soybean leaf proteins by two-dimensional gel electrophoresis and mass spectrometry. Phytochemistry. 2006;67:2431–40.
Acknowledgements
The authors express gratitude to Norzainizul Julaihi, Rina Tommy, and Datu Sajeli Kipli of Land Custody and Development Authority (PELITA) Sarawak for access to the sago palms used in this study. We are thankful to Universiti Malaysia Sarawak for the Zamalah scholarship awarded to Mehvish Nisar.
Funding
The authors acknowledged and express thanks to the Ministry of Education Malaysia FRGS Grant F07/FRGS/1611/2017 and UNIMAS Tun Openg Sago Chair Research Grant F07/TOC/1743/2018 for funding of this research.
Author information
Authors and Affiliations
Contributions
MN wrote the proposal, participated in sample and data collections, analyzed the data, and drafted the paper, HH approved the proposal, participated in sample collection, data collection and revised subsequent drafts of the paper. Both authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Nisar, M., Hussain, H. The protein extraction method of Metroxylon sagu leaf for high-resolution two-dimensional gel electrophoresis and comparative proteomics. Chem. Biol. Technol. Agric. 7, 14 (2020). https://doi.org/10.1186/s40538-020-00180-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40538-020-00180-w