Abstract
Background
Needle fungi in conifers have been extensively studied to explore their diversity, but environmental factors influencing the composition of fungal communities in Pinus radiata D.Don needles have received little attention. This study was conducted to examine the influence of the environment as defined by rainfall, elevation and temperature on the composition of fungal communities in pine needles at an age prior to that at which spring needle cast (SNC) is generally observed. Elucidating the entire fungal community in the needles is a first step towards understanding the cause of the disease.
Methods
Needle samples were collected from 5-year-old P. radiata trees, their age predating the onset of SNC, from 12 plantations in Tasmania. Interpolated data for the climate variables, including seasonal components for rainfall and temperature, were obtained from an enhanced climate data bank. Pooled needle samples were examined for the fungi they contained using DNA sequencing of cloned polymerase chain reaction (PCR) products. Clones were grouped into operational taxonomic units (OTUs) and identified to their lowest possible taxonomic level by comparison with reference isolates and public DNA databases.
Results
DNA sequencing revealed that needle fungal communities differed greatly, depending upon the total annual rainfall and needle age. Needle fungi that have been previously associated with pathogenic behaviour, such as Cyclaneusma minus, Dothistroma septosporum, Lophodermium pinastri, Strasseria geniculata and Sydowia polyspora, were all found in the needles in this study. Also, of 70 ascomycetous OTUs that were discriminated by their DNA sequences, 21 were identified as belonging to the Teratosphaeriaceae.
Conclusions
An overview of the endophytic fungi present in the needles of P. radiata has been obtained from trees whose age precedes the onset of SNC. All of the previously known pathogenic species for needle cast diseases were present, in addition to a range of unidentified OTUs belonging to the Teratosphaeriaceae.
Similar content being viewed by others
Background
As described in previous studies (Ganley & Newcombe 2006; Koukol et al. 2012; Terhonen et al. 2011), the Pinus phyllosphere (i.e. the environment for microorganisms on and below the needle surface) is a rich and varied microbial community. Its composition is strongly influenced by both genotypic and environmental factors (Botes et al. 1997; Van Maanen et al. 2000; Wang & Guo 2007). These factors can affect the complex interactions between microbial organisms, which in turn influence their interactions with their host plant (Giauque & Hawkes 2013). Abiotic or environmental factors influencing the composition of fungal communities associated with Pinus radiata D.Don needles have received little attention.
Studies have shown that endophyte communities in Pinus sylvestris L. needles can vary according to geographic location (Gourbière & Debouzie 2003) and elevation (Gourbière et al. 2001). Communities in Picea mariana (Mill.) Britton, Sterns & Poggenb. needles have been reported to vary with latitude (Sokolski et al. 2007). A study by Hata and Futai (1996) showed that the endophytic mycota associated with 45 pine species or varieties was similar for those pines belonging to the same taxonomic group. That study also concluded that endophytic colonisation might reflect the degree of adaptation of the host to local biotic and abiotic factors. However, considerable controversy exists about whether an endophyte, usually an organism such as a bacterium or a fungus, can become a pathogen. Some authors have suggested that endophytes may play a role in protecting trees from pathogens (Arnold et al. 2003; Bradshaw 2004; Gao et al. 2010), but others maintain that a commensal or mutualist fungal species can be triggered under certain environmental conditions to act as a virulent parasite (Krabel et al. 2013). The definition of an endophyte differs greatly, but in a comprehensive chapter on endophytic fungi, Stone et al. (2004) wrote that, in general, the term endophytic fungi applies to ‘fungi capable of symptomless occupation of apparently healthy plant tissue’. Broadly, this means that endophytes are viewed as colonising living plant tissues without causing them any immediately obvious negative effect. Nevertheless, others, such as Brown et al. (1998), who elucidated the endophytic fungal communities of the Musa acuminata Colla species (banana) complex and found 24 taxa of which almost all were ascomycetes, raised the question ‘Can endophytes of Musa spp. become pathogens?’ They noted the strong correlation between lists of pathogenic taxa of Musa spp. and frequently isolated tropical endophyte genera, but in the context of their study, they determined that it was not possible to conclude if banana pathogens isolated as species of endophytes in healthy leaves were the same pathogenic strains. As the fungi may be non-pathogenic strains, they concluded that experiments following Koch’s postulates were required to confirm any pathogenicity. Similarly, Larran et al. (2002) and Douanla-Meli et al. (2013) found that their lists of fungal species in the healthy leaves of soybean and citrus, respectively, were predominantly or almost exclusively made up of common plant pathogens, but each study concluded that further investigations were necessary to understand the precise role of these endophytes.
Spring needle cast (SNC) in Tasmania appears in pine plantations at canopy closure (age 6–7 years) and is manifested by the rapid browning of 1-year-old needles followed by the heavy casting of needles. Spring needle cast is not considered to be a classic needle blight disease caused by a primary fungal pathogen (Podger & Wardlaw 1990); instead, it is thought to be due to a suite of endophytic fungi that are triggered into secondary pathogenic activity by an environmental stress. In their study of 53 stands selected for climatic analysis, Podger and Wardlaw (1990) found that 23 of the sites were visually assessed as having SNC, and that these stands were restricted to a mean annual rainfall range of 1200–2000 mm and mean annual temperature range of 8–11.5 °C. Podger and Wardlaw (1990) putatively associated Cyclaneusma minus, Lophodermium pinastri and Strasseria geniculata with the disease in Tasmania, but there were many areas with severe SNC where Podger and Wardlaw (1990) found no fungal fruiting bodies of these species on attached needles, dead or alive. However, these three species of fungi were ubiquitous on fallen needles at sites both affected and unaffected by SNC. More recently, fungal communities were surveyed in a P. radiata family trial established in 1999 in Oonah, Tasmania. The trial was assessed at 8 years of age in 2007 using DNA extraction from needles, polymerase chain reaction (PCR) with fungal-specific primers and sequencing to detect the presence of fungal species (Prihatini et al. 2015a). Trees with contrasting levels of SNC disease severity were found to have significantly different needle fungal communities. The four fungi, L. pinastri, C. minus, S. geniculata and Dothistroma septosporum, previously reported as common pathogens or endophytes in P. radiata in Tasmania implicated in causing SNC (Podger 1984; Podger and Wardlaw 1990) were also identified in the Prihatini et al. (2015a) study. Of these, only L. pinastri was correlated with high levels of disease. Several species of Teratosphaeriaceae were also detected. Teratosphaeriaceae sp. 23 was detected in almost all of the needle samples and was presumed to be endophytic as it was associated with needles from healthy trees as well as those affected by SNC. In contrast, Teratosphaeriaceae sp. 3 was detected most frequently in needles from severely diseased trees and not at all in needles from trees with <5 % disease. An overview of the fungal species detected in these two studies (Prihatini et al. 2015b) highlighted the discrepancy between fungal associates of P. radiata detected by PCR and sequencing and those detected by isolation.
The present study was conducted in parallel with Prihatini et al. (2015a, b) to explore the types and diversity of needle fungi present in 5-year-old P. radiata in Tasmania, an age at which all trees have green, non-symptomatic needles, and to examine the influence of climate and landform as defined by rainfall, elevation and temperature on the composition of fungal communities. The motivation for this was to obtain a catalogue of fungal species from healthy tissues that could point towards the fungi that caused, or might be implicated in, SNC. This follows the approach of Petrini (1986) and Carroll (1988), who advocated systematic sampling of healthy plant tissue for internal fungi rather than the usual practice of the time of examining diseased or necrotic plant organs.
Methods
Sampling sites and environmental data
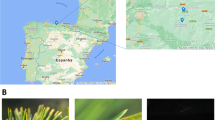
Twelve P. radiata plantations were chosen in Tasmania for the collection of needle samples from 5-year-old trees (Fig. 1). At each site, five 20-tree plots had been established for a previous study (unpublished), and needle samples were obtained from those plots. Interpolated estimates (data drill) of various aspects of rainfall and temperature data were downloaded from SILO (an enhanced climate data bank; Queensland Government 2014). These included average annual mean temperature, average annual minimum and maximum temperature, average winter minimum and maximum temperature, average annual rainfall and total monthly rainfall in winter and in summer. Rainfall was used both as a categorical predictor variable in a permutational analysis of variance and as a continuous variable in a permutational regression analysis (see the Statistical Analysis section below). For the analysis of variance, there were three replications (sampling sites), two classes for elevation (above and below 320 m) and three classes for mean annual rainfall: dry sites (<800 mm); intermediate sites (800–1100 mm) and ‘wet’ sites (>1100 mm) (Table 1).
Location of pine plantations in Tasmania from which materials were collected (latitude, longitude and other information of these sites are presented in Table 1)
Needle samples
Four composite needle samples were obtained from each site, corresponding to needles of four different age classes (1-year-old, 2-year-old, 3-year-old and fallen needles), as described in Prihatini et al (2015b), to reduce the number of cloning reactions without losing information about the effect of needle age on fungal communities. Needles were collected between December 2006 and April 2007. Five trees were randomly selected from each of the five 20-tree plots, and a single fascicle of each class was taken from the middle section of each tree. Fallen needles were collected from beneath each tree. Fascicles in the same class were pooled for each site, placed into paper bags and stored at 10 °C for 1–2 days before drying at 42 °C.
DNA extraction
Approximately 1 cm was cut from the middle part of each needle, placed into a 1.5-mL microcentrifuge tube and stored at −80 °C prior to grinding with a mortar and pestle Prihatini et al. (2015b). DNA extractions were performed using the procedure described in Glen et al. (2002).
Polymerase chain reaction and electrophoresis
To amplify the ITS region, PCR was carried out using primers ITS4A (Larena et al. 1999) and ITS5 (White et al. 1990) with the conditions as described in Prihatini et al. (2015b). Amplification products that were clearly visible on an agarose gel stained with ethidium bromide were cloned directly after purification with an UltraClean® PCR Clean-up Kit (MO BIO Laboratories, USA). Samples with insufficient product after the first round of PCR were diluted 1 in 5 and re-amplified by nested PCR using the primers ITS1 and ITS4 (White et al. 1990). Nested PCR products were purified and cloned as above. All 48 samples obtained by standard PCR or nested PCR were cloned.
Cloning of amplified DNA, screening and DNA sequencing
The PCR and nested PCR products were cloned using pGEM®-T Vector Systems (Promega) according to the technical manual provided. Clones were screened by PCR-restriction fragment length polymorphisms (PCR-RFLP), and three or four clones from each RFLP group from each needle sample were sequenced. Chromatograms were viewed in ChromasPro version 1.34 software and edited to remove poor quality sequences at each end; the sequence was then saved in FASTA format. Sequencing of PCR products was performed by Macrogen Inc. (Seoul, Korea).
Fungal DNA identification
A search of public DNA databases (BLAST, Altschul et al. 1990) retrieved sequences of high similarity. Sequences were grouped according to BLAST search results and aligned with sequences of high similarity using ClustalW (Larkin et al. 2007). If single nucleotide polymorphisms occurred between clones from the same operational taxonomic unit (OTU), chromatograms were rechecked to confirm these. Isolates or clones with less than 2 % sequence variation were grouped into OTUs. The OTUs were identified to the lowest possible taxonomic level based on sequence similarity to known fungi from public DNA databases and reference cultures (Prihatini et al. 2015b). Phylogenetic analysis helped to refine the discrimination and identification of OTUs. Several sequences were selected from the best matches retrieved by BLAST searches of public databases for each OTU and included in phylogenetic analysis for that OTU. One or two sequences from more distantly related taxa were also included as outgroups. All sequences for each OTU were aligned using the ClustalW program in BioEdit version 7.0.9.0 (Hall 1999) prior to phylogenetic analysis using DNAml from the PHYLIP package (Felsenstein 1989). Trees were drawn by TreeView software (Page 1996) and edited using MEGA4 software (Tamura et al. 2007).
Statistical analysis
All statistical analyses to study the community of needle fungi present were conducted using PRIMER v6 (Clarke & Gorley 2006), with its add-on PERMANOVA+ v1.0.2 (Anderson et al. 2008). The basic data matrix consisted of the presence or absence of 70 ascomycetous OTUs discriminated from the phylogenetic analysis on the 34 pooled samples that yielded clones. A Bray-Curtis resemblance (similarity) matrix constructed from all possible pairs of samples was the input for each of the routines used. Both permutational multivariate analysis of variance (PERMANOVA) and canonical analysis of principal coordinates (CAP) were used to test separately whether the factors needle age, elevation (low vs. high) and total annual rainfall (dry, intermediate, wet) produced significantly different fungal assemblages. The advantage of CAP is that it is specifically designed to find canonical axes that maximise differences amongst pre-defined groups in the data cloud, and it also tests individual samples for the correctness or otherwise of their group membership by cross-validation, employing a leave-one-out procedure. The third routine used, DistLM, modelled whether the climate variables elevation, rainfall and temperature could explain a significant proportion of the variation in the data cloud. Total annual and monthly rainfall data were available along with average annual and seasonal data for maximum, minimum and average temperatures. There were high correlations amongst them so principal component analysis (PCA) was first applied to the raw data, which was standardised to have unit variance. Subsequently, the DistLM modelling was conducted on the first three principal components, which accounted for almost all of the total variation, and the Bayesian selection criterion BIC was used as the ‘stopping rule’. All three routines of the PERMANOVA+ program that were used here employed 9999 permutations.
Results
Identification of fungi amplified from P. radiata needles
A total of 504 clones were sequenced but only 439 clones gave legible sequence results, allowing OTU assignment and identification. Overall, 407 ascomycetous sequences were identified. Sequences were grouped into OTUs with over 98 % sequence similarity. After phylogenetic analysis, 70 ascomycetous OTUs were discriminated (Table 2), of which 36 were detected from more than one sample. Seventeen species of hitherto unknown members of the Teratosphaeriaceae were identified as part of the P. radiata marker-aided selection trial (Prihatini et al. 2015a) run in parallel with the current study, and nine of these were also detected in the current study.
Fungal prevalence across sites
Clones for DNA identification were obtained for only 34 out of the 48 pooled samples. However, all 12 plantation sites were represented by at least one pooled sample that gave rise to clones. Each OTU observed in this study (Table 2) was scored as present or absent in each pooled sample. The ascomycetous OTUs present in at least three of the 34 pooled samples are listed in Table 3, except for S. geniculata and D. septosporum, which were less frequent but are also included in Table 3 because of their putative previous involvement as causal agents of needle cast disease. Forty OTUs were identified only from a single site, but seven of these were detected in more than one cloning sample at that site. The remaining 30 OTUs were present in more than one site, but only six OTUs occurred at six or more of the sites. Several of the commonly found OTUs were well-known pine associates belonging to the genera Lophodermium and Cyclaneusma, but their distribution varied amongst the 12 plantation sites, and none was found at all 12 sites. The two most dominant OTUs were Teratosphaeriaceae sp. 26, found in 15 pooled samples at seven sites, and L. pinastri, found in 14 pooled samples and at all but one of the 12 sites. C. minus ‘simile’ and C. minus ‘verum’ were each identified at half of the sites.
Fungal communities
Effect of climate
Permutational multivariate analysis of variance (PERMANOVA) produced a significant separation (P = 0.0217) of the fungal communities present at the three levels of annual rainfall (<800 mm, dry; 800–1200 mm, intermediate; >1200 mm, wet). Use of CAP provided further evidence of the importance of rainfall, strongly separating the three groups (P = 0.0048; see Fig. 2). Neither PERMANOVA nor CAP analyses of fungal communities provided evidence for an association between communities and elevation (P = 0.182 and 0.217, respectively). Similarly, using DistLM, no evidence was found of a correlation between fungal communities and temperature (P = 0.167 using a composite score from PCA), whereas there was evidence of an association with rainfall (P = 0.040 using a composite score from PCA), thereby supporting the results from PERMANOVA and CAP that rainfall is a significant environmental variable influencing fungal community assemblages.
Effect of needle age
Significant differences in fungal assemblages as a function of needle age were also obtained using permutational multivariate analysis of variance. From PERMANOVA, pairwise differences were indicated between 1-year-old needles and fallen needles (P = 0.0128) and between 2-year-old needles and fallen needles (P = 0.0022), with marginal evidence of an assemblage difference between 3-year-old needles and fallen needles (P = 0.0584). Using CAP was an alternative method of testing whether or not the multivariate means are different for the four different age classes. Results were unequivocal in indicating a significant difference between age classes (P = 0.0181 (trace test) or 0.006 (test based on the first squared canonical correlation)), with eight out of the 11 fallen needle samples being correctly classified, i.e. placed in the correct needle class by the cross-validation procedure of CAP. However, no 1-year-old needle sample was correctly classified; only two of the eight 2-year-old needle samples and only two of the eight 3-year-old needle samples were correctly classified. This indicates that fallen needles have a different fungal assemblage to those of the three age classes, with their OTUs being more distinct than those in the needles retained by the trees. The widespread species L. pinastri was present in 14 of the cloning samples, nine of which were in fallen needles. The sites at which L. pinastri was not present in the fallen needle sample were Tower Hill, Styx and Branches Creek, the three driest sites.
Discussion
The main purpose of the study was to produce an inventory of the fungi present in the needles of healthy trees at an age that precedes the onset of SNC disease. This was achieved with the identification of 70 distinct OTUs. A number of these species were also found as part of the P. radiata marker-aided selection trial (Prihatini et al. 2015a) run in parallel with the current study, including some members of the Teratosphaeriaceae. However, there is some doubt about the taxonomic placement of some of the OTUs listed in Table 2. For example, Mycosphaerella was originally thought to be monophyletic but has more recently been shown to be polyphyletic (Crous et al. 2007). Also, genera such as Davidiella and Teratosphaeria were split off from the genus and have formed the basis of new families. Despite this, the separation between Teratosphaeria and Mycosphaerella is not always clear, and many phylogenetically distinct taxa still remain in ‘Mycosphaerella’, ‘Teratosphaeria’ or in one of the associated asexual genera, with their correct generic disposition requiring clarification of their taxonomy and DNA phylogeny (Crous et al. 2009). Obtaining living cultures of previously described species, coupled with advances in molecular techniques, is assisting in the resolution of these generic complexes. With this in mind, some of the 21 OTUs of the present study listed in Table 2 as a ‘Teratosphaeriaceae sp.’ may prove to belong to another family.
The needle fungal communities varied significantly from site to site in the 12 different plantations of 5-year-old P. radiata that were investigated. Other studies also indicate similar significant variation in the needle fungal communities of another Pinus species (P. sylvestris) across different sites in the Massif Central in France, colonised by the same or similar needle cast fungi active in Tasmania, i.e. L. pinastri and C. minus (Gourbière et al. 2001; Gourbière and Debouzie 2003). Results from Prihatini et al. 2015a also indicated that needle fungal communities may be highly variable amongst trees at a single site. A key part of the original experimental design involved conducting a needle-health survey in the 12 sites sometime during the following 3 years, when the trees would be susceptible to SNC. Unfortunately, changing circumstances (particularly changes in ownership and management of the plots) prevented this from taking place. Therefore, no definitive conclusions can be drawn from the present study regarding the influence of needle fungal communities on subsequent development of SNC.
Of the two main variables reflecting climate, i.e. temperature and rainfall, only the latter influenced the composition of needle fungal communities in these 5-year-old trees. Podger and Wardlaw (1990) predicted that an annual rainfall range of 1200–2000 mm was a climatic characteristic of the disease distribution of SNC. Unpublished reports from Forestry Tasmania records suggest that the two wettest sites studied here (Inglis River and Oonah) subsequently suffered from SNC but further research is required to confirm these observations.
Conclusions
Many of the fungal species previously linked to needle-cast diseases were present in needles of young, healthy Pinus radiata. This indicates that environmental or host factors may have a significant role in the development of SNC. A range of previously unknown Teratosphaeriaceae species were detected in young pine needles, some of these appear to be almost ubiquitous endophytes.
References
Altschul, SF, Gish, W, Miller, W, Myers, EW, & Lipman, DJ. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215, 403–410.
Anderson, MJ, Gorley, RN, & Clarke, KR (2008). PERMANOVA+ for PRIMER: guide to software and statistical methods (p. 214). Plymouth, UK: PRIMER-E and Auckland, New Zealand: University of Auckland.
Arnold, AE, Mejía, LC, Kyllo, D, Rojas, EI, Maynard, Z, Robbins, N, Herre, EA. (2003). Fungal endophytes limit pathogen damage in a tropical tree. Proceedings of the National Academy of Sciences of the United States of America, 100, 15649–15654.
Botes, WM, Swart, WJ, & Crous, PW. (1997). Distribution of fungal endophytes in needles of Pinus radiata. South African Journal of Science, 93,v-vi. ISSN 00382353. Abstracts of the annual meeting of the Southern African Society for Plant Pathology, Badplaas.
Bradshaw, RE. (2004). Dothistroma (red-band) needle blight of pines and the dothistromin toxin: a review. Forest Pathology, 34, 163–185.
Brown, KB, Hyde, KD, & Guest, DI. (1998). Preliminary studies on endophytic fungal communities of Musa acuminata species complex in Hong Kong and Australia. Fungal Diversity, 1, 27–51.
Carroll, G. (1988). Fungal endophytes in stems and leaves: from latent pathogen to mutualistic symbiont. Ecology, 69, 2–9.
Clarke, KR, & Gorley, RN (2006). PRIMER v6: user manual/tutorial (p. 192). Plymouth, UK: PRIMER-E.
Crous, PW, Braun, U, & Groenewald, JZ. (2007). Mycosphaerella is polyphyletic. Studies in Mycology, 58, 1–32.
Crous, PW, Summerell, BA, Carnegie, AJ, Wingfield, MJ, Hunter, GC, Burgess, TI, Andjic, V, Barber, PA, & Groenewald, JZ. (2009). Unravelling Mycosphaerella: do you believe in genera? Persoonia, 23, 99–118.
Douanla-Meli, C, Langer, E, & Talontsi Mouafo, F. (2013). Fungal endophyte diversity and community patterns in healthy and yellowing leaves of Citrus limon. Fungal Ecology, 6, 212–222.
Felsenstein, J. (1989). PHYLIP—phylogeny inference package (version 3.2). Cladistics, 5, 164–166.
Ganley, RJ, & Newcombe, G. (2006). Fungal endophytes in seeds and needles of Pinus monticola. Mycological Research, 110, 318–327.
Gao, FK, Dai, CC, & Liu, XZ. (2010). Mechanisms of fungal endophytes in plant protection against pathogens. African Journal of Microbiology Research, 4, 1346–1351.
Giauque, H, & Hawkes, CV. (2013). Climate affects symbiotic fungal endophyte diversity and performance. American Journal of Botany, 100, 1435–1444.
Glen, M, Tommerup, IC, Bougher, NL, & O'Brien, PA. (2002). Are Sebacinaceae common and widespread ectomycorrhizal associates of Eucalyptus species in Australian forests? Mycorrhiza, 12, 243–247.
Gourbière, F, & Debouzie, D. (2003). Local variations in microfungal populations on Pinus sylvestris needles. Mycological Research, 107, 1221–1230.
Gourbière, F, van Maanen, A, & Debouzie, D. (2001). Associations between three fungi on pine needles and their variation along a climatic gradient. Mycological Research, 105, 1101–1109.
Hall, TA. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41, 95–98.
Hata, K, & Futai, K. (1996). Variation in fungal endophyte populations in needles of the genus Pinus. Canadian Journal of Botany, 74, 103–114.
Koukol, O, Kolařík, M, Kolářová, Z, & Baldrian, P. (2012). Diversity of foliar endophytes in wind-fallen Picea abies trees. Fungal Diversity, 54, 69–77.
Krabel, D, Morgenstern, K, & Herzog, S. (2013). Endophytes in changing environments—do we need new concepts in forest management? iForest – Biogeosciences and Forestry, 6, 109–112.
Larena, I, Salazar, O, González, V, Julián, MC, & Rubio, V. (1999). Design of a primer for ribosomal DNA internal transcribed spacer with enhanced specificity for ascomycetes. Journal of Biotechnology, 75, 187–194.
Larkin, MA, Blackshields, G, Brown, NP, Chenna, R, McGettigan, PA, McWilliam, H, Valentin, F, Wallace, IM, Wilm, A, Lopez, R, Thompson, JD, Gibson, TJ & Higgins, DG. (2007). ClustalW and ClustalX version 2. Bioinformatics, 23, 2947–2948.
Larran, S, Rollán, C, Ángeles, HB, Alippi, HE, & Urrutia, MI. (2002). Endophytic fungi in healthy soybean leaves. Investigacion Agraria Produccion y Proteccion Vegetales, 17(1), 173–178.
Page, RDM. (1996). TreeView: an application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences, 12, 357–358.
Petrini, O (1986). Taxonomy of endophytic fungi of aerial plant tissues. In NJ Fokkema & J van den Heuvel (Eds.), Microbiology of the phyllosphere (pp. 175–187). Cambridge: Cambridge University Press.
Podger, FD. (1984). Dothistroma in Tasmania. Australasian Plant Pathology, 13, 65.
Podger, FD, & Wardlaw, TJ. (1990). Spring needle-cast of Pinus radiata in Tasmania: I. Symptoms, distribution, and association with Cyclaneusma minus. New Zealand Journal of Forest Science, 20, 184–205.
Prihatini, I, Glen, M, Wardlaw, TJ, & Mohammed, CL (2015a). Lophodermium pinastri and an unknown species of Teratosphaeriaceae are associated with needle cast in a Pinus radiata selection trial. Forest Pathology 45, 281-289. doi:10.1111/efp.12169.
Prihatini, I, Glen, M, Wardlaw, TJ, & Mohammed, CL. (2015b). Diversity and identification of fungi associated with needles of Pinus radiata in Tasmania. Southern Forests (in press) http://dx.doi.org/10.2989/20702620.2015.1092345.
Queensland Government (2014). SILO climate data. Department of Science, Information Technology, Innovation and the Arts, The State of Queensland. http://www.longpaddock.qld.gov.au/. Accessed 2 January 2014.
Sokolski, S, Bernier-Cardou, M, Piché, Y, & Bérubé, JA. (2007). Black spruce (Picea marinara) foliage hosts numerous and potentially endemic fungal endophytes. Canadian Journal of Forest Research, 37, 1737–1747.
Stone, JK, Polishook, JD, & White, JF, Jr (2004). Endophytic fungi. In GM Mueller, GF Bills, & MS Foster (Eds.), Biodiversity of fungi: inventory and monitoring methods (pp. 241–270). Burlington: Elsevier Academic Press.
Tamura, K, Dudley, J, Nei, M, & Kumar, S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution, 24, 1596–1599.
Terhonen, E, Marco, T, Sun, H, Jalkanen, R, Kasanen, R, Vuorinen, M, & Asiegbu, F. (2011). The effect of latitude, season and needle-age on the mycota of Scots pine (Pinus sylvestris) in Finland. Silva Fennica, 45, 301–317.
Van Maanen, A, Debouzie, D, & Gourbière, F. (2000). Distribution of three fungi colonising fallen Pinus sylvestris needles along altitudinal transects. Mycological Research, 104, 1133–1138.
Wang, Y, & Guo, LD. (2007). A comparative study of endophytic fungi in needles, bark, and xylem of Pinus tabulaeformis. Canadian Journal of Botany, 85, 911–917.
White, TJ, Bruns, T, Lee, S, & Taylor, J (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In MA Innis, DH Gelfand, JJ Sninsky, & TJ White (Eds.), PCR protocols: a guide to methods and applications (pp. 315–322). London: Academic Press.
Acknowledgements
This study was part of a project funded by the Australian Research Council, Forestry Tasmania, Taswood Growers, Norske-Skog, Forest NSW and Hosking Ltd. New Zealand. We thank David Page for his help with the sampling. Istiana Prihatini is the recipient of a John Allwright Fellowship, ACIAR.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Author’s contributions
IP carried out the sampling and laboratory work, DNA sequence analyses, some statistical analyses and drafted the manuscript, MG supervised the laboratory work, DNA analyses and revised the manuscript, TJW selected the study sites, provided input to the design of the experiments and feedback on early drafts of the manuscript, DAR provided statistical advice, carried out the main statistical analyses, aided in their interpretation and assisted in revision of the manuscript, CLM conceived of the study, supervised some of the laboratory work and critically revised the manuscript. All authors read and approved the final manuscript.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Prihatini, I., Glen, M., Wardlaw, T.J. et al. Needle fungi in young Tasmanian Pinus radiata plantations in relation to elevation and rainfall. N.Z. j. of For. Sci. 45, 25 (2015). https://doi.org/10.1186/s40490-015-0055-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40490-015-0055-6