Abstract
Background
Squamous cell carcinoma of the external auditory canal (EACSCC) is an uncommon tumor and responsible for no more than 0.2% of all the head and neck malignancies. Although there is remarkable research evidence exhibiting that high-risk human papillomavirus (HPV) accounts for considerable head and neck malignancies, its role in the pathogenesis of EACSCC is yet to be determined.
Methods
We evaluated 16 patients with EACSCC treated at our department. We employed PCR to assay for high-risk subtypes of HPV. Two pathologists reviewed the histopathological staining via hematoxylin and eosin along with immunohistochemical staining of p16INK4a and Ki‑67.
Results
Detection of HPV DNA was done via PCR in 3 (18.75%) patients, and 8 (50%) positive (+) cases were determined via p16INK4a immunostaining. Besides, 3 (37.5%) individuals were HPV positive as per p16INK4a PCR results. In addition, all of the p16INK4a-positive specimens were diagnosed as moderately differentiated carcinomas.
Conclusions
Expression of Ki-67 was related to HPV status. This is the first report implicating high-risk HPV in squamous cell carcinoma of the external auditory canal. However, p16INK4a immunostaining is a suspectable approach for diagnosing HPV for EACSCC. In addition, HPV might enhance an elevated proliferation rate in EACSCC, illustrated via expression of Ki-67.
Graphical Abstract

Similar content being viewed by others
Introduction
External auditory canal squamous cell carcinoma (EACSCC) is rare, occurring in one to six cases in about 1,000,000 persons per year [1], and is responsible for not more than 0.2% of all head and neck malignancies [2]. The global annual prevalence is approximately 1.3 cases per million persons [3]. Because of its rarity, there is no established evidence-centered treatment approach for EACSCC [4]. Based on these, it is clinically, as well as biologically remarkable to explore the molecular properties of EACSCC to assess novel treatment targets.
EACSCC, manifesting histologically as a progressive squamous dysplasia, emanates from aggregated genetic and epigenetic alterations triggered by the exposure to solar ultraviolet light and inflammatory stimuli of long-standing chronic suppurative otitis media [5]. In addition, remarkable research evidence suggests that oncogenic human papillomaviruses (HPV) genotypes 16 and 18 play an indispensable role in head and neck squamous cell carcinoma (HNSCC): infection with high-risk forms of HPV is linked to the onset of 25% of head and neck cancers [6]. However, there are no reports on the role of HPV in the carcinogenesis of the external auditory canal. Elevated rates of HPV+ tumors improved outcomes for individuals with HPV+ and enhanced survival rates in contrast with HPV− cases, which have frequently been published in HNSCC studies [7, 8]. Considering this, the diagnosis of high-risk HPV in EACSCC is necessary and useful for stratifying patients, avoiding unnecessary overtreatment and incapacitating side effects.
Infection with HPV is frequently detected via p16INK4a IHC (immunohistochemistry). As a cyclin-dependent kinase repressor, the expression of p16INK4a is influenced by HPV-associated oncoproteins as E7 [9, 10]. There is research evidence implicating that an inverse feedback mechanism, modulating levels of p16INK4a in normal cells, gets disrupted via a decrease in the activity of pRb in growing squamous epithelial cells that are expressing HR-HPV E7 [11]. Hence, p16INK4a is utilized as an HPV-linked surrogate biologic signature in numerous study methods since the procedure is well-developed and saves time [12, 13]. However, it is reported that p16INK4a staining for detecting infection with HPV usually does not provide accurate results [14, 15]. Firstly, p16INK4a over-expression might also be seen in tubal metaplasia along with atrophic cells and in normal columnar cells from the cervix, resulting in poor specificity [16]. Besides, p16INK4a over-expression does not take place exclusively in HPV-triggered but additionally in an estimated 5–10% of HPV-negative HNSCC [13, 17, 18]. Moreover, numerous trends of p16INK4 expression result in diverse interpretations of "p16INK4a-positivity" [19,20,21]. Thus, this situation restricts the utilization of p16INK4a for diagnosing HPV infection in HNSCC.
Ki-67 constitutes a nuclear antigen as well as a biological signature of cellular proliferation, which is expressed in all stages of the cell cycle except G0 [22]. p16 and Ki-67 expression are mutually exclusive in normal cells. Recently, the simultaneous detection of p16INK4a and Ki-67 in a cell implies epithelial cell transformation that might progress to cancer [23]. p16INK4a has two diametrically opposed biological roles in cell cycle regulation. Conversely, p16INK4a expression could serve as a pivotal modulator of cellular senescence [24]. On the contrary, p16INK4a expression can be triggered upon signaling of HPV E7 oncogene [25]. Thus, p16INK4a /Ki-67 dual staining is useful to differentiate successfully cell cycle arrest from malignantly transformed p16INK4a-positive cells.
The objectives of this research work were twofold: first, to investigate the incidence of HPV in EACSCC; second, to explore the clinical performance of p16INK4a /Ki-67 as a marker for detecting HPV-induced transformed cells in EACSCC.
Materials and methods
Patients and specimens
Sixteen individuals with EACSCC were enrolled in this research work, with a mean age of 57 years, ranging from 1 to 76 years, with a male/female ratio of 10:6. The histopathologic assessment of all samples was confirmed and verified by two board-certified pathologists. The research work was granted approval by the Institutional Ethics and Review Committee of the Faculty of Clinical Medicine, Eye and ENT Hospital, Fudan University, Shanghai, China.
Immunohistochemistry
Immunohistochemical analysis was done on five μm-thick paraffin sections of EACSCC and adjacent normal skin tissue. The following deparaffinization with xylene, and subsequent rehydration via 96% ethanol, blocking of the activity of endogenous peroxides was done with 3% hydrogen peroxide for 10 min and rinsed in PBS. Exposing of the antigens was done by boiling the sections in citrate buffer (pH 6.0) for 15 min. After that, the primary antibody, anti-p16INK4a anti-Ki67, was introduced overnight. The slides were inoculated for 10 min with secondary antibody (Biotinylated Goat Anti-Polyvalent, NeoMarker, United States). The immunoperoxidase labeling was done; diaminobenzidine (DAB, Zymed, United States) was utilized as a chromogen for visualizing docking of the antibody. Finally, we stained the sections with Harris's haematoxylin, followed by clearing and mounting.
For p16INK4a, the presence of nuclear staining with or without cytoplasmic staining was regarded as a positive result. The tumoral p16INK4a expression trends were documented and categorized as per following criteria: (1) diffuse (> 10% of tumor cells harboring p16INK4a expression exhibiting clonal formation trend); (2) focal (numerous p16INK4a-positive cells not harboring criteria for diffuse p16INK4a expression).
Ki‑67 expression was stratified into four groups as per the distribution along with the fractions of cells with positive nuclear staining as shown: Score 0, < 10% of the cells; score 1, 10‑29% of the cells; score 2, 30‑69% of the cells; score 3, ≥ 70% epithelial cells [26].
Detection and genotyping of human papillomavirus DNA
Total genomic DNA was extracted from fresh frozen tissue of all samples using the TIANamp Genomic DNA Kit (Tiangen Biotech, Beijing, China). HPV detection and genotyping were done using the Human Papillomavirus (HPV) Genotyping Real-Time PCR Kit (Liferiver Bio-Tech, CA, United States). The Luminex bead-based genotyping approach allows for the diagnosis of 15 high-risk forms (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68, 73, and 82) and six low-risk types (6, 11, 42, 43, 44, and 70), according to the manufacturer’s instructions. The HPV genotypes were categorized as HR‑HPV or LR‑HPV based on the scheme opined by Dunne et al.
Statistical analysis
SPSS software (version 22.0; SPSS, Inc., Chicago, IL, USA) was used for statistical analysis. Fischer's tests and non-parametric Mann–Whitney's test were used for univariate analysis. P < 0.05 was considered to indicate a statistically significant difference (Table 1).
Results
HPV DNA status
3 (18.75%) patients were HPV + based on PCR results (Table 2). Only in 3 (18.75%) cases did the PCR result match the positive p16INK4a staining (Table 2). In 5 (31.25%) cases, p16INK4a staining was positive, but the PCR result was negative (Table 2). In summary, p16INK4a staining for diagnosing HPV is inadequately sensitive and specific.
Among the 3 HPV positive specimens, the most often diagnosed HPV type was HPV-16 (66.7%), followed by HPV-18 (33.3%). Low-risk type HPV DNA (HPV-6/11) was not found in all specimens.
Expression of p16INK4a and Ki-67 in EACSCC
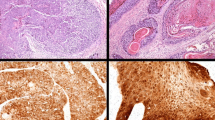
Complete p16INK4a expression absence was seen in 8 of 16 (50%) EACSCC (Table 1). The remaining EACSCC were scored with varying extent as well as the pattern of p16INK4a expression (Fig. 1 and Table 1). As demonstrated in Table 1, a diffuse p16INK4a expression pattern was frequently observed in EACSCC. The 16 EACSCC were categorized as well to moderately differentiated (Fig. 1) as follows: G 1 (eight cases), G 2 (eight cases). It is pivotal to mention that all eight moderately-differentiated (G 2) EACSCC displayed p16INK4a positivity (Table 3). Comparing the well (G1) with moderately differentiated (G2) EACSCC, it was noted that all cases diagnosed as G2 presented more frequent p16INK4a positivity, whereas the G1 cases were p16INK4a negative (Table 3).
Ki-67 expression was observed in 15 of 16 (93.7%) EACSCC (Table 1). The greater proliferation index was documented in HPV-positive specimens with 2.34 mean PI value, which was 1.0 in HPV-negative specimens (Fig. 1 and Table 2). The difference between groups was significant (p = 0.0383; p < 0.05). However, there was no remarkable difference in Ki-67 expression scores between the well (G1) and moderately differentiated (G2) of EACSCC. Furthermore, concomitant with diffuseness, a high Ki‑67 index was implicated in the HR‑HPV group.
Discussion
To investigate the presence of HPV DNA, we studied 16 cases of surgically removed primary SCC in the external auditory canal. We carried out a dual investigation: a molecular biology study (using PCR to diagnose the presence of HPV DNA) and immunohistochemical profiling (to investigate the expression of p16INK4a protein). Furthermore, 3 of 16 patients were found to have detectable HPV 16 and HPV 18 DNA. Our data align with the other research conducted by Masterson et al. [1]. Although the 18.75% detection rate of high-risk HPV-DNA in this research work represents a minority, the clinical significance may be apparent that high-risk HPV is implicated in squamous cell carcinoma of the external auditory canal. However, a potential limitation may be the lack of an adequate sample size because of its rarity.
It is reported that p16INK4a was an insensitive signature of HPV condition with a low estimation significance for HPV infection [27]. This supports that p16INK4a staining might be an inadequate HPV detection approach because we reported a high rate of failure in cases that were positive for p16INK4a but were actually HPV−. Through this method, the number of false-positive HPV+ cases would be 50 percent higher. Presently, ultraviolet along with ionizing radiation constitutes the only evidence-centered supported risk factors for TB SCC [28]. In addition, the previous investigations illustrated that the ultraviolet B irradiation could activate the CDKN2A (cyclin-dependent kinase inhibitor 2A) gene that encodes the p16INK4a protein in human keratinocytes [29,30,31,32]. On the other hand, activation of CDKN2A in keratinocytes seems to be related to tumor de-differentiation in head and neck areas [33]. In our study, activation of p16INK4a protein was exceeded in one-half of the EACSCCs. Notably, a strong relationship between p16INK4a over-expression and the degree of tumor differentiation is seen. These facts suggest that p16INK4a should be necessary to induce tumor progression and de-differentiation.
Ki‑67 constitutes a nuclear protein that is linked to RNA transcription along with the progress of cell cycle [11]. The present research work explored the efficacy of Ki‑67 in the EACSCC and compared the findings of HR‑HPV and HPV‑negative groups. The assessment of the data shows a considerable relationship between the detection of the HPV genome and the immune-histochemical expression of Ki-67. Hence, it appears that HPV can be an indispensable cofactor in the onset and progression of squamous carcinoma of EAC. In squamous carcinoma, HPV appears to increase direct changes in the mitosis along with multiplication phases of the cell cycle that could be estimated via the expression of Ki-67, triggering a higher cell proliferation index. Thus, HPV may increase the proliferation index in EACSCC, exhibited via the expression of Ki-67.
In summary, our data show that high-risk HPV 16 and HPV 18 are related to the SCC of EAC. p16INK4a immunostaining seems to be insufficient for HPV detection in EACSCC. However, a p16-dependent cascade might be implicated in tumor de-differentiation of EACSCC. Furthermore, we found an impressive link of the presence of HPV to Ki-67 expression. Because the number of patients in our study was limited, a larger study is necessary to address this issue.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Abbreviations
- EACSCC:
-
Squamous cell carcinoma of the external auditory canal
- HPV:
-
Human papillomavirus
- PCR:
-
Polymerase chain reaction
- IHC:
-
Immunohistochemistry
- HNSCC:
-
Head and neck squamous cell carcinoma
- PBS:
-
Phosphate buffered saline
- HR-HPV:
-
High-risk human papillomavirus
- LR-HPV:
-
Low-risk human papillomavirus
- TBSCC:
-
Temporal bone squamous cell carcinoma
- CDKN2A:
-
Cyclin-dependent kinase inhibitor 2A
References
Sato K, Komune N, Hongo T, Koike K, Niida A, Uchi R, Noda T, Kogo R, Matsumoto N, Yamamoto H, et al. Genetic landscape of external auditory canal squamous cell carcinoma. Cancer Sci. 2020;111(8):3010–9.
Moody SA, Hirsch BE, Myers EN. Squamous cell carcinoma of the external auditory canal: an evaluation of a staging system. Am J Otol. 2000;21(4):582–8.
Madsen AR, Gundgaard MG, Hoff CM, Maaremd C, Holmboe P, Knap M, Thomsen LL, Buchwald C, Hansen HS, Bretlau P, et al. Cancer of the external auditory canal and middle ear in Denmark from 1992 to 2001. Head Neck J Sci Spec. 2008;30(10):1332–8.
Lovin BD, Gidley PW. Squamous cell carcinoma of the temporal bone: a current review. Laryngosc Investig. 2019;4(6):684–92.
Allanson BM, Low TH, Clark JR, Gupta R. Squamous cell carcinoma of the external auditory canal and temporal bone: an update. Head Neck Pathol. 2018;12(3):407–18.
Mehanna HM, Rattay T, Smith J, McConkey CC. Treatment and follow-up of oral dysplasia: a systematic review and meta-analysis. Head Neck J Sci Spec. 2009;31(12):1600–9.
Keane FK, Chen YH, Neville BA, Tishler RB, Schoenfeld JD, Catalano PJ, Margalit DN. Changing prognostic significance of tumor stage and nodal stage in patients with squamous cell carcinoma of the oropharynx in the human papillomavirus era. Cancer Am Cancer Soc. 2015;121(15):2594–602.
Kolk A, Jubitz N, Mengele K, Mantwill K, Bissinger O, Schmitt M, Kremer M, Holm PS. Expression of Y-box-binding protein YB-1 allows stratification into long- and short-term survivors of head and neck cancer patients. Brit J Cancer. 2011;105(12):1864–73.
Liang CH, Marsit CJ, McClean MD, Nelson HH, Christensen BC, Haddad RI, Clark JR, Wein RO, Grillone GA, Houseman EA, et al. Biomarkers of HPV in head and neck squamous cell carcinoma. Cancer Res. 2012;72(19):5004–13.
Lee CC, Ho HC, Su YC, Yu CH, Yang CC. modified tumor classification with inclusion of tumor characteristics improves discrimination and prediction accuracy in oral and hypopharyngeal cancer patients who underwent surgery. Medicine. 2015;94(27):e1114.
Lim S, Lee MJ, Cho I, Hong R, Lim SC. Efficacy of p16 and Ki-67 immunostaining in the detection of squamous intraepithelial lesions in a high-risk HPV group. Oncol Lett. 2016;11(2):1447–52.
Malinowsky K, Wolff C, Ergin B, Berg D, Becker KF. Deciphering signaling pathways in clinical tissues for personalized medicine using protein microarrays. J Cell Physiol. 2010;225(2):364–70.
Lingen MW, Xiao WH, Schmitt A, Jiang B, Pickard R, Kreinbrink P, Perez-Ordonez B, Jordan RC, Gillison ML. Low etiologic fraction for high-risk human papillomavirus in oral cavity squamous cell carcinomas. Oral Oncol. 2013;49(1):1–8.
Mirghani H, Amen F, Moreau F, St Guily JL. Do high-risk human papillomaviruses cause oral cavity squamous cell carcinoma? Oral Oncol. 2015;51(3):229–36.
Marur S, D’Souza G, Westra WH, Forastiere AA. HPV-associated head and neck cancer: a virus-related cancer epidemic. Lancet Oncol. 2010;11(8):781–9.
Wentzensen N, Bergeron C, Cas F, Eschenbach D, Vinokurova S, Doeberitz MV. Evaluation of a nuclear score for p16(INK4a)-stained cervical squamous cells in liquid-based cytology samples. Cancer Cytopathol. 2005;105(6):461–7.
Reimers N, Kasper HU, Weissenborn SJ, Stutzer H, Preuss SF, Hoffmann TK, Speel EJM, Dienes HP, Pfister HJ, Guntinas-Lichius O, et al. Combined analysis of HPV-DNA, p16 and EGFR expression to predict prognosis in oropharyngeal cancer. Int J Cancer. 2007;120(8):1731–8.
Lewis JS, Thorstad WL, Chernock RD, Haughey BH, Yip JH, Zhang Q, El-Mofty SK. p16 positive oropharyngeal squamous cell carcinoma: an entity with a favorable prognosis regardless of tumor HPV status. Am J Surg Pathol. 2010;34(8):1088–96.
Begum S, Gillison ML, Ansari-Lari MA, Shah K, Westra WH. Detection of human papillomavirus in cervical lymph nodes: a highly effective strategy for localizing site of tumor origin. Clin Cancer Res. 2003;9(17):6469–75.
Hoffmann M, Tribius S, Quabius ES, Henry H, Pfannenschmidt S, Burkhardt C, Gorogh T, Halec G, Hoffmann AS, Kahn T, et al. HPV DNA, E6*I-mRNA expression and p16(INK4A) immunohistochemistry in head and neck cancer: how valid is p16(INK4A) as surrogate marker? Cancer Lett. 2012;323(1):88–96.
Pai RK, Erickson J, Pourmand N, Kong CS. p16(INK4A) immunohistochemical staining may be helpful in distinguishing branchial cleft cysts from cystic squamous cell carcinomas originating in the oropharynx. Cancer Cytopathol. 2009;117(2):108–19.
Scholzen T, Gerdes J. The Ki-67 protein: from the known and the unknown. J Cell Physiol. 2000;182(3):311–22.
Ikenberg H, Bergeron C, Schmidt D, Griesser H, Alameda F, Angeloni C, Bogers J, Dachez R, Denton K, Hariri J, et al. Screening for cervical cancer precursors with p16/Ki-67 dual-stained cytology: results of the PALMS study. JNCI J Natl Cancer I. 2013;105(20):1550–7.
Collado M, Blasco MA, Serrano M. Cellular senescence in cancer and aging. Cell. 2007;130(2):223–33.
McLaughlin-Drubin ME, Crum CP, Mungera K. Human papillomavirus E7 oncoprotein induces KDM6A and KDM6B histone demethylase expression and causes epigenetic reprogramming. Proc Natl Acad Sci USA. 2011;108(5):2130–5.
Reuschenbach M, Seiz M, Doeberitz CV, Vinokurova S, Duwe A, Ridder R, Sartor H, Kommoss F, Schmidt D, Doeberitz MV. Evaluation of cervical cone biopsies for coexpression of p16INK4a and Ki-67 in epithelial cells. Int J Cancer. 2012;130(2):388–94.
Walline HM, Komarck C, McHugh JB, Byrd SA, Spector ME, Hauff SJ, Graham MP, Bellile E, Moyer JS, Prince ME, et al. High-risk human papillomavirus detection in oropharyngeal, nasopharyngeal, and oral cavity cancers comparison of multiple methods. Jama Otolaryngol. 2013;139(12):1320–7.
Ungar OJ, Santos F, Nadol JB, Horowitz G, Fliss DM, Faquin WC, Handzel O. Invasion patterns of external auditory canal squamous cell carcinoma: a histopathology study. Laryngoscope. 2021;131(2):E590–7.
Abu Juba B, Sovrea A, Crisan D, Melincovici C, Coneac A, Badea M, Crisan M. Apoptotic markers in photoinduced cutaneous carcinoma. Rom J Morphol Embryo. 2013;54(3):741–7.
Chazal M, Marionnet C, Michel L, Mollier K, Dazard JE, Della Valle V, Larsen CJ, Gras MP, Basset-Seguin N. P16(INK4A) is implicated in both the immediate and adaptative response of human keratinocytes to UVB irradiation. Oncogene. 2002;21(17):2652–61.
Hodges A, Smoller BR. Immunohistochemical comparison of p16 expression in actinic keratoses and squamous cell carcinomas of the skin. Modern Pathol. 2002;15(11):1121–5.
Kaabipour E, Haupt HM, Stern EB, Kanetsky PA, Podolski VF, Martin AM. p16 expression in keratoacanthomas and squamous cell carcinomas of the skin: an immunohistochemical study. Arch Pathol Lab Med. 2006;130(1):69–73.
Ciortea CD, Jung I, Gurzu S, Kovecsi A, Turdean SG, Bara T. Correlation of angiogenesis with other immunohistochemical markers in cutaneous basal and squamous cell carcinomas. Rom J Morphol Embryo. 2015;56(2):665–70.
Acknowledgements
Not applicable.
Funding
This work was funded by Science Project of Shanghai Municipal Health Bureau (No. 20174Y0062).
Author information
Authors and Affiliations
Contributions
XL and CZ designed the study. ZC and PW performed the experiments, analyzed/interpreted data, and wrote the manuscript. MS and CM performed the statistical analysis. CZ checked English and drafted the manuscript. XL and LS performed the histopathologic assessment of all samples. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Informed consent was obtained from all participants. Human studies conformed to the principles of the Declaration of Helsinki and were approved by the Research Ethics Committee of the Eye and ENT Hospital of Fudan University. The patient signed the consent form for participation and publication.
Consent for publication
All presentations of case reports must have consent for publication.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Pan, W., Zhang, C., Chen, M. et al. Expression of Ki-67 and P16 are related with HPV in squamous cell carcinoma of the external auditory canal. J of Otolaryngol - Head & Neck Surg 51, 40 (2022). https://doi.org/10.1186/s40463-022-00592-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40463-022-00592-8