Abstract
Background
Pathogenic water dwelling protozoa such as Acanthamoeba spp., Hartmannella spp., Naegleria spp., Cryptosporidium spp. and Giardia spp. are often responsible for devastating illnesses especially in children and immunocompromised individuals, yet their presence and prevalence in certain environment in sub-Saharan Africa is still unknown to most researchers, public health officials and medical practitioners. The objective of this study was to establish the presence and prevalence of pathogenic free-living amoeba (FLA), Cryptosporidium and Giardia in Queen Elizabeth Protected Area (QEPA).
Methods
Samples were collected from communal taps and natural water sites in QEPA. Physical water parameters were measured in situ. The samples were processed to detect the presence of FLA trophozoites by xenic cultivation, Cryptosporidium oocysts by Ziehl-Neelsen stain and Giardia cysts by Zinc Sulphate floatation technique. Parasites were observed microscopically, identified, counted and recorded. For FLA, genomic DNA was extracted for amplification and sequencing.
Results
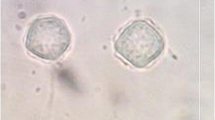
Both natural and tap water sources were contaminated with FLA, Cryptosporidium spp. and Giardia spp. All protozoan parasites were more abundant in the colder rainy season except for Harmannella spp. and Naegleria spp. which occurred more in the warmer months. The prevalence of all parasites was higher in tap water than in natural water samples. There was a strong negative correlation between the presence of Acanthamoeba spp., Hartmannella spp., Cryptosporidium spp. and Giardia spp. with Dissolved Oxygen (DO) (P < 0.05). The presence of Cryptosporidium spp. showed a significant positive correlation (P < 0.05) with conductivity, pH and Total Dissolved Solids (TDS); whereas the presence of Giardia spp. had only a strong positive correlation with TDS. Molecular genotyping of FLA produced 7 Acanthamoeba, 5 Echinamoeba, 2 Hartmannella, 1 Bodomorpha, 1 Nuclearia and 1 Cercomonas partial sequences.
Conclusions
All water collection sites were found to be contaminated with pathogenic protozoa that could possibly be the cause of a number of silent morbidities and mortalities among rural households in QEPA. This implies that water used by communities in QEPA is of poor quality and predisposes them to a variety of protozoan infections including the FLA whose public health importance was never reported, thus necessitating adoption of proper water safety measures.
Similar content being viewed by others
Multilingual abstracts
Please see Additional file 1 for translations of the abstract into the five official working languages of the United Nations.
Background
The factors that contribute to the emergence and re-emergence of infectious diseases originate from changes in the ecosystems; such as ecosystem variations of natural or anthropogenic origin with climatic and geographic influences on pathogens and vectors, or changes in the microbes themselves, or their host spectrum [1, 2]. Such changes in the ecosystem could be the cause of the emergence of infections such as Zika, Ebola, Marburg, Influenza and West Nile viruses as well as protozoan and other parasitic infections. Pathogenic waterborne protozoan parasites such as Acanthamoeba, Hartmannella, Naegleria, Cryptosporidium and Giardia are typically associated with poor and often marginalised communities, as observed mostly by studies done in America, Asia, Europe and the Middle East [3–5]. Lack of adequate information about waterborne parasites in sub-Saharan Africa especially on FLA has compelled the present study.
In many rural African households, untreated water is used for various purposes such as bathing, cooking, drinking and swimming, often exposing them to waterborne parasitic infections [3, 5]. More than 300 million people in sub-Saharan Africa have poor access to safe water, predisposing them to a number of infections directly or indirectly, the commonest being diarrhoeal infections which have been reported to be the second leading cause of death in children under 5 years [3, 6]. The commonly studied waterborne pathogens in poor rural households are those associated with typhoid, dysentery, cholera, hepatitis E and rotavirus in children and immunocompromised individuals [7–10]. Other waterborne parasites such as pathogenic free-living amoeba (FLA) that may cause severe health effects in humans and animals have not been widely studied in developing countries. However, they have been studied more in developed countries and are known to be associated with severe infections in humans [5]. In sub-Saharan Africa, due to lack of information, more studies are needed to establish the health importance of FLA [5].
Pathogenic FLA such as Acanthamoeba spp., Hartmanella vermiformis, Balamuthia mandrillaris, Naegleria spp. and Vahlkampfia avara are aerobic eukaryotic protists that can potentially cause infections in humans and animals [7]. They have been implicated in infections of the central nervous system, eye, nose and skin. Most pathogenic FLA are known to facilitate intracellular multiplication of Legionella pneumophila, Vibrio cholerae, Bacillus anthracis and Mycobacterium tuberculosis which are responsible for legionellosis, cholera, anthrax and tuberculosis, respectively [11, 12]. Acanthamoeba has been reported as a causative agent of granulomatous amoebic encephalitis (GAE), a fatal disease of the CNS and amoebic keratitis (AK), a painful sight-threatening disease of the eyes [11]. It has also been associated with cutaneous lesions and sinusitis in HIV/AIDS patients and other immunocompromised individuals [13]. A case of mixed keratitis infection has been reported due to Acanthamoeba spp. and thermotolerant H. vermiformis [14]. Hartmannella like Acanthamoeba is also a host of a number of human bacterial organisms such as Legionella pneumophila and Pseudomonas aeruginosa [15, 16]. Naegleria fowleri, N. australiensis and N. italica are associated with acute, fulminant, necrotizing and hemorrhagic primary amoebic meningoencephalitis (PAM) that often causes death in children and adults with a history of recent contact with fresh water [7]. Other FLA such as Cercozoa (Cercomonadida, Thaumatomonadida, Euglyphida, Desmothoracida, Gromiidae, Phaeodarea), Platymoeba and Echinamoeba (E. thermarum, E. exundans) have not yet been reported as pathogenic, but with ecosystem changes, climatic changes, geographic influences and mutations, they could as well cause infections in the near future.
Over the past decade, Cryptosporidium and Giardia have emerged as major waterborne pathogens [17] whose transmission occurs as a result of water contamination with animal and human faeces. These infections are attributed to poor hygiene followed by lack of clean potable water resulting in cryptosporidiosis and giardiasis which are characterised by vomiting, chills, headache, fever, profuse diarrhoea, abdominal pain and cramping [18, 19]. C. parvum and G. lamblia can cause severe clinical infections in young individuals [20]. Infections caused by Cryptosporidium and Giardia pose a significant environmental and public health concern because of their tremendous ability to be transmitted from animal to animal and from animal to humans [20].
Free-living amoeba, Cryptosporidium and Giardia are parasites commonly found in natural water resources and piped water systems, however not much is known about their occurrence, prevalence and distribution in both rural and urban communities in Uganda.
Methods
Study area
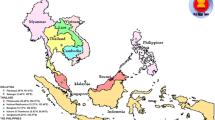
The study was conducted in Queen Elizabeth Protected Area (QEPA), Uganda (Fig. 1). It is located at 00 12S, 30 00E (Latitude: 0.2000; Longitude: 30.0000) and is 1 978 sq. km in size. This Protected Area (PA) harbours Lakes George and Edward joined by the 40 km long Kazinga Channel. QEPA is a UNESCO ‘Man and Biosphere Reserve’ with 11 village enclaves, all with a fast growing population of humans whose main economic activities are fishing and livestock production. Their source of water is direct natural water from Kazinga channel, River Kyambura, Lake George and Lake Edward or untreated piped tap water channelled directly from the natural sources and stored in reservoir tanks before it is supplied to them.
Sample collection, storage and transportation
The water sources considered were those from natural water sources and tap water systems. The sampling sites were purposively selected based on their benefit, convenience and importance to public health. They were based on certain landmarks that included the following: (1) along River Kyambura (R. Kyambura), (2) Kazinga channel banks (KCB), (3) Kazinga mid channel (KMC) (water in the middle of the channel), (4) Fish landing sites (FLS), and (5) Community piped tap water. Sampling was done within 1 year in the months of November, January, March, May, July and September. Important physical water parameters including; Dissolved Oxygen (DO) (mg/L), surface water temperature (°C), conductivity (μS/m), pH and Total Dissolved Solids (TDS)(g/L) were determined in situ using a Multi-parameter water sensor (Greenspan, USA). A total number of 408 (324 natural water and 84 tap water) water samples were collected using 50 ml sterile polypropylene falcon tubes (Falcon® Centrifuge Tubes, Discovery Labware, USA). All samples were stored at room temperature and transported to the Makerere University parasitology laboratory within 48 h.
Laboratory methods
Three methods were used for parasite detection. Culture in non-nutritive media for FLA, Modified Ziehl-Neelsen staining for Cryptosporidium and the Zinc Sulphate flotation technique for Giardia cysts. For the FLA positive samples, DNA extraction, amplification and sequencing were carried out.
Sample processing and growth in xenic cultures
The non-nutritive medium (Page Amoeba Saline solution of 2.5 mM NaCl, 1 mM KH2PO4, 0.5 mM Na2HPO4, 40 mM CaCl2 and 20 mM MgSO4) was seeded with 0.1 ml of a heat inactivated 48-h culture of E. coli BL2 [21]. Water samples collected in the 50 ml tubes were centrifuged at 1 000 × g for 15 min and supernatant poured off to expose the pellets. Using sterile Pasteur pipettes, the pellets were removed from all the tubes and carefully spread on pre-seeded NNA-EI agar plates. The plates were incubated at 32 °C overnight. The following day each plate was sealed with a plastic film and incubated upside down at 32 °C up to 7 days. After 3 days of incubation, the plates were monitored for detection of FLA trophozoites until the 7th day using an inverted microscope (Motic® AE2000 Binocular, TED PELLA Inc. USA). The number of FLA trophozoites were counted using a haemocytometer (MicrobeHunter, Germany) and recorded.
Modified Ziehl-Neelsen carbol fuchsin staining for cryptosporidium
To maximise recovery of oocysts, the samples were centrifuged at 1 000 × g for 10 min to concentrate the oocysts. A modified Ziehl-Neelsen’s (ZN) carbol-fuchsin stain [18] was used to identify the oocysts of Cryptosporidium spp. A few drops of water were placed on a slide and stained with ZN-carbol fuchsin stain for 2 min, rinsed with tap water, followed by rinsing with acid alcohol (3 % hydrochloric acid in 70 % ethanol) and subsequently with tap water again. The product was counterstained with Brilliant Green for 2 min before rinsing it with tap water. This was followed by air-drying the slide before viewing under a microscope.
Zinc sulphate floatation technique for Giardia
Zinc Sulphate floatation technique [22] was used to identify Giardia cysts. To maximise recovery of cysts, each water sample was concentrated by centrifugation at 1 000 × g for 1 min. The supernatant was poured off leaving a small pellet. A test tube was filled with Zinc Sulphate solution (33 % w/v, Specific gravity 1.18-1.2). An estimated 1-2 ml of the water was added to the test tube and stirred. The test tube was then filled to the brim with Zinc Sulphate solution. A grease-free cover slip was put on the full to the brim test tube and left for 15 min to give time for the cysts to float. After 15 min, the cover slip was carefully lifted off the test tube, placed face down on a microscope slide and viewed under a microscope to identify the cysts.
Identification and counting parasites at genus level
To determine the genus of each protozoan parasite, its movement and structural properties were examined [23]. Representatives trophozoites/(oo)cysts of FLA, Cryptosporidium and Giardia were counted using a haemocytometer (Bright-Line™, Sigma-Aldrich Co. LLC, USA) and recorded.
Deoxyribonucleic acid (DNA) extraction
Genomic DNA was extracted from only FLA culture positive plates by chemical lysis and purification with phenol/chloroform/isoamyl alcohol extraction method [24]. This involved adding 500 μl of STE buffer (0.1 M NaCl, 1 mM EDTA, 10 mM Trischloride, pH 8, 1 % SDS) and 10 μl proteinase K (10 mg/ml) directly to each sample in Eppendorf tubes. The samples were incubated at 56 °C for one hour and then cooled before phenol extraction was started. Equal volumes of phenol-chloroform (521 μl) were added to the samples, mixed by vortexing and centrifuged at maximum speed (13 200 rpm) for 10 min. The aqueous layers from each tube were recovered and transferred to new Eppendorf tubes. This step was repeated to make two phenol-chloroform extractions. The aqueous layers were subjected to another chloroform extraction, recovered by centrifugation and transferred to new Eppendorf tubes, after which 1 000 μl of absolute alcohol (96-100 %) was added to each sample. The samples were then put in a freezer at -80 °C for precipitation overnight. The next day, samples were removed from the freezer and centrifuged at 13 200 rpm for 30 min. Absolute alcohol was poured off. The pellet in each tube was then washed with 1 000 μl of 70 % alcohol, centrifuged at 13 200 rpm for 15 min and alcohol poured off to expose the pellet. Finally, the pellet was air dried and dissolved in 50 μl of TE buffer.
DNA amplification
Amplification of the partial 18S ribosomal DNA (18S rDNA) gene from FLA was performed using primer pairs JDP1/JDP2 and CRN5/1137 [25, 26]. The JDP1/JDP2 primer was specific to Acanthamoeba organisms whereas CRN5/1137 obtained amplimers from any eukaryote, aiding amplification of the 18S Ribosomal DNA gene from Hartmanella, Cercozoa, Bodomorpha, Echinamoeba and several other groups of FLA.
Amplification reactions were performed using a DreamTaq PCR kit (Thermoscientific DreamTaq, USA). We used a 25 μl reaction volume containing 12.5 μl DreamTaq Green PCR Master Mix (2X), 0.5 μM forward primer, 0.5 μM reverse primers, 9 μl nuclease free water and 2.5 μl DNA template (50 pg concentration). The PCR was done under the following conditions: Initial denaturation at 94 °C for 3 min then 35 cycles with denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s, extension at 72 °C for 30 s and a final extension at 72 °C for 5 min. A sample of 5 μl of each PCR reaction was screened for successful amplification on a 2.5 % (W/V) agarose gel stained with ethidium bromide and run against 1 kb DNA ladder (Finnzymes, Finland). Electrophoresis was performed at 100 V of current and buffer used was 1 × TAE containing 0.5 μg/ml of ethidium bromide. Once enough electrophoretic separation was realised, the agarose gel was observed using a UV gel documentation system (Wagtec, UK). The gel images were captured and a soft copy stored.
Nucleic acid sequencing and analysis
The samples that showed the strongest positive bands between 400-600 bp with JDP1/JDP2, and 1-1 475 bp with CRN5/1137) were extracted from the gel and the DNA was purified using QIAquick gel extraction kit (Qiagen Inc. Sample and Assay Technologies, Netherlands). The 18S rDNA segment from each of the FLA isolates was subjected to cycle sequencing using Dyenamic Terminator Cycle sequencing kit with JDP1/JDP2 and CRN5/1137 as sequencing primers [26]. The sequencing included 2 μl of PCR product, 5 × BigDye Buffer, and 2 pmol primer. Sequencing was done in 30 cycles with step 1 at 94 °C for 30 s, step 2 at 55 °C for 15 s and step 3 at 65 °C for 4 min. The sequence files were checked for quality and base trimming carried out using the Seqbuilder software (Dnastar, USA). For each of the nucleotide query sequences, a search for homologues in the NCBI database was carried out using the blastn tool. Homologues with query coverage >75 %, identity >70 % and low E values were considered. The molecular phylogenetic analysis was then completed by using the Maximum Likelihood Method in MEGA6 [27].
Statistical analysis
Data was analysed using IBM SPSS version 22. Numerical variables were summarised using mean and standard error of the mean (SEM). Univariate analysis to compare the prevalence of parasites across sampling sites was done using cross-tabulation with a Chi-square or Fisher’s exact test. Variables with a P-value of ≤ 0.05 were taken to be significant. Correlation analysis between environmental variables and waterborne parasite presence was done using Pearson correlation coefficient (r), a P-value of ≤0.05 was considered statistically significant.
Results
Prevalence of the organisms
The water samples were collected during cold rainy (November, March & July) and cool dry (January, May & September) seasons. Overall, protozoan parasite prevalence was higher during the rainy season except for Hartmannella and Naegleria spp. that were higher in the dry season (Fig. 2). The prevalence and means (SEM) of the parasites from different sources are shown in Table 1. Both natural and tap water sources were contaminated with FLA, Cryptosporidium spp. and Giardia spp. The prevalence of all the parasites was higher in tap water, whereas as mean (SEM) was higher in natural water. The prevalence and mean of the parasites from the natural water sites were highest at KCB. The number of organisms isolated was significantly influenced by the sampling site.
Physical parameters
The means (SEM) of physical parameters are presented in Table 2. The mean DO (mg/L) from highest to lowest was as follows; KCM (4.56 ± 0.26), Tap water (4.08 ± 0.27), River Kyambura (4.03 ± 0.15), FLS (1.84 ± 0.21) and KCB (1.74 ± 0.15). All water sources had an alkaline condition (pH 7.99 ± 0.11- pH 9.31 ± 0.04) at 21.43 ± 0.14 (°C) - 5.76 ± 20 (°C). The mean TDS (g/L) was lowest at KCM with no significant difference at all the other points. Conductivity (μS/m) did not exhibit much difference.
Correlation between waterborne parasites and physical parameters
Linear correlation analysis showed a strong inverse correlation between the presence of Acanthamoeba spp., Hartmannella spp., other FLA, Cryptosporidium spp. and Giardia spp. with DO (P < 0.05). The presence of these parasites showed a weak negative correlation with temperature. The presence of Cryptosporidium spp. showed a significant positive correlation (P < 0.05) with conductivity, pH and TDS. Similarly, the presence of Giardia spp. showed a strong positive correlation with TDS. Detailed analysis is presented in Table 3.
Molecular identification of the FLA isolates and phylogenetic analysis
Thirty-one representative samples scored positive with PCR, 10 with JDP-PCR and 20 with CRN5/1137-PCR. The FLA isolates that produced the strongest positive bands (Figs. 3 and 4) were further sequenced and investigated by phylogenetic analysis. Following sequence blasting and comparison with the GenBank results from NCBI (Fig. 5, Table 4 and Appendix: Table 6), the following species was identified: Acanthamoeba spp., Acanthamoeba polyphaga, Hartmannella vermiformis, Nuclearia pattersoni, Echinamoeba exundans, Bodomorpha minima and Cercomonas agilis. The Acanthamoeba sequences got belonged to the group of sequence types T1, T4, and T11. All the parasites identified in this study were matched with the reported diseases they cause in humans (Table 5).
Discussion
The availability of potable water in poor countries is a challenge as it causes serious health problems. Polluted water sources are the major causes for the prevalence of waterborne infections which sometimes results in severe morbidities and mortalities [3, 28]. Diseases such as cholera, campylobacteriosis, shigellosis, salmonellosis and a variety of fungal and parasitic infections are known to affect a number of rural communities in sub-Saharan Africa [29], but infections due to FLA, Cryptosporidium and Giardia due to contaminated water sources are less reported. Acanthamoebiasis (brain, eye and skin infection), Hartmannellosis (respiratory tract infections or generalised fatal meningoencephalitis) and Naegleriasis (brain-eating amoeba infection) are not mentioned as possible infectious diseases in Uganda, and yet they may be silently occurring among individuals causing severe pathogenic effects. Cryptosporidiosis and Giardiasis which are diseases of poverty have over-time been neglected. They are known to be prevalent among communities which lack access to clean potable water supply [7, 9, 30]. Most communities in QEPA, like any other protected wildlife areas in Africa, are surrounded by poor communities with an average income of less than US$1 per person per day [31]. This level of income limits the communities from accessing privately owned water resources that provide safe water in the area as the price for this essential services is costly.
In the present study, all parasites except Hartmannella spp. and Naegleria spp. were more prevalent in the rainy season than the dry season, possibly due to contamination of the water sources with sewage or faeces through flooding. Previous studies have indicated that most waterborne parasites, especially FLA are more prevalent in cold rainy months of the year [32], whereas some like Hartmannella spp and Naegleria spp. are more common in warmer months [33]. Tap water had a higher prevalence of FLA than the natural water source. Considering specific natural water sites, prevalence and mean numbers of FLA were higher in KCB, FLS and R. Kyambura compared to KCM. These high numbers could be explained by high TDS due to organic matter from rotting leaves, animal and human faeces which are from the run-off from the land. Water from KCM appeared clear with less organic matter, the reason for few parasite presence. Tap water, on the other hand, had more FLA prevalence likely because all taps are supplied by the same natural water source with high TDS and organic matter, consequently, resulting in more biofilm formation along the piped water network. This is consistent with findings from other studies which explain that microorganisms settle on the inner surfaces of water pipes later becoming a source of secondary microbial contamination [34]. Acanthamoeba, Hartmannella, Naegleria and other FLA are known to thrive in areas containing high bacterial content which provide them with nutrition [11, 35]. The prevalence of Cryptosporidium and Giardia in these water sources were not significant but their mere presence is a health concern. The presence of Cryptosporidium and Giardia is due to human and animal faecal contamination of the water sources. Water sources for the communities in QEPA are all exposed to contamination from various points such as (1) hotels whose drainage enters directly into the water bodies, (2) fishing communities living nearby using shallow latrines from which excreta goes directly into the water bodies, (3) wild and domestic animal excreta also directly or indirectly ending up into water used by communities, and (4) Domestic refuse, rotten plants and soil entering into the water.
The mean DO (mg/L) at all the water sources did not meet the minimal requirement of 4.90 mg/L set by Uganda National Bureau of Standards (UNBS). Reduced DO is due to higher levels of TDS in the water. Decreased DO levels can be indicative of a large number bacteria and an excess amount of biological oxygen demand (BOD) due to untreated sewage, organic discharges and anoxic discharges, which deplete DO [36, 37]. Total Dissolved Solids are known to facilitates parasite proliferation [17, 38]. Linear correlation analysis showed that DO level had a negative correlation with the presence of Acanthamoeba spp., Hartmannella spp., other FLA, Cryptosporidium spp. and Giardia spp. Sometimes more parasites were found at slightly lower DO and less at higher DO. Previous studies have demonstrated that FLA, Cryptosporidium and Giardia can survive at low levels of DO [39]. In general, these parasites are reported to be more common in organic matter contaminated, bacteria-rich water irrespective of the physical parameters of the water [32]. However, often, they may be influenced by fluctuations in different physical parameters [40].
This study found pathogenic and non-pathogenic FLA in the water samples from the study sites. The potentially pathogenic FLA identified was Acanthamoeba spp. (T1, T4 and T11), A. polyphaga and H. vermiformis. A. polyphaga is one of the many Acanthamoeba spp. that cause GAE, AK, sinusitis and cutaneous infections especially in HIV/AIDS patients and other immunocompromised individuals [13, 41]. The inclusion of H. vermiformis on the list of human parasites was previously challenged by scientists citing lack of substantial evidence. However, recently H. vermiformis has consistently been mentioned as possible human parasites following its isolation in various mixed human AK infections [14, 41, 42] and as a result of their experimental effects in animal models [43]. A case of mixed keratitis infection reported due to H. vermiformis [14, 41] and another recent report regarding Vahlkampfia avara in Iran, in a mixed infection with Acanthamoeba spp. [44] should place more attention on other FLA not originally considered pathogenic. This means that it is possible for more FLA found in water, soil, dust, and other areas to become virulent anytime, given a conducive environment.
C. parvum and G. lamblia are the most common waterborne pathogens associated with diarrhoea in developing countries [45] and yet there is little concern by authorities in mitigating their effects. The presence of FLA, Cryptosporidium and Giardia in all the water sources in QEPA possess a high risk to the communities. In QEPA, water used by households, park staff, and restaurants comes from River Kyambura, Kazinga channel, Lake George and Edward, where most wild animals inhabit or water themselves. All the human communities living at the periphery of QEPA have their homes located close to the water bodies from which they fetch water for domestic use. During the rainy season, wash off of human and animal faecal matter from land into the water bodies is severe, with an eventual concentration of a large portion of the organic matter at points where communities fetch water from. This, coupled with inadequate water treatment, poor hygiene practices (irregular washing of water vessels), drinking unboiled water and ignorance, predisposes community members to a variety of parasites. Most water parasites are highly abundant during the rainy seasons [46] with a resultant concentration of parasites to points where animals and humans have access to water, dictating a continuous pattern of infection. The QEPA communities whose population is increasing at an alarming rate continues to compete for the same resources with the wildlife. They use the same water for their livestock and families. Sharing of this resource by wildlife and domestic animals with no definitive disease control measures exposes them to a continued cycle of infection. The fact that certain potentially pathogenic waterborne parasites like FLA, Cryptosporidium and Giardia are not often mentioned as possible infectious agents easily transmitted from water and contaminated food poses a risk of silent infections and death [47].
High-level community awareness, policy formulations and regular surveillance is needed in order to mitigate the pathogenic effects of waterborne parasites. This can be possible through “One Health” initiatives involving multidisciplinary teams of animal health workers, medical workers, politicians, quality control officers, local key informants and opinion leaders sharing ideas on the best way forward in improving water safety.
Conclusion
The study shows that the protozoan parasites such as FLA, Cryptosporidium spp. and Giardia spp. found in the water resources in QEPA are a public health concern. This evidence shows the need for the relevant authorities in Uganda to initiate programmes for the control and prevention of diseases caused by these parasites. There must be a commitment from the government to supply clean potable water to the communities and this initiative must be of high priority. Community programmes must be initiated to educate the people on water safety measures, personal hygiene and water treatment processes. This study will, therefore, help in engaging the government in providing the necessary resources to improve community livelihoods of QEPA communities.
Abbreviations
+ve, Positive; °C, Celsius; AK, amoebic keratitis; CNS, Central Nervous System; FLA, free-living amoeba; FLS, fish landing site; g, Grams; g, gravitational force; GAE, granulomatous amoebic encephalitis; HIV/AIDS, Human Immunodeficiency Virus/Acquired Immune Deficiency Syndrome; KCB, Kazinga channel bank; KCM, Kazinga channel mid; L, Liter; mg, Milligrams; ml, Milliliters; mM, millimolar; NCBI, National Center for Biotechnology Information; No., Number; PA, Protected Area; PAM, Primary amoebic meningoencephalitis; pg, picogram; pmol, picomoles; Prev, Prevalence; QEPA, Queen Elizabeth Protected Area; rpm, revolutions per minute; UNESCO, United Nations Educational, Scientific and Cultural Organization; V, Volts; μS, Microseconds.
References
Bengis RG, Leighton FA, Fischer JR, Artois M, Morner T, Tate CM. The role of wildlife in emerging and re-emerging zoonoses. Rev Sci Tech (International Office of Epizootics). 2004;23(2):497–511.
Mathews F. Zoonoses in wildlife integrating ecology into management. Adv Parasitol. 2009;68:185–209.
World Health Organization, The International Network to Promote Household Water Treatment and Safe Storage. Combating waterborne disease at the household level. Geneva: World Health Organization; 2007.
Centers for Disease Control and Prevention, USA. Neglected Parasitic Infections (NPIs) in the United States. Atlanta, USA: Centers for Disease Control and Prevention, United States of America; 2014.
Centers for Disease Control and Prevention, USA. Domestic Water, Sanitation, and Hygiene Epidemiology. Atlanta, USA: Centers for Disease Control and Prevention, United States of America; 2015.
Gebru T, Taha M, Kassahun W. Risk factors of diarrhoeal disease in under-five children among health extension model and non-model families in Sheko district rural community, Southwest Ethiopia: comparative cross-sectional study. BMC Public Health. 2014;14:395.
Visvesvara GS, Moura H, Schuster FL. Pathogenic and opportunistic free-living amoebae: Acanthamoeba spp., Balamuthia mandrillaris, Naegleria fowleri, and Sappinia diploidea. FEMS Immunol Med Microbiol. 2007;50(1):1–26.
Yongsi HB. Pathogenic microorganisms associated with childhood diarrhea in low-and-middle income countries: case study of Yaounde - Cameroon. Int J Environ Res Public Health. 2008;5(4):213–29.
Yongsi HB. Suffering for water, suffering from water: access to drinking-water and associated health risks in Cameroon. J Health Popul Nutr. 2010;28(5):424–35.
Choy SH, Al-Mekhlafi HM, Mahdy MA, Nasr NN, Sulaiman M, Lim YA, Surin J. Prevalence and associated risk factors of Giardia infection among indigenous communities in rural Malaysia. Sci Rep. 2014;4:6909.
Amir S. Acanthamoeba castellanii as a host and model to study bacterial virulence. Solna: Karolinska Universitetssjukhuset Huddinge; 2009.
Dey R, Hoffman PS, Glomski IJ. Germination and amplification of an Appl Environ Microbiol. Appl Environ Microbiol. 2012;78(22):8075–81.
Torno Jr MS, Babapour R, Gurevitch A, Witt MD. Cutaneous acanthamoebiasis in AIDS. J Am Acad Dermatol. 2000;42(2 Pt 2):351–4.
Solgi R, Niyyati M, Haghighi A, Mojarad EN. Occurrence of Thermotolerant Hartmannella vermiformis and Naegleria spp. in hot springs of Ardebil Province, Northwest Iran. Iran J Parasitol. 2012;7(2):47–52.
Buse HY, Lu J, Ashbolt NJ. Exposure to synthetic gray water inhibits amoeba encystation and alters expression of Legionella pneumophila virulence genes. Appl Environ Microbiol. 2015;81(2):630–39.
Cateau E, Imbert C, Rodier M. Hartmanella vermiformis can be permissive for Pseudomonas aeruginosa. Lett Appl Microbiol. 2008;47(5):475–7.
Carmena D: Waterborne transmission of Cryptosporidium and Giardia: detection, surveillance and implications for public health. In: Current Research, Technology and Education topics in Applied Microbiology and Microbial Biotechnology. Edited by Mendez-Vilas. London: MRC Clinical Sciences Centre, Faculty of Medicine, Imperial College, Hammersmith Hospital Campus; 2010: 1-14.
Gideon AA, Njine T, Nola M, Menbohan SF, Ndayo MW. Measuring resistant forms of two pathogenic protozoa (Giardia spp. and Cryptosporidium spp.) in two aquatic biotopes in Yaounde (Cameroon). Sante. 2007;17(3):167–72 (in French).
Bowman DD, Lucio-Forster A. Cryptosporidiosis and giardiasis in dogs and cats: veterinary and public health importance. Exp Parasitol. 2010;124(1):121–7.
Munoz FM, Attias E, Czinn SJ, Black J. Paediatrician and caregiver awareness of cryptosporidiosis and giardiasis in children: US survey responses. Zoonoses Public Health. 2015. doi:10.1111/zph.12246.
Schuster FL, Visvesvara GS. Free-living amoebae as opportunistic and non-opportunistic pathogens of humans and animals. Int J Parasitol. 2004;34(9):1001–27.
Cheesbrough M. District Laboratory Practice in Tropical Countries. 2nd ed. New York, USA: Cambridge University Press; 2006.
Smirnov AV, Goodkov AV. An illustrated list of basic morphotypes of Gymnamoebia (Rhizopoda, Lobosea). Paraitology. 1999;11:20–9.
Pelandakis M, Pernin P. Use of multiplex PCR and PCR restriction enzyme analysis for detection and exploration of the variability in the free-living amoeba Naegleria in the environment. Appl Environ Microbiol. 2002;68(4):2061–5.
Schroeder JM, Booton GC, Hay J, Niszl IA, Seal DV, Markus MB, Fuerst PA, Byers TJ. Use of subgenic 18S ribosomal DNA PCR and sequencing for genus and genotype identification of acanthamoebae from humans with keratitis and from sewage sludge. J Clin Microbiol. 2001;39(5):1903–11.
Booton GC, Kelly DJ, Chu YW, Seal DV, Houang E, Lam DS, Byers TJ, Fuerst PA. 18S ribosomal DNA typing and tracking of Acanthamoeba species isolates from corneal scrape specimens, contact lenses, lens cases, and home water supplies of Acanthamoeba keratitis patients in Hong Kong. J Clin Microbiol. 2002;40(5):1621–5.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–9.
Landrigan PJ, Fuller R. Global health and environmental pollution. Int J Public Health. 2015;60(7):761–2.
Zamxaka A, Pironcheva G, Muyima G. Bacterial community patterns of domestic water sources in the Gogogo and Nkonkobe areas of the Eastern Cape Province, South Africa. Water SA. 2004;30(3):341–6.
World Health Organization. Report of World Health Organization. Geneva: World Health Organisation; 2014.
Munthali S. Transfrontier conservation areas: Integrating biodiversity and poverty alleviation in Southern Africa. Nat Res Forum. 2007;31:51–60.
Bonilla-Lemus P, Caballero Villegas AS, Carmona Jiménez J, Lugo Vázquez A: Occurrence of free-living amoebae in streams of the Mexico Basin. Exper Parasitol. 2014, 145, Supplement:S28-S33.
Sifuentes LY, Choate BL, Gerba CP, Bright KR. The occurrence of Naegleria fowleri in recreational waters in Arizona. J Environ Sci Health A Tox Hazard Subst Environ Eng. 2014;49(11):1322–30.
Rozej A, Cydzik-Kwiatkowska A, Kowalska B, Kowalski D. Structure and microbial diversity of biofilms on different pipe materials of a model drinking water distribution systems. World J Microbiol Biotechnol. 2015;31(1):37–47.
Khan NA. Acanthamoeba: biology and increasing importance in human health. FEMS Microbiol Rev. 2006;30(4):564–95.
Reyher S: A water Quality analysis of the Altamaha River with a focused study in the vicinity of Jesup, Georgia. Bachelor thesis: Georgia Southwestern State University, Geology Department; 2013.
Mehari AK, Gebremedhin S, Ayele B. Effects of bahir dar textile factory effluents on the water quality of the head waters of Blue Nile River, Ethiopia. Int J Anal Chem. 2015;2015:905247.
Siddiqui R, Khan NA. Biology and pathogenesis of Acanthamoeba. Parasit Vectors. 2012;5:6.
Onichandran S, Kumar T, Salibay CC, Dungca JZ, Tabo HA, Tabo N, Tan TC, Lim YA, Sawangjaroen N, Phiriyasamith S, et al. Waterborne parasites: a current status from the Philippines. Parasit Vectors. 2014;7:244.
Al-Gahwari YAK. Physicochemical parameters and microorganisms as water quality indicators of Teluk Bahang Reservoir and Batu Ferringhi Treatment Plant. PhD thesis. Universiti Sains Malaysia. 2007. http://eprints.usm.my/10093/1/PHYSICO-CHEMICAL_PARAMETERS_AND_MICROORGANISMS_AS_WATER_QUALITY_INDICATORS_OF_TELUK_BAHANG_RESERVOIR_AND_BATU_FERRINGHI_TREATMENT_PLANT.pdf.
Lorenzo-Morales J, Martinez-Carretero E, Batista N, Alvarez-Marin J, Bahaya Y, Walochnik J, Valladares B. Early diagnosis of amoebic keratitis due to a mixed infection with Acanthamoeba and Hartmannella. Parasitol Res. 2007;102(1):167–9.
Abedkhojasteh H, Niyyati M, Rahimi F, Heidari M, Farnia S, Rezaeian M. First report of Hartmannella keratitis in a cosmetic soft contact lens wearer in Iran. Iran J Parasitol. 2013;8(3):481–5.
Kinnear F. Cytopathogenicity of Acanthamoeba, Vahlkampfia and Hartmannella: Quantative & qualitative in vitro studies on keratocytes. J Infect. 2003;46(4):228–37.
Niyyati M, Lorenzo-Morales J, Rezaie S, Rahimi F, Martin-Navarro CM, Mohebali M, Maghsood AH, Farnia S, Valladares B, Rezaeian M. First report of a mixed infection due to Acanthamoeba genotype T3 and Vahlkampfia in a cosmetic soft contact lens wearer in Iran. Exper Parasitol. 2010;126(1):89–90.
Yakoob J, Abbas Z, Beg MA, Naz S, Khan R, Islam M, Jafri W. Prevalences of Giardia lamblia and Cryptosporidium parvum infection in adults presenting with chronic diarrhoea. Ann Trop Med Parasitol. 2010;104(6):505–10.
Bonilla-Lemus P, Caballero Villegas AS, Carmona Jiménez J, Lugo Vázquez A: Occurrence of free-living amoebae in streams of the Mexico Basin. Exp Parasitol. 2014, Suppl:S28-33. doi: 10.1016/j.exppara.2014.07.001.
Trabelsi H, Dendana F, Sellami A, Sellami H, Cheikhrouhou F, Neji S, Makni F, Ayadi A. Pathogenic free-living amoebae: epidemiology and clinical review. Pathol Biol (Paris). 2012;60(6):399–405.
Ledee DR, Iovieno A, Miller D, Mandal N, Diaz M, Fell J, Fini ME, Alfonso EC. Molecular identification of t4 and t5 genotypes in isolates from Acanthamoeba keratitis patients. J Clin Microbiol. 2009;47(5):1458–62.
Zhao G, Sun S, Zhao J, Xie L. Genotyping of Acanthamoeba isolates and clinical characteristics of patients with Acanthamoeba keratitis in China. J Med Microbiol. 2010;59(Pt 4):462–6.
Mirjalali H, Niyyati M, Abedkhojasteh H, Babaei Z, Sharifdini M, Rezaeian M. Pathogenic assays of Acanthamoeba belonging to the t4 genotype. Iran J Parasitol. 2013;8(4):530–5.
Obi CL, Potgieter N, Bessong PO, Matsaung G. Scope of potential bacterial agents of diarrhoea and microbial assessment of quality of river water sources in rural Venda communities in South Africa. Water Sci Technol. 2003;47(3):59–64.
Acknowledgements
The researchers would like to thank WaterNet, SIG-RISE and CARNEGIE-Next Generation of African Academics (NGAA-Carnegie), for supporting and funding this research, and Molecular Biology Laboratory, Makerere University for the equipment used in the diagnostics.
Authors’ contributions
CS carried out all the field data collection, culturing, DNA extraction and drafting of the manuscript. JE, MO and IN supervised the data collection, laboratory work and the development of the manuscript. PKM and SO carried out organism cultures and molecular diagnostic studies. BGN developed the field study designs. JM participated in sequence alignment and drafting of the manuscript. CKD carried out the statistical analysis. All authors read and approved the final version of the manuscript.
Authors’ information
CS is a wildlife Veterinarian/public health professional currently an Assistant Lecturer. JE is a Veterinary microbiologist and Associate Professor. MO is an epidemiologist, department Chair and Professor. IN is water safety expert and Lecturer. PKM and SO are seniour laboratory technologists. BGN is a medical Doctor attached to Uganda Virus Institute. JM is a molecular biologist, biochemist and Lecturer. CKD is an Assistant Lecturer and statistician. GS is a laboratory assistant and Masters Student. KM is a laboratory assistant and Masters Student.
Competing interests
The authors declare that they have no competing interests.
Ethics approval and consent to participate
This study does not require ethical ‘approval’.
Author information
Authors and Affiliations
Corresponding author
Additional file
Additional file 1:
Multilingual abstracts in the five official working languages of the United Nations. (PDF 566 kb)
Appendix
Appendix
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Sente, C., Erume, J., Naigaga, I. et al. Prevalence of pathogenic free-living amoeba and other protozoa in natural and communal piped tap water from Queen Elizabeth protected area, Uganda. Infect Dis Poverty 5, 68 (2016). https://doi.org/10.1186/s40249-016-0162-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40249-016-0162-5