Abstract
Background
The role of the human microbiome in health and disease is an emerging and important area of research; however, there is a concern that African populations are under-represented in human microbiome studies. We, therefore, conducted a systematic survey of African human microbiome studies to provide an overview and identify research gaps. Our secondary objectives were: (i) to determine the number of peer-reviewed publications; (ii) to identify the extent to which the researches focused on diseases identified by the World Health Organization [WHO] State of Health in the African Region Report as being the leading causes of morbidity and mortality in 2018; (iii) to describe the extent and pattern of collaborations between researchers in Africa and the rest of the world; and (iv) to identify leadership and funders of the studies.
Methodology
We systematically searched Medline via PubMed, Scopus, CINAHL, Academic Search Premier, Africa-Wide Information through EBSCOhost, and Web of Science from inception through to 1st April 2020. We included studies that characterized samples from African populations using next-generation sequencing approaches. Two reviewers independently conducted the literature search, title and abstract, and full-text screening, as well as data extraction.
Results
We included 168 studies out of 5515 records retrieved. Most studies were published in PLoS One (13%; 22/168), and samples were collected from 33 of the 54 African countries. The country where most studies were conducted was South Africa (27/168), followed by Kenya (23/168) and Uganda (18/168). 26.8% (45/168) focused on diseases of significant public health concern in Africa. Collaboration between scientists from the United States of America and Africa was most common (96/168). The first and/or last authors of 79.8% of studies were not affiliated with institutions in Africa. Major funders were the United States of America National Institutes of Health (45.2%; 76/168), Bill and Melinda Gates Foundation (17.8%; 30/168), and the European Union (11.9%; 20/168).
Conclusions
There are significant gaps in microbiome research in Africa, especially those focusing on diseases of public health importance. There is a need for local leadership, capacity building, intra-continental collaboration, and national government investment in microbiome research within Africa.
Video Abstract
Similar content being viewed by others
Microbiome research in Africa
What is known about this topic? | What are the gaps? | What does this study add to our knowledge? |
There is an exponential growth of microbiome studies in North America and Europe. | The number of African countries where microbiome studies were conducted is unknown. | Microbiome studies were conducted in 61% of the countries in Africa, with the top three being South Africa, Kenya, and Uganda. |
Most of these microbiome studies are dedicated to understanding diseases of public health importance (e.g. cancers, irritable bowel disorder, diabetes, etc.) in these countries. | The extent to which these studies focused on diseases of public health significance in Africa remains uninvestigated. | Only 26.8% (45/168) of the studies focused on diseases of the highest public health importance in Africa, with HIV accounting for 64.4% (29/45). |
The leadership and pattern of collaboration in African human microbiome studies are unknown. | Non-Africans led 79.8% of all the studies, and the most collaborative efforts were between the United States of America and African scientists. There is the need for local leadership, capacity building, intra-continental collaboration, and national government investment in microbiome research within Africa. |
Introduction
The human microbiome plays pivotal roles in immune and brain development, nutrition, and metabolism [1, 2]. Imbalances in the gut microbiome have been associated with impairment and diseases of many organ systems [1] including cancers [3, 4], obesity [5], asthma [6, 7], allergy, inflammatory bowel disease, and metabolic diseases [1]. More recent reports have added sickle cell disease [8], brain disorders, and behaviors to the growing list of diseases [9]. Although the causal basis for many microbiome associations is unknown, the microbiome is likely to be key to precision medicine approaches [10].
In order for the microbiome field to contribute effectively to personalized medicine, it is imperative to draw an accurate picture of the human microbiome in health and disease. Almost all research into human health is dependent on context. This is particularly true for microbiome research as gut microbiomes, for example, vary extensively based on geography, age, diet, ethnicity, genetics, disease, medication, climate, and other environmental factors [1]. Consequently, there is an urgent need to characterize the microbiome of as many unique populations as possible.
The microbiomes of western populations have been extensively characterized; however, information regarding the microbiome of residents of Africa is considerably sparser. Microbiome studies extending our understanding of important diseases must be replicated in Africa due to context-specific factors [11]. In particular, environmental determinants may vary [12,13,14,15], and genomic heterogeneity [16] within the human population is more marked compared to other continents. Important environmental exposures include diet, geography, climate, infectious diseases, urbanization, living conditions, and pollution [11,12,13,14]. These variabilities preclude the generalization of microbiome studies conducted in one specific population in Africa to the entire continent. Therefore, the representation of diverse African participants in microbiome studies is a priority.
Although non-communicable diseases, including cancers, diabetes, and cardiovascular diseases, have emerged as public health threats in both developed and developing countries, Africa has an additional burden of infectious diseases [17]. Infections account for at least 70% of all deaths on the continent [18], including malaria, tuberculosis, HIV/AIDS, and neglected tropical diseases (Buruli ulcer, trypanosomiasis, schistosomiasis, and guinea worm) [17]. Lower respiratory infections, HIV/AIDS, diarrheal diseases, malaria, preterm birth complications, tuberculosis, neonatal sepsis/infections, stroke, and ischaemic heart diseases are responsible for the highest morbidity and mortality in Africa [19]. Health-related research in Africa, including microbiome-based research, must address the diseases that are of foremost public health importance.
A number of human microbiome studies have been conducted in Africa. Although Brewster and colleagues [14] have provided a survey of microbiome research conducted in Africa, this addressed only gut microbiome studies. Currently, no study has summarized all human microbiome research conducted in Africa in order to identify knowledge gaps and areas for further research. We, therefore, undertook a systematic survey of human microbiome studies involving African participants to provide an overview of and to identify research gaps in the field. Our secondary objectives were: (i) to determine the overall number of peer-reviewed publications; (ii) to identify the extent to which the researches focused on diseases identified by the WHO State of Health in the African Region Report 2018 as being the leading causes of morbidity and mortality [19]; (iii) to provide information on the extent and pattern of collaboration between researchers in Africa and the rest of the world; and (iv) to identify leadership and the main funders of these studies.
Materials and methods
Search terms and strategy
This review followed the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) guidelines [20]. A comprehensive literature search was undertaken from inception through to 1st April 2020 using the following databases: Medline via PubMed, Scopus, ISI Web of Science (Web of Knowledge), and Academic Search Premier, Africa-Wide Information, and CINAHL via EBSCOhost according to the search strategy outlined in (Supplementary Table S1). No filters were applied to any of the searches. All citations were exported into ENDNOTE (X9; Thomson Reuters). The search was independently conducted by two reviewers IA and REA. The reference lists of reviews were searched for eligible papers that were not recovered by the search terms.
Study selection criteria
Studies were included only if they meet all of the following criteria: (i) human studies involving residents of Africa only or as part of a multinational study regardless of age, sex, health status, study design, or care setting; (ii) published in English or French; and (iii) described either bacteria, archaea, fungi, viruses, or parasites identified from any human samples using next-generation sequencing (NGS) including both shotgun metagenomics and targeted amplicon sequencing. Our exclusion criteria were: (i) studies that did not include any human participants from African; (ii) those that utilized publicly available data on African participants; (iii) studies that did not characterize the microbiome; and (iii) studies that did not utilize NGS to characterize the microbiome or those that targeted only specific microorganisms in their analysis.
Screening of studies
Records retrieved from the literature search of the six databases were independently downloaded into ENDNOTE (X9; Thomson Reuters) by two reviewers (IA and REA). These reviewers independently removed duplicates, reviews, commentaries, editorials, notes, news, and opinions. They then screened the title and abstract of residual articles against the inclusion and exclusion criteria. The full texts of the studies that passed this stage were retrieved. The reviewers proceeded to independently review these full texts based on the eligibility criteria. At each stage of the process, the two reviewers compared their results and disagreements were resolved by mutual discussion.
Data extraction and synthesis
Once consensus was reached on which articles to include in the study, IA and REA independently extracted data into a predesigned data extraction table in Microsoft ExcelTM. The data extracted included the country of origin of the samples; techniques used to analyze the microbiome, disease of focus, type of sample, participants metadata (number, age, gender, ethnicity, geographic region of the participants), aims and conclusions of the studies, whether the participants were from rural or urban settings, source of funding for the studies, country location of institutions to which the participating scientists were affiliated, name of the journal, first and last author’s information, and information on data availability. The extracted data were compared for accuracy and merged. IA and REA analyzed the merged data separately, and the results were compared for accuracy.
Under funding, any institute under the National Institute of Health (NIH) and European Union (EU) were captured as NIH and EU, respectively during the analysis. Furthermore, only agencies that directly funded the studies via project-specific grants were captured. Those that indirectly supported the research by providing training grants, scholarships, or fellowships to specific authors were not reported as funders. This is because our objective was to highlight organizations that directly funded human microbiome studies in Africa and we were not able to directly determine whether the funds from these sources were directly invested in the microbiome project reported.
Where studies were multinational, we captured only the number of participants from the African cohort. In this situation as well, we listed all the countries involved but highlighted the African countries in bold typeface. Information not specified in the full-text article or its supplementary data were captured as “NA.” Rural/urban designation of the sample’s origin was only indicated when specified in the article using words such as “rural” (rural), “village” (rural), “city” (urban), and “town” (semi-urban or peri-urban or semi-rural). The Human Microbiome Project (HMP) classification of body sites was used to categorize the sample types. The age range was divided into four categories; young children (0 to 5 years), older children (6 to 12 years), adolescents (13 to 17 years), and adults (18 years and above). We determined the article’s accessibility to African researchers by checking if the paper is designated open-access at the journal website or if the journal itself is open-access or if the paper can be obtained from PubMed Central.
To determine the extent to which the studies focused on diseases of high public health importance in Africa, we analyzed the number of studies that focus on any of the following conditions identified in the World Health Organization [WHO] State of Health in the African Region 2018 Report as being in the top 10 causes of morbidity and mortality in Africa: lower respiratory infections, HIV/AIDS, diarrheal diseases, malaria, preterm birth complications, tuberculosis, neonatal sepsis/infections, stroke, and ischemic heart diseases.
Results
Results of the search
The search yielded 5515 records (including three articles from additional sources [hand-searching]) with 3066 remaining after removing duplicates. From these records, 2811 were excluded because of ineligibility, and 255 full-text articles were further assessed for eligibility. After a full-text review, a total of 168 eligible human microbiome studies were obtained. Figure 1 shows the PRISMA flowchart summarizing the steps followed in the selection of the final subset of papers used in the analysis.
Human microbiome research publications in Africa
We found 168 published articles that utilized NGS technology to characterize the human microbiome among African participants. Five broad study designs were used, with cross-sectional studies being the most common (46.4%, 78/168) (Tables 1, 2, and 3). Other designs utilized in the studies were case-control (20.8%, 35/168), randomized control trial (14.3%, 24/168), longitudinal (8.9%, 15/168), and cohort design (8.9%, 15/168). One study involved both longitudinal and cross-sectional designs. The majority of the studies (73.2%, 123/168) involved only one sampling time point. The studies were published in 86 different peer-reviewed journals. The most frequent journal of publication was PLoS One (13.1%, 22/168) followed by Scientific Reports (4.8%, 8/168), mBio (3.6%, 6/168), Microbiome (3.0%, 5/168), and PLOS Neglected Tropical Diseases (3.0%, 5/168). More than half of all the studies (67.8%, 114/168) were only published between January 2017 and March 2020 (Fig. 2). A total of 140/168 (83.3%) studies were published as open-access in subscription-based journals or open-access journals or available via PubMed Central and are therefore accessible to researchers based in Africa.
Distribution of studies across Africa
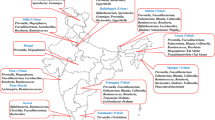
We analyzed the African countries of sample origin for all 168 eligible papers. The included studies collected samples from participants residing in 33 of the 54 countries in Africa (61%) (Fig. 3). The countries with the highest number of studies were South Africa (16.1%, 27/168), Kenya (13.7%, 23/168), and Uganda (10.7%, 18/168). Tanzania (7.1%, 12/168), Malawi (7.1%, 12/168), and Nigeria (6.5%, 11/168) also had a moderate number of studies conducted in them. The 27 remaining countries had less than ten studies each. Regionally, most of the studies were conducted in East Africa (39.9%, 67/168) followed by Southern (29.8%, 50/168), West (29.2%, 49/168), Central (7.7%, 13/168), and North Africa (6.5%, 11/168). The region with the highest coverage of countries was West Africa, where studies were conducted in 11/15 countries (73%). This was followed by Central Africa 4/7 (57%), East Africa 8/14 (57%), and North Africa 4/7 (57%) and finally Southern Africa 6/11 (55%).
Body sites, sample types, methodology, and data archiving
The gastrointestinal tract (GIT) was the most studied body site (52.4%, 88/168, Table 1 and Table S2a), followed by the urogenital tract (24.4%, 41/168, Table 2 and Table S2b) while the eye (1.2%, 2/168, Table 3 and Table S2c) and placenta (0.6%, 1/168, Table 3 and Table S2c) were the least studied sites (Fig. 4). Similarly, the predominant sample type studied was stool (47.6%, 80/168) followed by vaginal samples (16.1%, 27/168). Placenta and fecal membrane samples were the least frequently studied (0.6%, 1/168). A total of 144 studies investigated the bacterial component, while 14 characterized the virome of the human microbiota. One study each focused on only fungi and only protozoa. While two studies investigated both bacteria and viruses, one each focused on bacteria and fungi collectively, and then bacteria and protozoa. Two studies explored viruses, bacteria, and the protozoal component of the microbiota while the remaining two studies investigated helminths in addition to the former three.
The majority of studies characterizing the bacteriome used only 16S rRNA amplicon sequencing (73.8%, 124/168) while 14/168 (8.3%) used only shotgun metagenomic sequencing. Eleven studies (6.5%) used both methods. One study targeted the cpn60 gene in place of the 16S rRNA gene for bacteriome characterization. For virome studies, shotgun metagenomics was used in seven studies while targeted methods including RNA sequencing, phage sequencing, and VirCapSeq-VERT were used in four studies. Targeted sequencing of the ITS1 of 18S rRNA and 5.8S conserved fungal region was used to characterize the mycobiome in one study. One study conducted full-length 16S rRNA sequencing while two utilized both 16S rRNA sequencing and ITS2. One study included both MALDI-TOF culturomics and 16S rRNA sequencing technologies. Two studies failed to specify the method used; however, they were included in the final analysis because they utilized high throughput sequencing. The platform most commonly used was Illumina MiSeq (57.1%, 96/168), followed by Roche 454 pyrosequencer (22.6%, 38/168) and Illumina HiSeq (14.3%, 24/168). Of the 168 studies, 64 (38.1%) did not indicate whether their data are publicly available (Tables S2a-S2c). However, for those that did, most (29.8%, 50/168) deposited their sequence data in the National Center for Biotechnology Institute Sequence Read Archive (NCBI-SRA). Similarly, 20/168 (11.9%) data sets were archived in The European Nucleotide Archive (ENA), 9/168 (5.4%) in GenBank, 5/168 (3.0%) in Metagenomics Rapid Annotation using Subsystems Technology (MG-RAST), 3/168 (1.8%) in Open Science Framework (OSF), and 2/168 (1.2%) in the DNA Data Bank of Japan, and 1/168 (0.6%) in NCBI Gene Expression Omnibus (GEO). Ten studies (6.0%) deposited data in more than one of the repositories mentioned above, while four studies (2.4%) indicated that they would make their data available upon request.
Study participant information
More studies (42.2%, 71/168) investigated adult [≥ 18 years] microbiomes than those of young children [0 to 5 years] (23.8%, 40/168). No study focused on only older children [6 to 12 years old] or adolescents [13 to 17 years] (Fig. 5). However, 31.5% (53/168) of the studies compared the microbiomes of more than one age group. While 51.8% (87/168) of the studies included both males and females, 24.4% (41/168) and 11.3% (19/168) included only females and only males, respectively. The sex of participants was not specified in 19 (11.3%) studies. The two (1.2%) remaining studies included mothers and their infants however, the sex of the infants was undefined. A total of 84.5% (142/168) of the studies did not specify the ethnicity of their participants. While 31% (52/168) of studies focused on participants in rural settings and 4.8% (8/168) investigated microbiomes of urban dwellers, five studies (3%) collected samples from residents in peri-urban communities and 54.7% (92/168) did not specify whether their participants were from rural or urban settings. Only two studies compared microbiomes of participants from rural, urban and semi-urban settings. Eight studies compared rural and urban while one study compared rural and semi-urban residents’ microbiomes. Most studies [60% (101/168)] included less than 100 participants, while 31.5% (59/168) studies enrolled 100 to 499 participants. Six studies included 500 to 999 participants. Only two studies involved 1000 or more people (Fig. 5). In total, Nigeria, the Gambia, Kenya, Malawi, South Africa, and Uganda had the microbiome of more than 1000 residents characterized. Additionally, several of the studies were derived from the same cohort of people [21] and [22,23,24] and [25,26,27,28] and [29, 30] and [31, 32] and [33]. Figure 6 and Figure 7 summarize the gut and urogenital studies in Africa.
African map with pie charts showing the age categories and the number of participants included in human microbiome studies per country. The size of the divisions within the pie charts corresponds to the proportion of studies that included each age category (young children (0 to 5 years), older children (6 to 17 years), and adults (≥ 18 years)). The size of the pie chart represents the cumulative number of participants from all studies conducted in the country
African map with pie charts showing the age categories and the number of participants included in human gut microbiome studies per country. The size of the divisions within the pie charts corresponds to the proportion of studies that included each age category (young children (0 to 5 years), older children (6 to 17 years), and adults (≥ 18 years)). The size of the pie chart represents the cumulative number of participants from all studies conducted in the country
African map with pie charts showing the age categories and the number of participants included in human urogenital microbiome studies per country. The size of the divisions within the pie charts corresponds to the proportion of studies that included each age category (adolescents (13 to 17 years) and adults (≥ 18 years)). The size of the pie chart represents the cumulative number of participants from all studies conducted in the country
Diseases of focus of the studies
To identify the extent to which the studies focused on diseases of major public health importance in Africa, we analyzed the diseases of focus. Of the 168 eligible studies, 38.1% (64/168) did not focus on any specific disease (Fig. 4). Of the remaining 61.9% (104/168) that investigated the microbiome in the context of a specific disease, 45 studies focused on the top nine diseases responsible for the highest morbidity and mortality in Africa. They are as follows: lower respiratory infections (4), HIV/AIDS (29), diarrheal diseases (6), malaria (2), preterm birth complications (1), tuberculosis (1), and neonatal sepsis/infections (2). Other diseases that were frequently studied included malnutrition (8/104), bacterial vaginosis (5/104), obesity only (2/104), diabetes only (2/104), obesity and diabetes (1/104), and metabolic syndrome (1/104). Under neglected tropical diseases, only one study investigated Buruli ulcer, two each focused on trachoma and schistosomiasis and four on other parasitic infections (helminths and blastocystis). Fifteen studies examined non-communicable diseases (cancers, anemia, atopic dermatitis, environmental enteric dysfunction, and toxic blood metal levels).
Intercontinental and intra-continental collaborations among study co-authors
We analyzed the countries of institutional affiliation of all authors on each manuscript in order to understand the extent and pattern of collaborations between researchers in Africa and the rest of the world. For within-country collaborations, 17 studies had all the collaborating scientists based within the same country [Egypt (6), USA (5), South Africa (3), France (2), and Germany (1)]. Out of these, Egypt and South Africa were the only African countries where the collaborating scientists were from the same country. Furthermore, seven of the studies that involved researchers collaborating from more than one country did not include any African scientists as an author (Table 4). Asian countries whose scientists collaborated with African scientists included China, India, Bangladesh, Indonesia, Thailand, and Vietnam. Scientists from South America who collaborated with African scientists were based in Colombia, Brazil, Puerto Rico, Venezuela, and Chile. A total of 85.7% (144/168) of the studies involved intercontinental collaborations between one African country and one or more non-African countries (Fig. 8). Among these studies, the most significant collaborative efforts were between scientists in the USA and African countries, mainly South Africa (13/168), Uganda (12/168), Kenya (10/168), and Malawi (10/168). Intercontinental collaboration was also common between African scientists and researchers based in the UK, Canada, and the Netherlands.
Leadership in microbiome studies
To determine the extent to which these studies were led by African scientists, we analyzed the countries of institutional affiliations of the first (Fig. 9A), and the senior (last) authors of the studies (Fig. 9B) as proxies. Among first authors with a single country of institutional affiliation, 43.5% (73/168) were from the USA, 6.5% (11/168) from South Africa, 4.8% (8/168) from Canada, and France and 3% (5/168) from Germany. A total of 12.5% (21/168) were affiliated with institutions in more than one country. Out of these 21 studies, the first authors of 13/21 were affiliated to both an African institution and a non-African institution while 8/21 were affiliated to two institutions from different non-African countries. Only one study had the first author affiliated to institutions in two African countries (South Africa and Zimbabwe).
South Africa 6.5% (11/168), Egypt 3.6% (6/168), and Nigeria 1.8% (3/168) were the only African countries that had a scientist with a single African country of institutional affiliation as the first author.
In contrast, other first authors with affiliations to institutions in Africa (Nigeria, Kenya, Mali, Botswana, Malawi, Morocco, Zambia, and Niger) also concurrently held affiliations to institutions in non-African countries mainly the USA and the UK. Regionally, the majority of these authors were from Southern and East Africa (5/13 from Southern Africa and 4/13 from East Africa compared to 3/13 from West Africa, one from North, and none from Central Africa).
The affiliations of the last authors followed a similar pattern (Fig. 9B); 42.3% (71/168) were from the USA, followed by Canada 6% (10/168), France 6% (10/168), South Africa 4.8% (8/168) and Germany, Switzerland and the UK (3.6% (6/168) each). Thirteen percent (22/168) were affiliated to institutions in more than one country, mainly the USA, the UK, and Australia. Similar to the observation made with the first authors, South Africa (8/168), Nigeria (2/168), and Egypt (6/168), and, in this case, Morocco (1/168) were the only African countries that had a scientist with a single African country of institutional affiliation as the last author.
African researchers from South Africa, Zimbabwe, Kenya, Malawi, Uganda, and The Gambia were also simultaneously affiliated to institutions in other countries outside the African continent, mainly the USA and the UK. Regionally, the majority of these authors were from Southern and East Africa (11/17 from Southern Africa and 4/17 from East Africa compared to 2/17 from West Africa and none from North and Central Africa). Using the first and last authors as proxies for the leadership of studies, we found that 79.8% of all the studies had first and/or last authors affiliated to institutions outside Africa.
Funding
We analyzed the agencies that directly supported the studies by awarding research grants. We found that funding from USA sources predominated (Fig. 2), with more than 70% of the studies partially or fully funded by American governmental institutions, foundations, and agencies. These included the National Institute of Health (NIH) 76/168 (45.2%), Bill and Melinda Gates Foundation 30/168 (17.8%), the United States National Science Foundation 8/168 (4.8%), and Blood Systems Research Institutes 5/168 (3.0%). This was followed by the European Union 20/168 (11.9%), through the European and Developing Countries Clinical Trials Partnership (EDCTP) (4/20), European Research Council (5/20), European Union Regional development fund (2/20), European Union’s Seventh Framework program (3/20) and other European Union agencies (6/20). Other funding sources included the Wellcome Trust (UK) 14/168 (8.3%), and the Canadian Institute of Health Research (CIHR) 8/168 (4.8%). It is noteworthy that South Africa is the only African country to have funded a published microbiome study on the continent.
Discussion
We conducted a systematic survey of studies that utilized NGS to characterize the human microbiome of residents of Africa. Our results revealed that up to 1st April 2020, 168 published studies utilized NGS to characterize the human microbiome of African participants. Of the 61.9% (104) of studies that examined the microbiome in the context of disease, less than half (43.3%, 45/104) focused on diseases that are responsible for the highest morbidity and mortality in Africa with HIV/AIDS accounting for 29/45 studies alone. With regard to collaboration, partnerships between the USA and African scientists were most common. However, the leadership of these studies (first and last authorship) was mainly assumed by the American scientists.
African human microbiome publications
With the advances in NGS extending over a decade, it is interesting that half of all the studies were only published within the past 3 years (Fig. 2). It is, however, disappointing that only 168 studies investigated the African human microbiome using this technology. Considering that Africa is made up of 54 independent countries with extremely diverse genetic backgrounds and cultures and is also the second most populous continent with a population of 1.3 billion (2018 estimates [34]), Africa is under-represented in the global microbiome literature. More than half of the studies involved less than 100 participants further suggestive of reduced coverage. Additionally, several of the publications were derived from the same cohort of people [21] and [22,23,24] and [25,26,27,28] and [29, 30] and [31, 32] and [33], which further reduces the diversity and coverage of African people included in microbiome studies. Since the literature search extended only to April 2020, the numbers reflected for this year are lower.
Most studies (73.2%) involved a single sampling time point. Cross-sectional designs are appropriate for studies that aim to describe the microbiome signatures associated with a particular outcome of interest [35]. However, owing to high within-subject and between-subject variability and the influence of environmental factors, longitudinal study designs with multiple temporally-separated sampling points are recommended for more robust and reproducible results [35]. Cross-sectional designs were common probably because of the following factors: budgetary constraints, invasiveness of sampling procedure, participant compliance to study protocol, and availability of samples in the case of retrospective studies [35]. With regard to budgetary constraints, multiplexing techniques [36, 37] allow multiple samples from the same or even different origins to be processed and sequenced together. This technique substantially reduces sequencing costs.
Most (83.3%) of the studies were either published in open access journals or as open access articles in subscription-based journals or freely accessible through PubMed Central. This may be due to the open-access revolution that has gained ground in the scientific world [38]. Furthermore, the open access publishing policies adopted by the top funders (NIH, Bill and Melinda Gates Foundation, and Wellcome Trust) of the studies may also explain this observation [39]. Open-access publishing of studies conducted in Africa is crucial because the majority of libraries in African universities struggle to afford expensive subscriptions to prominent publishing companies. Although journal access initiatives such as WHO Health InterNetwork Access to Research Initiative (HINARI) [40, 41] allow access to some of these subscription-based journals, some vital research articles remain behind a paywall. Open access publishing will, therefore, improve access to studies conducted on the continent to researchers, students, and the general public. Access to research already conducted in Africa will inform, equip and encourage African scientists to engage in microbiome research. It will also encourage intra-continental collaboration by increasing the visibility of African researchers who already have the capacity to undertake microbiome research.
Similarly, 61.9% of the studies mentioned storing their sequence data in publicly available repositories mainly NCBI-SRA. The increase in data archiving for public access is fuelled in part by funder [39] and journal requirements [42]. This will allow the secondary use of the data by other researchers, particularly those in Africa who may not have the funding, capacity, and facilities to generate such data. The preference for NCBI may be influenced by the fact that most of the studies were led by America scientists who may be more familiar with NCBI-SRA than the other repositories.
The countries where most studies were conducted were in East and Southern Africa. This may be influenced by the fact that most of the first and last authors who had multiple affiliations (from both African and non-African institutions) were from East and Southern Africa. Therefore, these scientists have more opportunities through their North American/European affiliations to foster collaborations outside Africa and also secure funding for microbiome studies in these specific regions of the continent. Another reason for the over-representation of Eastern and Southern Africans in the microbiome studies may be the higher prevalence of HIV in these parts of Africa (20 million in Eastern and Southern Africa compared to 6 million in West, Central, and North Africa collectively in 2018 [43]. As a high proportion of studies focused on HIV/AIDS (29/168 compared to less than 10 for any other disease), it follows that more of such studies will be situated in these two regions to permit the recruitment of required large numbers. However, Africans have widely different genetic and cultural backgrounds [16] and this diversity may affect their microbiomes [1, 35, 44]. This variability argues for broader coverage of residents of Africa from all regions in microbiome studies.
Most of the studies reported very little metadata related to participants. For instance, 54.7% of the studies did not specify whether participants are from rural or urban areas. Other studies mentioned the hospitals where the patients were recruited without specifying any further details about the location of residence of the participants themselves. This specification is important because Africans in cities are increasingly adopting western diets and lifestyles compared to those in rural areas [12, 45]. This change in lifestyle can confound microbiome associations found in studies and must, therefore, be accounted for. Indeed, Lokmer et al. found that Cameroonians along an urbanization gradient differed by diet, habitat, and socio-cultural conditions, and this affected their gut and salivary microbiomes [46]. This difference further underscores the importance of collecting as much metadata as possible for microbiome studies.
Ethnicity information was not collected in 84.5% of the studies. Ethnicity may directly impact the microbiome, but more importantly, it is frequently strongly associated with a specific culture, lifestyle, and diet, which in turn affect the microbiome [12, 47, 48]. Failure to collect this information may be because ethnicity is not always easy to define. Also, in studies in localized geographic areas, ethnicity may be relatively homogenous and therefore not the focus of the research. Furthermore, ethnicity may be confounded by the increasing frequency of inter-marriage. Additional metadata that would add value to studies include disease status, medication exposure, family history, socio-economic status, and lifestyle (diet, smoking, alcohol consumption, physical activity) [49].
Most of the studies (124/168) utilized 16S rRNA metagenomic sequencing to profile the bacterial component of the microbiome. This limits the number of studies that have looked at the fungi, viral, and eukaryotic components of the African microbiome [50]. These other components are also important in health and disease [51, 52] and therefore warrant attention. The extensive use of 16S rRNA metagenomic sequencing limits the resolution of microbial profiles to genus level [50]. It also fails to provide the genomic as well as functional contexts of the bacteria identified [50]. Decreasing the cost of shotgun metagenomic sequencing and simplifying bioinformatic analysis techniques will tip the scale toward this superior methodology.
African human microbiome studies focusing on diseases of significant public health concern
Apart from HIV/AIDS, which was the focus of 29 studies, few studies focused on diseases among the top 10 diseases of public health importance in Africa. Human microbiome studies focusing on diseases including malaria, diarrheal diseases, pneumonia, tuberculosis were limited. This may be due to the perceived relative low contribution of the microbiome to each of these diseases. However, the role of the microbiome in these conditions cannot be completely ruled out as limited research has been conducted in these areas. Metabolic diseases including obesity and diabetes that are mediated by the microbiome were also sparsely studied. These conditions are also highly prevalent in Africa and warrant microbiome-based investigation [17, 53]. While 38.1% of studies did not characterize the microbiome in the context of any particular disease, research on healthy individuals are important to establish what the “normal” or “healthy” microbiome is for comparative purposes. Additionally, although certain conditions such as bacterial vaginosis are not part of the top 10 diseases of public health importance, they are still relevant health issues in Africa, particularly for reproductive health outcomes which are a focus of the United Nation’s Sustainable Development goals.
The extent and pattern of collaboration with researchers in Africa and the rest of the world
African scientists collaborated most commonly with American scientists on microbiome research projects with the latter often assuming leadership. Reasons for this observation are not known but could be speculated. One factor may be that the American partners were the principal investigators of the grants funding the studies. They may also have conducted the laboratory investigations, data analysis, and drafting of the manuscripts. The African collaborator’s primary role may have only been recruitment and sample collection [54]. The practice of scientists from the global north using African scientists as conduits to obtain samples, then shipping them away without building the capacity of their African partners or directly benefiting the continent is commonly known as “helicopter research”. To address this phenomenon, the H3Africa consortium ethics working group developed a guideline in 2018 on the ethical handling of genomic samples from Africa [55]. It calls for investigators from the global north to build the capacity of their African collaborators to equip them to work independently post projects [55]. The guideline also invites western researchers to allow for substantial intellectual contribution from African scientists on studies that draw on samples recovered from the continent [55]. For this guideline to effectively combat “helicopter research,” funding agencies could specify local capacity building as a condition for awarding grants to western scientists who partner with African scientists. Institutional review boards in Africa could consider making capacity building a requirement in studies that involve international collaboration.
Additionally, African governments must recognize the importance of research and invest in microbiome studies. Apart from South Africa, through the Department of Science and Technology, no other African country directly funded any of the microbiome research projects identified here. This factor may also contribute to African scientists’ inability to initiate and therefore lead microbiome studies. Intra-continental collaboration within Africa was almost non-existent, possibly hampered by lack of funding and language barrier. Similar findings were made by Boshoff, who investigated intra-regional research collaboration among countries within the Southern African Development Community (SADC) [56]. This author found only 3% and 5% of intra-regional and continental collaboration respectively in contrast to 47% inter-continental collaboration with high-income countries [56]. Onyancha et al. also observed a similar pattern for research collaborations in sub-Saharan Africa [57] where intra-continental collaboration was minimal compared to inter-continental north-south partnerships. To encourage intra-continental collaborations, Onyancha recommended regional conferences as well as student and staff exchanges [57]. However, these exchanges will have a limited impact if researchers cannot access funding to conduct projects. Lack of pathways to independent funding necessitates outside collaboration and is therefore likely to be a key limitation for African leadership on articles and grants. Access to independent funding streams is the most important factor that should be tackled to address low African leadership of microbiome studies. High reagent costs associated with microbiome studies in Africa also frequently result in the shipping of samples out of the region. Microbiome research in Africa would be greatly improved by efforts to reduce the cost per sample for assays such as 16S amplicon sequencing.
International collaborations with non-African partners followed colonial ties [54] as well as commonality of a national language, with African scientists from Francophone countries collaborating with French scientists, while English-speaking western countries partnered with Anglophone African researchers. An African scientist who collaborates with a western scientist increases his/her chances of securing funds for research, and this may explain the preference for international collaboration. Indeed, several funding agencies specifically make international collaboration, usually with a partner from the funder’s own country, a requirement for funding. This requirement further discourages intra-continental collaboration.
Computational resources to handle bioinformatics analysis are also scarce on the continent, making inter-continental partnerships important. However, the H3Africa Consortium [58] through its subsidiary, the H3ABioNet [59] has launched many initiatives to build capacities for African scientists to lead and conduct microbiome research in Africa. Additional efforts [59], including workshops by other agencies, are being made to further build bioinformatics capacity in Africa [11].
Conclusion
Residents in Africa are under-represented in human microbiome studies. There is a need to build capacity for microbiome research in Africa, increase collaboration among scientists within Africa, and ensure equitable partnerships with international collaborators. African governments and research funding agencies should identify microbiome research as a priority area for investigation and funding.
Limitations
Certain studies utilized the same cohort resulting in multiple counting of the same individuals. Funding information was sometimes difficult to extract as some authors did not clearly distinguish personal funding from project funds. Some African researchers may receive internal funding from within their research institutions, which may not be captured in our review. African scientists may travel abroad for educational purposes, and during this period may be affiliated with non-African institutions Although they may still return to Africa in leadership positions, this could not be assessed in this review. The number of studies that focused on priority health care areas of Africa may be underestimated due to the exclusion of publications that did not employ NGS technology.
Availability of data and materials
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
Change history
19 January 2022
A Correction to this paper has been published: https://doi.org/10.1186/s40168-022-01226-x
Abbreviations
- AD:
-
Atopic dermatitis
- AIDS:
-
Acquired immune deficiency syndrome
- BV:
-
Bacterial vaginosis
- BVAB:
-
Bacterial vaginosis-associated bacteria
- CAPRISA:
-
Centre for the AIDS Program of Research in South Africa
- CDC:
-
Centers for Disease Control and Prevention
- CIHR:
-
Canadian Institute of Health Research
- CINAHL:
-
Current Nursing and Allied Health Literature
- CIPHER:
-
Collaborative Initiative for Pediatric HIV Education and Research
- Cpn60 UT:
-
Chaperonin-60 Universal Target
- EDCTP:
-
European and Developing Countries Clinical Trials Partnership
- EED:
-
Environmental enteric dysfunction
- ENA:
-
European Nucleotide Archive
- ETH Global:
-
Swiss Federal Institute of Technology
- EU:
-
European Union
- GEMS:
-
Global Enterics Multicenter Study
- GOS:
-
Galacto-oligosaccharides
- H. pylori :
-
Helicobacter pylori
- HCV:
-
Hepatitis C virus
- HIV:
-
Human immunodeficiency virus infection
- HMOs:
-
Human milk oligosaccharides
- HMP:
-
Human Microbiome Project
- hrHPV:
-
High-risk human papillomavirus infection
- IAVI:
-
International AIDS Vaccine Initiative
- Ion Torrent PGM:
-
Ion Torrent Personal Genome Machine
- MALDI-TOF:
-
Matrix-assisted laser desorption/ionization-time-of-flight
- MG-RAST:
-
Metagenomic rapid annotations using subsystems technology
- MNP:
-
Micronutrient powder
- MTCT:
-
Mother-to-child transmission
- NA:
-
Not available
- NCBI-SRA:
-
National Center for Biotechnology Institute Sequence Read Archive
- NGS:
-
Next-Generation Sequencing
- NIH:
-
National Institute of Health
- NSF:
-
National Science Foundation
- OSF:
-
Open Science Framework
- P. falciparum :
-
Plasmodium falciparum
- PCR:
-
Polymerase chain reaction
- pH:
-
Potential of hydrogen
- PHiD-CV:
-
Pneumococcal nontypeable Haemophilus influenzae protein conjugate vaccine
- PNAS:
-
Proceedings of the National Academy of Sciences
- PrEP:
-
Partners pre-exposure prophylaxis study
- PRISMA:
-
Preferred Reporting Items for Systematic Reviews and Meta-analyses
- rDNA:
-
Ribosomal deoxyribonucleic acid
- rRNA:
-
Ribosomal ribonucleic acid
- SAM:
-
Severe acute malnutrition
- STI:
-
Sexually transmitted infections
- UK:
-
United Kingdom
- USA:
-
United States of America
- USAID:
-
United States Agency for International Development
- WHO:
-
World Health Organization
References
Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV, Knight R. Current understanding of the human microbiome. Nat Med. 2018;24:392–400.
Kho ZY, Lal SK. The human gut microbiome – a potential controller of wellness and disease. Front Microbiol. 2018;9:1835.
Allali I, Boukhatem N, Bouguenouch L, Hardi H, Boudouaya HA, Cadenas MB, et al. Gut microbiome of Moroccan colorectal cancer patients. Med Microbiol Immunol. 2018;207:211–25.
Sepich-Poore GD, Zitvogel L, Straussman R, Hasty J, Wargo JA, Knight R. The microbiome and human cancer. Science. 2021;371:eabc4552.
Cani PD, Van Hul M. Gut microbiota and obesity: causally linked? Expert Rev Gastroenterol Hepatol. 2020;14:401–3.
Stokholm J, Blaser MJ, Thorsen J, Rasmussen MA, Waage J, Vinding RK, et al. Maturation of the gut microbiome and risk of asthma in childhood. Nat Commun. 2018;9:141.
Huang YJ, Nariya S, Harris JM, Lynch SV, Choy DF, Arron JR, et al. The airway microbiome in patients with severe asthma: associations with disease features and severity. J Allergy Clin Immunol. 2015;136:874–84.
Lim SH, Morris A, Li K, Fitch AC, Fast L, Goldberg L, et al. Intestinal microbiome analysis revealed dysbiosis in sickle cell disease. Am J Hematol. 2018;93:E91–3.
Iannone LF, Preda A, Blottière HM, Clarke G, Albani D, Belcastro V, et al. Microbiota-gut brain axis involvement in neuropsychiatric disorders. Expert Rev Neurother. 2019;19:1037–50.
Cheng M, Cao L, Ning K. Microbiome big-data mining and applications using single-cell technologies and metagenomics approaches toward precision medicine. Front Genet. 2019;10:109–18.
Reid G, Nduti N, Sybesma W, Kort R, Kollmann TR, Adam R, et al. Harnessing microbiome and probiotic research in sub-Saharan Africa: recommendations from an African workshop. Microbiome. 2014;2.
Gupta VK, Paul S, Dutta C. Geography, ethnicity or subsistence-specific variations in human microbiome composition and diversity. Front Microbiol. 2017;8:1162.
De Filippo C, Di Paola M, Ramazzotti M, Albanese D, Pieraccini G, Banci E, et al. Diet, environments, and gut microbiota. A preliminary investigation in children living in rural and urban Burkina Faso and Italy. Front Microbiol. 1979;2017:8.
Brewster R, Tamburini FB, Asiimwe E, Oduaran O, Hazelhurst S, Bhatt AS. Surveying gut microbiome research in Africans: toward improved diversity and representation. Trends Microbiol. 2019;27:824–35.
Fragiadakis GK, Smits SA, Sonnenburg ED, Van Treuren W, Reid G, Knight R, et al. Links between environment, diet, and the hunter-gatherer microbiome. Gut Microbes. 2019;10:216–27.
Choudhury A, Aron S, Sengupta D, Hazelhurst S, Ramsay M. African genetic diversity provides novel insights into evolutionary history and local adaptations. Hum Mol Genet. 2018;27.
Gouda HN, Charlson F, Sorsdahl K, Ahmadzada S, Ferrari AJ, Erskine H, et al. Burden of non-communicable diseases in sub-Saharan Africa, 1990–2017: results from the Global Burden of Disease Study 2017. Lancet Glob Health. 2019;7:e1375–87.
Young F, Critchley JA, Johnstone LK, Unwin NC. A review of co-morbidity between infectious and chronic disease in Sub Saharan Africa: TB and diabetes mellitus, HIV and metabolic syndrome, and the impact of globalization. Glob. Health. 2009;5.
World Health Organization (WHO). The state of health in the WHO Africa Region: an analysis of the status of health, health services and health systems in the context of the Sustainable Development Goals. 2018. https://apps.who.int/iris/handle/10665/275292. Accessed 15 July 2020.
Moher D, Liberati A, Tetzlaff J, Altman DG, Altman D, Antes G, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6:e1000097.
Bisanz JE, Enos MK, Mwanga JR, Changalucha J, Burton JP, Gloor GB, et al. Randomized Open-label pilot study of the influence of probiotics and the gut microbiome on toxic metal levels in Tanzanian pregnant women and school children. mBio. 2014;5:e01580–14.
Bisanz JE, Enos MK, PrayGod G, Seney S, Macklaim JM, Chilton S, et al. Microbiota at multiple body sites during pregnancy in a rural tanzanian population and effects of Moringa-supplemented probiotic yogurt. Appl Environ Microbiol. 2015;81:4965–75.
Lennard KS, Dabee S, Barnabas SL, Havyarimana E, Jaumdally SZ, Botha G, et al. High rates of bacterial vaginosis and chlamydia in a low-income, high-population-density community in Cape Town. Suid-Afr Tydskr Vir Natuurwetenskap En Tegnol. 2017;36:a1495.
Lennard K, Dabee S, Barnabas SL, Havyarimana E, Blakney A, Jaumdally SZ, et al. Microbial Composition predicts genital tract inflammation and persistent bacterial vaginosis in South African adolescent females. Infect Immun. 2018;86.
Lennard K, Dabee S, Barnabas SL, Havyarimana E, Blakney A, Jaumdally SZ, et al. Vaginal microbiota varies by geographical location in South African women. Suid-Afr Tydskr Vir Natuurwetenskap En Tegnol. 2019;38:41-9.
Liu CM, Hungate BA, Tobian AA, Serwadda D, Ravel J, Lester R, et al. Male circumcision significantly reduces prevalence and load of genital anaerobic bacteria. mBio. 2013;4:e00076.
Liu CM, Hungate BA, Tobian AAR, Ravel J, Prodger JL, Serwadda D, et al. Penile microbiota and female partner bacterial vaginosis in Rakai, Uganda. mBio. 2015;6:e00589.
Liu CM, Prodger JL, Tobian AAR, Serwadda D, Galiwango RM, Nalugoda F, et al. Genital anaerobic bacterial overgrowth and the PrePex male circumcision device, Rakai, Uganda. J Infect Dis. 2016;214:595–8.
Liu CM, Prodger JL, Tobian AAR, Abraham AG, Kigozi G, Hungate BA, et al. Penile anaerobic dysbiosis as a risk factor for HIV infection. mBio. 2017;8:e00996–17.
Hummelen R, Fernandes AD, Macklaim JM, Dickson RJ, Changalucha J, Gloor GB, et al. Deep sequencing of the vaginal microbiota of women with HIV. PLoS One. 2010;5:e12078.
McMillan A, Rulisa S, Sumarah M, Macklaim JM, Renaud J, Bisanz JE, et al. A multi-platform metabolomics approach identifies highly specific biomarkers of bacterial diversity in the vagina of pregnant and non-pregnant women. Sci Rep. 2015;5:14174.
Rampelli S, Schnorr SL, Consolandi C, Turroni S, Severgnini M, Peano C, et al. Metagenome sequencing of the Hadza Hunter-gatherer gut microbiota. Curr Biol. 2015;25:1682–93.
Schnorr SL, Candela M, Rampelli S, Centanni M, Consolandi C, Basaglia G, et al. Gut microbiome of the Hadza hunter-gatherers. Nat Commun. 2014;5:3654.
Nations, Department of Economic and Social Affairs, Population Division (2019). World Population Prospects 2019: Highlights. https://population.un.org/wpp/Publications/Files/WPP2019_Highlights.pdf. Accessed 24 July 2020.
Goodrich JK, Di Rienzi SC, Poole AC, Koren O, Walters WA, Caporaso JG, et al. Conducting a microbiome study. Cell. 2014;158:250–62.
Armanhi JSL, de Souza RSC, de Araújo LM, Okura VK, Mieczkowski P, Imperial J, et al. Multiplex amplicon sequencing for microbe identification in community-based culture collections. Sci Rep. 2016;6:29543.
Fadrosh DW, Ma B, Gajer P, Sengamalay N, Ott S, Brotman RM, et al. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome. 2014;2:6–6.
Beal J. What the open-acess movement doesn’t want you to know. Academe. 2015;101, 37(3):–40.
Glover SW, Webb A, Gleghorn C. Open access publishing in the biomedical sciences: could funding agencies accelerate the inevitable changes? Health Inf Libr J. 2006;23:197–202.
Aronson B. WHO’s Health InterNetwork Access to Research Initiative (HINARI). Health Inf Libr J. 2002;19:164–5.
Aronson B, Glover S. Journal access initiatives: where are they? Why the World Health Organization? and What is HINARI? Ser Rev. 2005;31:279–83.
Sholler D, Ram K, Boettiger C, Katz DS. Enforcing public data archiving policies in academic publishing: a study of ecology journals. Big Data Soc. 2019;6:205395171983625.
Mahy M, Marsh K, Sabin K, Wanyeki I, Daher J, Ghys PD. HIV estimates through 2018. AIDS. 2019;33:S203–11.
Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555:210–5.
Arowolo D. The effects of western civilisation and culture on Africa. Afro Asian. J Soc Sci. 2010;1:1–3.
Lokmer A, Aflalo S, Amougou N, Lafosse S, Froment A, Tabe FE, et al. Response of the human gut and saliva microbiome to urbanization in Cameroon. Sci Rep. 2020;10:2856.
Gaulke CA, Sharpton TJ. The influence of ethnicity and geography on human gut microbiome composition. Nat Med. 2018;24:1495–6.
Hansen MEB, Rubel MA, Bailey AG, Ranciaro A, Thompson SR, Campbell MC, et al. Population structure of human gut bacteria in a diverse cohort from rural Tanzania and Botswana. Genome Biol. 2019;20:16.
Kim D, Hofstaedter CE, Zhao C, Mattei L, Tanes C, Clarke E, et al. Optimizing methods and dodging pitfalls in microbiome research. Microbiome. 2017;5:52.
Poretsky R, Rodriguez-R LM, Luo C, Tsementzi D, Konstantinidis KT. Strengths and limitations of 16S rRNA gene amplicon sequencing in revealing temporal microbial community dynamics. PLoS One. 2014;9:e93827.
Filyk HA, Osborne LC. The Multibiome: The Intestinal ecosystem’s influence on immune homeostasis, health, and disease. EBioMedicine. 2016;13:46–54.
Sartor RB, Wu GD. Roles for intestinal bacteria, viruses, and fungi in pathogenesis of inflammatory bowel diseases and therapeutic approaches. Gastroenterology. 2017;152:327–339.e4.
Institute for Health Metrics and Evaluation, Human Development Network, The World Bank. The Global Burden of Disease: Generating Evidence, Guiding Policy – Sub-Saharan Africa Regional Edition. 2013. http://www.healthdata.org/sites/default/files/files/data_for_download/2013/WorldBank_SubSaharanAfrica/IHME_GBD_WorldBank_SubSaharanAfrica_FullReport.pdf. Accessed 04 Aug 2020.
Boshoff N. Neo-colonialism and research collaboration in Central Africa. Scientometrics. 2009;81:413–34.
Nordling L. African scientists call for more control of their continent’s genomic data. Nature. 2018. Available from: http://www.nature.com/articles/d41586-018-04685-1. Accessed 07 Aug 2020.
Boshoff N. South–South research collaboration of countries in the Southern African Development Community (SADC). Scientometrics. 2010;84:481–503.
Onyancha OB, Maluleka JR. Knowledge production through collaborative research in sub-Saharan Africa: how much do countries contribute to each other’s knowledge output and citation impact? Scientometrics. 2011;87:315–36.
Adoga MP, Fatumo SA, Agwale SM. H3Africa: a tipping point for a revolution in bioinformatics, genomics and health research in Africa. Source Code Biol Med. 2014;9:10.
Mulder NJ, Adebiyi E, Alami R, Benkahla A, Brandful J, Doumbia S, et al. H3ABioNet, a sustainable pan-African bioinformatics network for human heredity and health in Africa. Genome Res. 2016;26:271–7.
Seck EH, Senghor B, Merhej V, Bachar D, Cadoret F, Robert C, et al. Salt in stools is associated with obesity, gut halophilic microbiota and Akkermansia muciniphila depletion in humans. Int J Obes. 2019;43:862–71.
Phan TG, Vo NP, Bonkoungou IJO, Kapoor A, Barro N, O’Ryan M, et al. Acute diarrhea in West African children: diverse enteric viruses and a novel parvovirus genus. J Virol. 2012;86:11024–30.
Oldenburg CE, Sié A, Coulibaly B, Ouermi L, Dah C, Tapsoba C, et al. Indirect effect of azithromycin use on the intestinal microbiome diversity of untreated children: a randomized trial. Open Forum. Infect Dis. 2019;6.
De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U A. 2010;107:14691–6.
Oldenburg CE, Sié A, Coulibaly B, Ouermi L, Dah C, Tapsoba C, et al. Effect of commonly used pediatric antibiotics on gut microbial diversity in preschool children in Burkina Faso: a randomized clinical trial. Open Forum. Infect Dis. 2018;5.
Lokmer A, Cian A, Froment A, Gantois N, Viscogliosi E, Chabe M, et al. Use of shotgun metagenomics for the identification of protozoa in the gut microbiota of healthy individuals from worldwide populations with various industrialization levels. PLoS One. 2019;14:e0211139.
Morton ER, Lynch J, Froment A, Lafosse S, Heyer E, Przeworski M, et al. Variation in rural African gut microbiota is strongly correlated with colonization by Entamoeba and subsistence. PLOS Genet. 2015;11:e1005658.
Yinda CK, Vanhulle E, Conceição-Neto N, Beller L, Deboutte W, Shi C, et al. Gut virome analysis of cameroonians reveals high diversity of enteric viruses, including potential interspecies transmitted viruses. mSphere. 2019;4:e00585–18.
Vonaesch P, Morien E, Andrianonimiadana L, Sanke H, Mbecko J-R, Huus KE, et al. Stunted childhood growth is associated with decompartmentalization of the gastrointestinal tract and overgrowth of oropharyngeal taxa. Proc Natl Acad Sci. 2018;115:E8489–98.
Gomez A, Petrzelkova KJ, Burns MB, Yeoman CJ, Amato KR, Vlckova K, et al. Gut microbiome of coexisting BaAka Pygmies and Bantu reflects gradients of traditional subsistence patterns. Cell Rep. 2016;14:2142–53.
Gomez A, Sharma AK, Mallott EK, Petrzelkova KJ, Jost Robinson CA, Yeoman CJ, et al. Plasticity in the human gut microbiome defies evolutionary constraints. mSphere. 2019;4:e00271–19.
Campbell TP, Sun X, Patel VH, Sanz C, Morgan D, Dantas G. The microbiome and resistome of chimpanzees, gorillas, and humans across host lifestyle and geography. ISME J. 2020;14:1584–99.
Brazier L, Elguero E, Koumavor CK, Renaud N, Prugnolle F, Thomas F, et al. Evolution in fecal bacterial/viral composition in infants of two central African countries (Gabon and Republic of the Congo) during their first month of life. PLoS One. 2017;12:e0185569.
Schneeberger PHH, Becker SL, Pothier JF, Duffy B, N’Goran EK, Beuret C, et al. Metagenomic diagnostics for the simultaneous detection of multiple pathogens in human stool specimens from Côte d’Ivoire: a proof-of-concept study. Infect Genet Evol. 2016;40:389–97.
Schneeberger PHH, Coulibaly JT, Panic G, Daubenberger C, Gueuning M, Frey JE, et al. Investigations on the interplays between Schistosoma mansoni, praziquantel and the gut microbiome. Parasit Vectors. 2018;11:168.
Aly AM, Adel A, El-Gendy AO, Essam TM, Aziz RK. Gut microbiome alterations in patients with stage 4 hepatitis C. Gut Pathog. 2016;8:42.
Sahly N, Moustafa A, Zaghloul M, Salem TZ. Effect of radiotherapy on the gut microbiome in pediatric cancer patients: a pilot study. PeerJ. 2019;7:e7683.
Salah M, Azab M, Ramadan A, Hanora A. New insights on obesity and diabetes from gut microbiome alterations in Egyptian adults. Omics. 2019;23:477–85.
Shankar V, Gouda M, Moncivaiz J, Gordon A, Reo NV, Hussein L, et al. Differences in gut metabolites and microbial composition and functions between Egyptian and U.S. children are consistent with their diets. mSystems. 2017;2:e00169–16.
Aiemjoy K, Altan E, Aragie S, Fry DM, Phan TG, Deng X, et al. Viral species richness and composition in young children with loose or watery stool in Ethiopia. BMC Infect Dis. 2019;19:53.
Altan E, Aiemjoy K, Phan TG, Deng X, Aragie S, Tadesse Z, et al. Enteric virome of Ethiopian children participating in a clean water intervention trial. PLoS One. 2018;13:e0202054.
Lane AA, McGuire MK, McGuire MA, Williams JE, Lackey KA, Hagen EH, et al. Household composition and the infant fecal microbiome: the INSPIRE study. Am J Phys Anthropol. 2019;169:526–39.
Davis JCC, Lewis ZT, Krishnan S, Bernstein RM, Moore SE, Prentice AM, et al. Growth and morbidity of Gambian infants are influenced by maternal milk oligosaccharides and infant gut microbiota. Sci Rep. 2017;7:40466.
Dugas LR, Bernabé BP, Priyadarshini M, Fei N, Park SJ, Brown L, et al. Decreased microbial co-occurrence network stability and SCFA receptor level correlates with obesity in African-origin women. Sci Rep. 2018;8:17135.
Easton AV, Quiñones M, Vujkovic-Cvijin I, Oliveira RG, Kepha S, Odiere MR, et al. The impact of anthelmintic treatment on human gut microbiota based on cross-sectional and pre- and postdeworming comparisons in western Kenya. mBio. 2019;10:e00519.
Jaeggi T, Kortman GA, Moretti D, Chassard C, Holding P, Dostal A, et al. Iron fortification adversely affects the gut microbiome, increases pathogen abundance and induces intestinal inflammation in Kenyan infants. Gut. 2015;64:731–42.
Mandal RK, Crane RJ, Berkley JA, Gumbi W, Wambua J, Ngoi JM, et al. Longitudinal analysis of infant stool bacteria communities before and after acute febrile malaria and artemether-lumefantrine treatment. J Infect Dis. 2019;220:687–98.
Mosites E, Sammons M, Otiang E, Eng A, Noecker C, Manor O, et al. Microbiome sharing between children, livestock and household surfaces in western Kenya. PLoS One. 2017;12:e0171017.
Schellenberg JJ, Links MG, Hill JE, Dumonceaux TJ, Kimani J, Jaoko W, et al. Molecular definition of vaginal microbiota in East African commercial sex workers. Appl Environ Microbiol. 2011;77:4066–74.
Paganini D, Uyoga MA, Kortman GAM, Cercamondi CI, Moretti D, Barth-Jaeggi T, et al. Prebiotic galacto-oligosaccharides mitigate the adverse effects of iron fortification on the gut microbiome: a randomised controlled study in Kenyan infants. Gut. 2017;66:1956–67.
Paganini D, Uyoga MA, Kortman GAM, Cercamondi CI, Winkler HC, Boekhorst J, et al. Iron-containing micronutrient powders modify the effect of oral antibiotics on the infant gut microbiome and increase post-antibiotic diarrhoea risk: a controlled study in Kenya. Gut. 2019;68:645–53.
Pop M, Walker AW, Paulson J, Lindsay B, Antonio M, Hossain M, et al. Diarrhea in young children from low-income countries leads to large-scale alterations in intestinal microbiota composition. Genome Biol. 2014;15:R76.
Tang M, Frank D, Hendricks A, Ir D, Esamai F, Liechty E, et al. Iron in micronutrient powder promotes an unfavorable gut microbiota in Kenyan infants. Nutrients. 2017;9:776.
Rosa BA, Supali T, Gankpala L, Djuardi Y, Sartono E, Zhou Y, et al. Differential human gut microbiome assemblages during soil-transmitted helminth infections in Indonesia and Liberia. Microbiome. 2018;6:33.
Cheung YB, Xu Y, Mangani C, Fan Y-M, Dewey KG, Salminen SJ, et al. Gut microbiota in Malawian infants in a nutritional supplementation trial. Trop Med Int Health. 2016;21:283–90.
Kortekangas E, Kamng’ ona AW, Fan Y, Cheung YB, Ashorn U, Matchado A, et al. Environmental exposures and child and maternal gut microbiota in rural Malawi. Paediatr Perinat Epidemiol. 2020;34:161–70.
Kortekangas E, Young R, Cheung YB, Fan YM, Jorgensen JM, Kamng’ ona AW, et al. A prospective study on child morbidity and gut microbiota in rural Malawi. J Pediatr Gastroenterol Nutr. 2019;69:431–7.
Ordiz MI, May TD, Mihindukulasuriya K, Martin J, Crowley J, Tarr PI, et al. The effect of dietary resistant starch type 2 on the microbiota and markers of gut inflammation in rural Malawi children. Microbiome. 2015;3:37.
Ordiz MI, Stephenson K, Agapova S, Wylie KM, Maleta K, Martin J, et al. Environmental enteric dysfunction and the fecal microbiota in Malawian children. Am J Trop Med Hyg. 2017;96:473–6.
Popovic A, Bourdon C, Wang PW, Guttman DS, Voskuijl W, Grigg ME, et al. Design and application of a novel two-amplicon approach for defining eukaryotic microbiota. Microbiome. 2018;6:228.
Reyes A, Blanton LV, Cao S, Zhao G, Manary M, Trehan I, et al. Gut DNA viromes of Malawian twins discordant for severe acute malnutrition. Proc Natl Acad Sci U A. 2015;112:11941–6.
Smith MI, Yatsunenko T, Manary MJ, Trehan I, Mkakosya R, Cheng J, et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science. 2013;339:548–54.
Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–7.
Yooseph S, Kirkness EF, Tran TM, Harkins DM, Jones MB, Torralba MG, et al. Stool microbiota composition is associated with the prospective risk of Plasmodium falciparum infection. BMC Genomics. 2015;16:631.
Kodio A, Coulibaly D, Koné AK, Konaté S, Doumbo S, Guindo A, et al. Blastocystis colonization is associated with increased diversity and altered gut bacterial communities in healthy Malian children. Microorganisms. 2019;7:649.
Liu J, Almeida M, Kabir F, Shakoor S, Qureshi S, Zaidi A, et al. Direct detection of shigella in stool specimens by use of a metagenomic approach. J Clin Microbiol. 2018;56.
Maiga M, Cohen K, Baya B, Srikrishna G, Siddiqui S, Sanogo M, et al. Stool microbiome reveals diverse bacterial ureases as confounders of oral urea breath testing for Helicobacter pylori and Mycobacterium tuberculosis in Bamako. Mali. J Breath Res. 2016;10:036012.
Doan T, Arzika AM, Ray KJ, Cotter SY, Kim J, Maliki R, et al. Gut microbial diversity in antibiotic-naive children after systemic antibiotic exposure: a randomized controlled trial. Clin Infect Dis. 2017;64:1147–53.
Doan T, Hinterwirth A, Arzika AM, Cotter SY, Ray KJ, O’Brien KS, et al. Mass azithromycin distribution and community microbiome: a cluster-randomized trial. Open Forum. Infect Dis. 2018;5.
Doan T, Hinterwirth A, Worden L, Arzika AM, Maliki R, Abdou A, et al. Gut microbiome alteration in MORDOR I: a community-randomized trial of mass azithromycin distribution. Nat Med. 2019;25:1370–6.
Ray KJ, Cotter SY, Arzika AM, Kim J, Boubacar N, Zhou Z, et al. High-throughput sequencing of pooled samples to determine community-level microbiome diversity. Ann Epidemiol. 2019;39:63–8.
Tidjani Alou M, Million M, Traore SI, Mouelhi D, Khelaifia S, Bachar D, et al. Gut bacteria missing in severe acute malnutrition, can we identify potential probiotics by culturomics? Front Microbiol. 2017;8:899.
Doumatey AP, Adeyemo A, Zhou J, Lei L, Adebamowo SN, Adebamowo C, et al. Gut Microbiome profiles are associated with type 2 diabetes in urban Africans. Front Cell Infect Microbiol. 2020;10:63.
Afolayan AO, Ayeni FA, Moissl-Eichinger C, Gorkiewicz G, Halwachs B, Högenauer C. Impact of a nomadic pastoral lifestyle on the gut microbiome in the Fulani living in Nigeria. Front Microbiol. 2019;10:2138.
Ayeni FA, Biagi E, Rampelli S, Fiori J, Soverini M, Audu HJ, et al. Infant and adult gut microbiome and metabolome in rural Bassa and urban settlers from Nigeria. Cell Rep. 2018;23:3056–67.
Cinek O, Kramna L, Mazankova K, Odeh R, Alassaf A, Ibekwe MU, et al. The bacteriome at the onset of type 1 diabetes: a study from four geographically distant African and Asian countries. Diabetes Res Clin Pract. 2018;144:51–62.
Nowak RG, Bentzen SM, Ravel J, Crowell TA, Dauda W, Ma B, et al. Rectal microbiota among HIV-uninfected, untreated HIV, and treated HIV-infected in Nigeria. AIDS. 2017;31:857–62.
Nowak RG, Bentzen SM, Ravel J, Crowell TA, Dauda W, Ma B, et al. Anal microbial patterns and oncogenic human papillomavirus in a pilot study of Nigerian men who have sex with men at risk for or living with HIV. AIDS Res Hum Retroviruses. 2019;35:267–75.
Colson P, Fancello L, Gimenez G, Armougom F, Desnues C, Fournous G, et al. Evidence of the megavirome in humans. J Clin Virol. 2013;57:191–200.
Lagier J-C, Armougom F, Million M, Hugon P, Pagnier I, Robert C, et al. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect. 2012;18:1185–93.
Krishnamoorthy S, Coetzee V, Kruger J, Potgieter H, Buys EM. Dysbiosis signatures of fecal microbiota in South African infants with respiratory, gastrointestinal, and other diseases. J Pediatr. 2020;218:106–113.e3.
Mahdavinia M, Rasmussen HE, Engen P, Van den Berg JP, Davis E, Engen K, et al. Atopic dermatitis and food sensitization in South African toddlers: role of fiber and gut microbiota. Ann Allergy Asthma Immunol. 2017;118:742–743.e3.
Ocvirk S, Wilson AS, Posma JM, Li JV, Koller KR, Day GM, et al. A prospective cohort analysis of gut microbial co-metabolism in Alaska Native and rural African people at high and low risk of colorectal cancer. Am J Clin Nutr. 2020;111:406–19.
Junhai O, Carbonero F, Zoetendal EG, DeLany JP, Mei W, Newton K, et al. Diet, microbiota, and microbial metabolites in colon cancer risk in rural Africans and African Americans. Am J Clin Nutr. 2013;98:111–20.
Wood LF, Brown BP, Lennard K, Karaoz U, Havyarimana E, Passmore J-ASAS, et al. Feeding-related gut microbial composition associates with peripheral T-cell activation and mucosal gene expression in African infants. Clin Infect Dis. 2018;67:1237–46.
Claassen-Weitz S, Gardner-Lubbe S, Nicol P, Botha G, Mounaud S, Shankar J, et al. HIV-exposure, early life feeding practices and delivery mode impacts on faecal bacterial profiles in a South African birth cohort. Sci Rep. 2018;8:5078.
Smits SA, Leach J, Sonnenburg ED, Gonzalez CG, Lichtman JS, Reid G, et al. Seasonal cycling in the gut microbiome of the Hadza hunter-gatherers of Tanzania. Science. 2017;357:802–5.
Monaco CL, Gootenberg DB, Zhao G, Handley SA, Ghebremichael MS, Lim ES, et al. Altered virome and bacterial microbiome in human immunodeficiency virus-associated acquired immunodeficiency syndrome. Cell Host Microbe. 2016;19:311–22.
Kristensen KHS, Wiese M, Rytter MJH, Özçam M, Hansen LH, Namusoke H, et al. Gut Microbiota in children hospitalized with oedematous and non-oedematous severe acute malnutrition in Uganda. PLoS Negl Trop Dis. 2016;10:e0004369.
Carruthers LV, Moses A, Adriko M, Faust CL, Tukahebwa EM, Hall LJ, et al. The impact of storage conditions on human stool 16S rRNA microbiome composition and diversity. PeerJ. 2019;7:e8133.
Atukunda P, Muhoozi GKM, van den Broek TJ, Kort R, Diep LM, Kaaya AN, et al. Child development, growth and microbiota: follow-up of a randomized education trial in Uganda. J Glob Health. 2019;9:10431.
Ellis RJ, Bruce KD, Jenkins C, Stothard JR, Ajarova L, Mugisha L, et al. Comparison of the distal gut microbiota from people and animals in Africa. PLoS One. 2013;8:e54783.
Flygel TT, Sovershaeva E, Classen-Weitz S, Hjerde E, Mwaikono KS, Odland JØ, et al. Composition of gut microbiota of children and adolescents with perinatal HIV infection taking antiretroviral therapy in Zimbabwe. J Infect Dis. 2019;221:483–92.
Bourke CD, Gough EK, Pimundu G, Shonhai A, Berejena C, Terry L, et al. Cotrimoxazole reduces systemic inflammation in HIV infection by altering the gut microbiome and immune activation. Sci Transl Med. 2019;11:eaav0537.
Kay GL, Millard A, Sergeant MJ, Midzi N, Gwisai R, Mduluza T, et al. Differences in the faecal microbiome in Schistosoma haematobium Infected children vs. uninfected children. PLoS Negl Trop Dis. 2015;9:e0003861.
Frank DN, Manigart O, Leroy V, Meda N, Valea D, Zhang W, et al. Altered vaginal microbiota are associated with perinatal mother-to-child transmission of HIV in African women from Burkina Faso. J Acquir Immune Defic Syndr. 2012;60:299–306.
Mehta SD, Green SJ, Maclean I, Hu H, Bailey RC, Gillevet PM, et al. Microbial diversity of genital ulcer disease in men enrolled in a randomized trial of male circumcision in Kisumu, Kenya. PLoS One. 2012;7:e38991.
Sivapalasingam S, McClelland RS, Ravel J, Ahmed A, Cleland CM, Gajer P, et al. An effective intervention to reduce intravaginal practices among HIV-1 uninfected Kenyan women. AIDS Res Hum Retroviruses. 2014;30:1046–54.
Wessels JM, Lajoie J, Vitali D, Omollo K, Kimani J, Oyugi J, et al. Association of high-risk sexual behaviour with diversity of the vaginal microbiota and abundance of Lactobacillus. PLoS One. 2017;12:e0187612.
Wessels JM, Lajoie J, Cooper MIJH, Omollo K, Felker AM, Vitali D, et al. Medroxyprogesterone acetate alters the vaginal microbiota and microenvironment in a Kenyan sex worker cohort and is also associated with increased susceptibility to HIV-1 in humanized mice. Dis Model Mech. 2019;12.
Masha SC, Owuor C, Ngoi JM, Cools P, Sanders EJ, Vaneechoutte M, et al. Comparative analysis of the vaginal microbiome of pregnant women with either Trichomonas vaginalis or Chlamydia trachomatis. PLoS One. 2019;14:e0225545.
McClelland RS, Lingappa JR, Srinivasan S, Kinuthia J, John-Stewart GC, Jaoko W, et al. Evaluation of the association between the concentrations of key vaginal bacteria and the increased risk of HIV acquisition in African women from five cohorts: a nested case-control study. Lancet Infect Dis. 2018;18:554–64.
Korhonen CJ, Srinivasan S, Huang D, Ko DL, Sanders EJ, Peshu NM, et al. Semen bacterial concentrations and HIV-1 RNA shedding among HIV-1-seropositive Kenyan men. J Acquir Immune Defic Syndr. 2017;74:250–7.
Joag V, Obila O, Gajer P, Scott MC, Dizzell S, Humphrys M, et al. Impact of standard bacterial vaginosis treatment on the genital microbiota, immune milieu, and ex vivo human immunodeficiency virus susceptibility. Clin Infect Dis. 2019;68:1675–83.
Dareng EO, Ma B, Famooto AO, Adebamowo SN, Offiong RA, Olaniyan O, et al. Prevalent high-risk HPV infection and vaginal microbiota in Nigerian women. Epidemiol Infect. 2016;144:123–37.
Adebamowo SN, Ma B, Zella D, Famooto A, Ravel J, Adebamowo C. Mycoplasma hominis and Mycoplasma genitalium in the vaginal microbiota and persistent high-risk human papillomavirus infection. Front Public Health. 2017;5:140.
Adebayo AS, Survayanshi M, Bhute S, Agunloye AM, Isokpehi RD, Anumudu CI, et al. The microbiome in urogenital schistosomiasis and induced bladder pathologies. PLoS Negl Trop Dis. 2017;11:e0005826.
Verwijs MC, Agaba SK, Darby AC, van de Wijgert JHHM. Impact of oral metronidazole treatment on the vaginal microbiota and correlates of treatment failure. Am J Obstet Gynecol. 2020;222:157.e1–157.e13.
Benning L, Golub ET, Anastos K, French AL, Cohen M, Gilbert D, et al. Comparison of lower genital tract microbiota in HIV-infected and uninfected women from Rwanda and the US. PLoS One. 2014;9:e96844.
McMillan A, Rulisa S, Gloor GB, Macklaim JM, Sumarah M, Reid G. Pilot assessment of probiotics for pregnant women in Rwanda. PLoS One. 2018;13:e0195081.
Onywera H, Williamson A-L, Mbulawa ZZA, Coetzee D, Meiring TL. Factors associated with the composition and diversity of the cervical microbiota of reproductive-age Black South African women: a retrospective cross-sectional study. PeerJ. 2019;7:e7488.
Onywera H, Williamson A-L, Mbulawa ZZA, Coetzee D, Meiring TL. The cervical microbiota in reproductive-age South African women with and without human papillomavirus infection. Papillomavirus Res. 2019;7:154–63.
Feng Y, Jaratlerdsiri W, Patrick SM, Lyons RJ, Haynes A, Collins CC, et al. Metagenomic analysis reveals a rich bacterial content in high‐risk prostate tumors from African men. Prostate. 2019;79:1731–8.
Balle C, Lennard K, Dabee S, Barnabas SL, Jaumdally SZ, Gasper MA, et al. Endocervical and vaginal microbiota in South African adolescents with asymptomatic Chlamydia trachomatis infection. Sci Rep. 2018;8:11109.
Anahtar MN, Byrne EH, Doherty KE, Bowman BA, Yamamoto HS, Soumillon M, et al. Cervicovaginal bacteria are a major modulator of host inflammatory responses in the female genital tract. Immunity. 2015;42:965–76.
Klatt NR, Cheu R, Birse K, Zevin AS, Perner M, Noël-Romas L, et al. Vaginal bacteria modify HIV tenofovir microbicide efficacy in African women. Science. 2017;356:938–45.
Gosmann C, Anahtar MN, Handley SA, Farcasanu M, Abu-Ali G, Bowman BA, et al. Lactobacillus-deficient cervicovaginal bacterial communities are associated with increased HIV acquisition in young South African women. Immunity. 2017;46:29–37.
Klein C, Gonzalez D, Samwel K, Kahesa C, Mwaiselage J, Aluthge N, et al. Relationship between the Cervical Microbiome, HIV Status, and Precancerous Lesions. mBio. 2019;10:e02785–18.
Price LB, Liu CM, Johnson KE, Aziz M, Lau MK, Bowers J, et al. The effects of circumcision on the penis microbiome. PLoS One. 2010;5:e8422.
Liu CM, Packman ZR, Abraham AG, Serwadda DM, Nalugoda F, Aziz M, et al. The effect of antiretroviral therapy initiation on the vaginal microbiome in HIV-infected women. Open Forum. Infect Dis. 2019;6.
Price JT, Vwalika B, Hobbs M, Nelson JAE, Stringer EM, Zou F, et al. Highly diverse anaerobe-predominant vaginal microbiota among HIV-infected pregnant women in Zambia. PLoS One. 2019;14:e0223128.
Gudza-Mugabe M, Havyarimana E, Jaumdally S, Garson KL, Lennard K, Tarupiwa A, et al. Human immunodeficiency virus infection is associated with preterm delivery independent of vaginal microbiota in pregnant African women. J Infect Dis. 2020;221:1194–203.
Pickering H, Palmer CD, Houghton J, Makalo P, Joof H, Derrick T, et al. Conjunctival microbiome-host responses are associated with impaired epithelial cell health in both early and late stages of trachoma. Front Cell Infect Microbiol. 2019;9:297.
Zhou Y, Holland MJ, Makalo P, Joof H, Roberts CH, Mabey DC, et al. The conjunctival microbiome in health and trachomatous disease: a case control study. Genome Med. 2014;6:99.
Kelly MS, Surette MG, Smieja M, Pernica JM, Rossi L, Luinstra K, et al. The nasopharyngeal microbiota of children with respiratory infections in Botswana. Pediatr Infect Dis J. 2017;36:e211–8.
Kelly MS, Surette MG, Smieja M, Rossi L, Luinstra K, Steenhoff AP, et al. Pneumococcal colonization and the nasopharyngeal microbiota of children in Botswana. Pediatr Infect J. 2018;37:1176–83.
Feazel LM, Santorico SA, Robertson CE, Bashraheil M, Scott JA, Frank DN, et al. Effects of vaccination with 10-valent pneumococcal non-typeable Haemophilus influenza protein D conjugate vaccine (PHiD-CV) on the nasopharyngeal microbiome of Kenyan toddlers. PLoS One. 2015;10:e0128064.
Ahmed N, Mahmoud NF, Solyman S, Hanora A. Human nasal microbiome as characterized by metagenomics differs markedly between rural and industrial communities in Egypt. OMICS J Integr Biol. 2019;23:573–82.
Camarinha-Silva A, Jáuregui R, Chaves-Moreno D, Oxley APA, Schaumburg F, Becker K, et al. Comparing the anterior nare bacterial community of two discrete human populations using Illumina amplicon sequencing. Environ Microbiol. 2014;16:2939–52.
Rylance J, Kankwatira A, Nelson DE, Toh E, Day RB, Lin H, et al. Household air pollution and the lung microbiome of healthy adults in Malawi: a cross-sectional study. BMC Microbiol. 2016;16:182.
Masekela R, Vosloo S, Venter SN, de Beer WZ, Green RJ. The lung microbiome in children with HIV-bronchiectasis: a cross-sectional pilot study. BMC Pulm Med. 2018;18:87.
Shenoy MK, Iwai S, Lin DL, Worodria W, Ayakaka I, Byanyima P, et al. Immune response and mortality risk relate to distinct lung microbiomes in patients with HIV and pneumonia. Am J Respir Crit Care Med. 2017;195:104–14.
Li J, Nasidze I, Quinque D, Li M, Horz H-P, André C, et al. The saliva microbiome of Pan and Homo. BMC Microbiol. 2013;13:204.
Nasidze I, Li J, Schroeder R, Creasey JL, Li M, Stoneking M. High diversity of the saliva microbiome in Batwa Pygmies. PLoS One. 2011;6:e23352.
Tawfik SA, Azab MM, Ahmed AAA, Fayyad DM. Illumina MiSeq sequencing for preliminary analysis of microbiome causing primary endodontic infections in Egypt. Int J Microbiol. 2018;2018:1–15.
Kidd JM, Sharpton TJ, Bobo D, Norman PJ, Martin AR, Carpenter ML, et al. Exome capture from saliva produces high quality genomic and metagenomic data. BMC Genomics. 2014;15:262.
Anderson AC, Al-Ahmad A, Elamin F, Jonas D, Mirghani Y, Schilhabel M, et al. Comparison of the bacterial composition and structure in symptomatic and asymptomatic endodontic infections associated with root-filled teeth using pyrosequencing. PLoS One. 2013;8:e84960.
Drago L, Toscano M, De Grandi R, Grossi E, Padovani EM, Peroni DG. Microbiota network and mathematic microbe mutualism in colostrum and mature milk collected in two different geographic areas: Italy versus Burundi. ISME J. 2017;11:875–84.
Meehan CL, Lackey KA, Hagen EH, Williams JE, Roulette J, Helfrecht C, et al. Social networks, cooperative breeding, and the human milk microbiome. Am J Hum Biol. 2018;30:e23131.
Ojo-Okunola C-W, Mwaikono G-L, Stein Z, et al. Influence of socio-economic and psychosocial profiles on the human breast milk bacteriome of South African Women. Nutrients. 2019;11:1390.
Boix-Amorós A, Puente-Sánchez F, du Toit E, Linderborg KM, Zhang Y, Yang B, et al. Mycobiome profiles in breast milk from healthy women depend on mode of delivery, geographic location, and interaction with bacteria. Appl Environ Microbiol. 2019;85:e02994–18.
Gómez-Gallego C, Morales J, Monleón D, du Toit E, Kumar H, Linderborg K, et al. Human breast milk NMR metabolomic profile across specific geographical locations and its association with the milk microbiota. Nutrients. 2018;10:1355.
Gómez-Gallego C, Kumar H, García-Mantrana I, du Toit E, Suomela J-P, Linderborg KM, et al. Breast milk polyamines and microbiota interactions: impact of mode of delivery and geographical location. Ann Nutr Metab. 2017;70:184–90.
Kumar H, du Toit E, Kulkarni A, Aakko J, Linderborg KM, Zhang Y, et al. Distinct patterns in human milk microbiota and fatty acid profiles across specific geographic locations. Front Microbiol. 2016;7:1619.
Decuypere S, Meehan CJ, Van Puyvelde S, De Block T, Maltha J, Palpouguini L, et al. Diagnosis of bacterial bloodstream infections: a 16S metagenomics approach. PLoS Negl Trop Dis. 2016;10:e0004470.
Luk K-C, Berg MG, Naccache SN, Kabre B, Federman S, Mbanya D, et al. Utility of metagenomic next-generation sequencing for characterization of HIV and Human pegivirus diversity. PLoS ONE. 2015;10:ee0141723.
Ngoi CN, Siqueira J, Li L, Deng X, Mugo P, Graham SM, et al. The plasma virome of febrile adult Kenyans shows frequent parvovirus B19 infections and a novel arbovirus (Kadipiro virus). J Gen Virol. 2016;97:3359–67.
Stremlau MH, Andersen KG, Folarin OA, Grove JN, Odia I, Ehiane PE, et al. Discovery of novel rhabdoviruses in the blood of healthy individuals from West Africa. PLoS Negl Trop Dis. 2015;9:e0003631.
Williams SH, Cordey S, Bhuva N, Laubscher F, Hartley M-A, Boillat-Blanco N, et al. Investigation of the plasma virome from cases of unexplained febrile illness in Tanzania from 2013 to 2014: a comparative analysis between unbiased and VirCapSeq-VERT high-throughput sequencing approaches. mSphere. 2018;3:e00311–8.
Li L, Deng X, Linsuwanon P, Bangsberg D, Bwana MB, Hunt P, et al. AIDS Alters the commensal plasma virome. J Virol. 2013;87:10912–5.
Eibach D, Hogan B, Sarpong N, Winter D, Struck NS, Adu-Sarkodie Y, et al. Viral metagenomics revealed novel betatorquevirus species in pediatric inpatients with encephalitis/meningoencephalitis from Ghana. Sci Rep. 2019;9:2360.
Hospodsky D, Pickering AJ, Julian TR, Miller D, Gorthala S, Boehm AB, et al. Hand bacterial communities vary across two different human populations. Microbiology. 2014;160:1144–52.
Van Leuvenhaege C, Vandelannoote K, Affolabi D, Portaels F, Sopoh G, de Jong BC, et al. Bacterial diversity in Buruli ulcer skin lesions: challenges in the clinical microbiome analysis of a skin disease. PLoS One. 2017;12:e0181994.
Ramadan M, Solyman S, Yones M, Abdallah Y, Halaby H, Hanora A. Skin microbiome differences in atopic dermatitis and healthy controls in Egyptian children and adults, and association with serum immunoglobulin E. Omics. 2019;23:247–60.
Manus MB, Yu JJ, Park LP, Mueller O, Windsor SC, Horvath JE, et al. Environmental influences on the skin microbiome of humans and cattle in rural Madagascar. Evol Med Public Health. 2017;2017:144–53.
Yu JJ, Manus MB, Mueller O, Windsor SC, Horvath JE, Nunn CL. Antibacterial soap use impacts skin microbial communities in rural Madagascar. PLoS One. 2018;13:e0199899.
van Zyl LJ, Abrahams Y, Stander EA, Kirby-McCollough B, Jourdain R, Clavaud C, et al. Novel phages of healthy skin metaviromes from South Africa. Sci Rep. 2018;8:12265.
Lackey KA, Williams JE, Meehan CL, Zachek JA, Benda ED, Price WJ, et al. What’s normal? Microbiomes in human milk and infant feces are related to each other but vary geographically: the INSPIRE study. Front Nutr. 2019;6:45.
Fei N, Bernabe BP, Lie L, Baghdan D, Bedu-Addo K, Plange-Rhule J, et al. The human microbiota is associated with cardiometabolic risk across the epidemiologic transition. PLoS One. 2019;14:e0215262.
Sande CJ, Njunge JM, Mwongeli Ngoi J, Mutunga MN, Chege T, Gicheru ET, et al. Airway response to respiratory syncytial virus has incidental antibacterial effects. Nat Commun. 2019;10:2218.
Smits SL, Zijlstra EE, van Hellemond JJ, Schapendonk CME, Bodewes R, Schürch AC, et al. Novel cyclovirus in human cerebrospinal fluid, Malawi, 2010–2011. Emerg Infect Dis. 2013;19:1511–3.
Doyle RM, Harris K, Kamiza S, Harjunmaa U, Ashorn U, Nkhoma M, et al. Bacterial communities found in placental tissues are associated with severe chorioamnionitis and adverse birth outcomes. PLoS ONE. 2017;12:e0180167.
Rocafort M, Noguera-Julian M, Rivera J, Pastor L, Guillén Y, Langhorst J, et al. Evolution of the gut microbiome following acute HIV-1 infection. Microbiome. 2019;7:73.
Kafetzopoulou LE, Pullan ST, Lemey P, Suchard MA, Ehichioya DU, Pahlmann M, et al. Metagenomic sequencing at the epicenter of the Nigeria 2018 Lassa fever outbreak. Science. 2019;363:74–7.
Piantadosi A, Freije CA, Gosmann C, Ye S, Park D, Schaffner SF, et al. Metagenomic sequencing of HIV-1 in the blood and female genital tract reveals little quasispecies diversity during acute infection. J Virol. 2019;93.
Shenoy MK, Fadrosh DW, Lin DL, Worodria W, Byanyima P, Musisi E, et al. Gut microbiota in HIV-pneumonia patients is related to peripheral CD4 counts, lung microbiota, and in vitro macrophage dysfunction. Microbiome. 2019;7:37.
Ramesh A, Nakielny S, Hsu J, Kyohere M, Byaruhanga O, de Bourcy C, et al. Metagenomic next-generation sequencing of samples from pediatric febrile illness in Tororo, Uganda. PLoS One. 2019;14:e0218318.
Cummings MJ, Tokarz R, Bakamutumaho B, Kayiwa J, Byaruhanga T, Owor N, et al. Precision surveillance for viral respiratory pathogens: virome capture sequencing for the detection and genomic characterization of severe acute respiratory infection in Uganda. Clin Infect Dis. 2019;68:1118–25.
Nakagawa S, Inoue S, Kryukov K, Yamagishi J, Ohno A, Hayashida K, et al. Rapid sequencing‐based diagnosis of infectious bacterial species from meningitis patients in Zambia. Clin Transl Immunol. 2019;8:e01087.
Paganini D, Uyoga MA, Kortman GAM, Boekhorst J, Schneeberger S, Karanja S, et al. Maternal human milk oligosaccharide profile modulates the impact of an intervention with iron and galacto-oligosaccharides in Kenyan infants. Nutrients. 2019;11:2596.
Acknowledgements
We will like to acknowledge Dr. Mamadou Kaba and Ms. Michelle Ngwarai of the Division of Medical Microbiology of the University of Cape Town for their assistance.
Funding
H3ABioNet is supported by the National Institutes of Health Common Fund grant number (U41HG006941). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. REA acknowledges the financial support of the Swedish International Development Cooperation Agency (SIDA), Organisation of Women in Science for the developing world (OWSD) PhD Fellowship, Margaret McNamara Education Grants, and L'Oréal UNESCO For Women in Science Fellowship. The funders had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
IA and REA contributed toward study conceptualization, conducted the literature search and screening, extracted and analyzed the data, and wrote the draft of the manuscript. All other authors have substantively revised the manuscript and approved the submitted version.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: Supplementary Data was missing from this article and has now been uploaded.
Supplementary Information
Additional file 1:
Table S1. Details of the search terms used in the respective databases. Table S2a. Additional summary of African Gut Microbiome studies. Table S2b. Additional summary of African Urogenital Microbiome studies. Table S2c. Additional summary of African Microbiome studies of other body sites.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Allali, I., Abotsi, R.E., Tow, L.A. et al. Human microbiota research in Africa: a systematic review reveals gaps and priorities for future research. Microbiome 9, 241 (2021). https://doi.org/10.1186/s40168-021-01195-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40168-021-01195-7