Abstract
Background
Biologically annotated neural networks (BANNs) are feedforward Bayesian neural network models that utilize partially connected architectures based on SNP-set annotations. As an interpretable neural network, BANNs model SNP and SNP-set effects in their input and hidden layers, respectively. Furthermore, the weights and connections of the network are regarded as random variables with prior distributions reflecting the manifestation of genetic effects at various genomic scales. However, its application in genomic prediction has yet to be explored.
Results
This study extended the BANNs framework to the area of genomic selection and explored the optimal SNP-set partitioning strategies by using dairy cattle datasets. The SNP-sets were partitioned based on two strategies–gene annotations and 100 kb windows, denoted as BANN_gene and BANN_100kb, respectively. The BANNs model was compared with GBLUP, random forest (RF), BayesB and BayesCπ through five replicates of five-fold cross-validation using genotypic and phenotypic data on milk production traits, type traits, and one health trait of 6,558, 6,210 and 5,962 Chinese Holsteins, respectively. Results showed that the BANNs framework achieves higher genomic prediction accuracy compared to GBLUP, RF and Bayesian methods. Specifically, the BANN_100kb demonstrated superior accuracy and the BANN_gene exhibited generally suboptimal accuracy compared to GBLUP, RF, BayesB and BayesCπ across all traits. The average accuracy improvements of BANN_100kb over GBLUP, RF, BayesB and BayesCπ were 4.86%, 3.95%, 3.84% and 1.92%, and the accuracy of BANN_gene was improved by 3.75%, 2.86%, 2.73% and 0.85% compared to GBLUP, RF, BayesB and BayesCπ, respectively across all seven traits. Meanwhile, both BANN_100kb and BANN_gene yielded lower overall mean square error values than GBLUP, RF and Bayesian methods.
Conclusion
Our findings demonstrated that the BANNs framework performed better than traditional genomic prediction methods in our tested scenarios, and might serve as a promising alternative approach for genomic prediction in dairy cattle.
Similar content being viewed by others
Background
Genomic selection [1] has significantly shortened the generation interval and increased the annual genetic gain of economic traits in dairy cattle [2,3,4] with breeding costs reduced by 92% compared to traditional progeny testing [5]. Statistical models serve as one of the key factors affecting the accuracy of genomic selection, consequently exerting an impact on genetic progress. Currently, the most commonly used models for genomic prediction in dairy cattle include the best linear unbiased prediction (BLUP) models that incorporate genomic information [e.g., the genomic BLUP (GBLUP) and single-step GBLUP (ssGBLUP) methods], executed through solving the mixed model equations (MME), as well as the Bayesian methods with various priors that use Markov chain Monte Carlo (MCMC) to estimate the required genetic parameters. However, the utilization of these linear models is often limited by their assumption that genetic variants influence phenotypes only in an additive manner and fail to capture interactions. The exponential growth of large-scale genomic databases provides a unique opportunity to move beyond traditional linear regression frameworks.
Machine learning (ML) algorithms can build complex nonlinear models and allow interaction between features (i.e., markers). Therefore, ML has been considered an effective tool for interpretating massive genomic datasets [6]. Recently, several studies showed that nonlinear ML algorithms typically exhibited higher predictive accuracy than conventional methods such as GBLUP and Bayesian approaches [6,7,8,9], especially for complex traits with broad-sense heritability driven by non-additive genetic variation (e.g., gene-gene interactions) [10]. In dairy science, ML has been successfully applied to predict a whole range of different traits, such as milk production [11, 12], mastitis [13], and methane production [14]. Ensemble methods are a category of advanced ML algorithms. Random forest (RF), as an ensemble method, is model specification free and may account for non-additive effects [15]. Moreover, it remains a relatively fast algorithm in ensemble methods even when dealing with a large number of covariates and interactions, making it suitable for both classification and regression problems [15]. Therefore, RF has been widely employed in genomic prediction [9, 15, 16]. Furthermore, to comprehensively capture interactions between markers and non-additive effects, an increasing body of research is being devoted to neural networks [17,18,19], which reflect the nonlinear relationships between variables by exploiting nonlinear activation functions between network layers. However, conventional neural networks often do not consider the varying influences of different genomic regions on traits, and thus lack certain biological interpretability. Studies have shown that genetic variants do not contribute equally to genetic variance, and genetic variations of large effect on a trait are often distributed within specific genomic regions [20,21,22]. Based on this framework, new prediction methods have been developed, including BayesRC [23], BayesRS [24], BayesRR-RC [25], NN-Bayes and NN-MM [26].
Most recently, Demetci et al. [27] developed the biologically annotated neural networks (BANNs), a nonlinear probabilistic framework for association mapping in genome-wide association studies (GWAS). BANNs are a class of feedforward Bayesian models that integrate predefined SNP-set annotations, and the BANNs framework has achieved better performance than state-of-the-art methods in the area of GWAS by using prior defined biology information [27]. BANNs employ variational inference for parameter estimation, which is an optimization method that can leverage modern optimization techniques such as Stochastic Gradient Descent (SGD), to find an approximation to the posterior distribution. Consequently, variational inference is often more efficient than MCMC sampling, as the latter requires extensive sampling to estimate the full posterior distribution [28]. Philosophically, compared to traditional linear models, the BANNs framework considers the heterogeneity of the function of SNP-sets according to annotations. BANNs take into account the interactions between markers through setting of neural network layers, which seems theoretically more in line with the biological process of complex traits. However, the existing BANNs framework has not been applied to genomic prediction.
The objectives of this study were to: (i) extend the BANNs framework to the field of dairy cattle genomic selection by exploring the optimal SNP-set partitioning strategies; and (ii) assess the predictive ability of the BANNs framework by comparing it with GBLUP, RF and Bayesian methods.
Materials and methods
Statistical models
BANNs
As an interpretable neural network, the BANNs framework models SNP effects in the input layers and SNP-set effects in the hidden layers separately. BANNs utilized sparse prior distributions to select variables for network weights. The weights and connections of the network are treated as random variables that present genetic effects at various genomic scales. Moreover, BANNs fall into the category of Bayesian Network (BN) models. BN models can be viewed as a non-conjugate form of Bayesian linear regression, because they automatically learn hyperparameters for priors from the data, making them generally more flexible and better suitable for capturing complex data structures [29].
The model representation for the BANNs framework is as follows:
where \(\varvec{y}\) is the vector of the response variable, that is, standardized de-regressed proofs (DRPs); \({\varvec{X}}_{g}=\left[{\varvec{x}}_{1},\dots ,{\varvec{x}}_{\left|{S}_{g}\right|}\right]\) is a subset of SNPs for SNP-set \(g\); \({\varvec{\theta }}_{g}=\left({\theta }_{1},\dots ,{\theta }_{\left|{S}_{g}\right|}\right)\) are the corresponding inner layer weights; \(h\left(\bullet \right)\) denotes the nonlinear activations defined for the neurons in the hidden layer; \({\varvec w}=\left({w}_{1},\cdots ,{w}_{G}\right)\) are the weights of the G-predefined SNP-sets in the hidden layer; \({\varvec{b}}^{\left(1\right)}=\left({b}_{1}^{\left(1\right)},\cdots ,{b}_{G}^{\left(1\right)}\right)\) and \({b}^{\left(2\right)}\) are deterministic biases generated during the training phase of the network in the input and hidden layers, respectively; 1 is an N-dimensional vector of ones. For convenience, the genotype matrix (column-wise) and the trait of interest are assumed to be mean-centered and standardized. In this study, \(h\left(\bullet \right)\) is defined as the Leaky rectified linear unit (Leaky ReLU) activation function. If x > 0, then \(h\left(x\right)=x\), otherwise, we define \(h\left(x\right)=0.01x\).
The weights of the input layer (\(\varvec{\theta }\)) and the hidden layer (\({\varvec w}\)) were treated as random variables, allowing simultaneous multi-scale genomic inference on both SNPs and SNP-sets. SNP-level effects are assumed to follow a sparse K-mixed normal distribution:
where \({\pi }_{\theta }\) represents the total proportion of SNPs that have a non-zero effect on the trait; \({\varvec{\eta }}_{\theta }=\left({\eta }_{\theta 1},\dots ,{\eta }_{\theta k}\right)\) denotes the marginal (unconditional) probability that a randomly selected SNP belongs to the k-th mixture component and that \({\sum }_{k}{\eta }_{\theta \kappa }\)=1; \({\varvec{\sigma }}_{\theta }^{2}=\left({\sigma }_{\theta 1}^{2},\dots ,{\sigma }_{\theta K}^{2}\right)\) are the variance of the K non-zero mixture components; and \({\delta }_{0}\) is a point mass at the zero point. The present study follows previous studies and lets K = 3, indicating that SNPs may have large, moderate and small non-zero effects on phenotypic variation [30,31,32]. To infer the hidden layer, it was assumed that the enriched SNP-sets contain at least one non-zero effect SNP by placing a spike and slab prior to the hidden weights:
Due to the lack of prior knowledge regarding the proportion of relevant SNPs and SNP-sets with non-zero weights, an assumption was made on relatively uniform priors on \(\text{log}\left({\pi }_{\theta }\right)\) and \(\text{log}\left({\pi }_{w}\right)\) [27]:
where \({\pi }_{\theta }\) denotes the total proportion of SNPs with a non-zero effect on the trait of interest, \(J\) denotes the number of SNPs, and \({\pi }_{w}\) denotes the total proportion of annotated SNP-sets enriched for the trait of interest. In addition, the variational Bayesian algorithm was used to estimate all model parameters. In the BANNs framework, the posterior inclusion probabilities (PIPs) provide statistical evidence for the importance of each variant in explaining the overall genetic architecture of a trait. These quantities are defined as the posterior probability that the weight of a given connection in the neural network is non-zero:
where \(j\) and \(g\) represent a specific SNP and a specific SNP-set, respectively.
In addition, the variational expectation-maximization (EM) algorithm was utilized for estimating the parameters of the neural network, and parameters in the variational EM algorithm were initialized through random draws from their assumed prior distributions. The iteration within the algorithm terminates upon meeting one of the following two stopping criteria: (i) the difference between the lower bounds of two consecutive updates falls within the range of 1 × 10−4, or (ii) the maximum iteration count of 10,000 is reached [27]. In addition, the initial values of variance \({\sigma }_{0}^{2}\) and the number of models L were set to 0.01 and 20, respectively. In summary, the Bayesian formulation in the BANNs framework makes network sparsity a goal for genomic selection applications through the context-driven sparse shrinkage prior distribution in Eqs. (1–4).
The original BANNs model partitioned SNP-sets according to gene-annotated SNPs. Two strategies were considered in this study to group the SNPs into different sets. Firstly, biological annotations were considered (denoted as BANN_gene). The cattle genome annotation file was obtained from the NCBI website (https://ftp.ensembl.org/pub/release-94/gtf/bos_taurus/) for mapping SNPs to their nearest neighboring genes and aptly annotating them with relevant gene information. Unannotated SNPs located within the same genomic region were denoted as “intergenic regions” between two genes. A total of G = 16,857 SNP-sets were analyzed, consisting of 9,369 intergenic SNP-sets and 7,488 annotated genes. Secondly, 100 kb windows were used to divide SNPs on each chromosome into different groups (denoted as BANN_100kb). A total of G = 22,626 SNP-sets were analyzed using this strategy. On note, the choice of a 100 kb window was based on our testing of the predictive ability with different SNP division intervals (50 kb, 100 kb, 200 kb, 300 kb, 400 kb, 600 kb, 800 kb, 1,000 kb), where we found that dividing based on a 100 kb window yielded better results (results not shown).
GBLUP
The model of the GBLUP is given as:
in which \({\varvec y}\) is also the vector of standardized DRPs, \(\mu\) is the overall mean, \({\boldsymbol 1}\) is a vector of ones, \({\varvec g}\) is the vector of genomic breeding values, \({\varvec e}\) is the vector of random residuals, and Z is an incidence matrix allocating records to \(\varvec g\). The assumptions of random effects were:
in which G is the genomic relationship matrix (G matrix), D is a diagonal matrix with \({d}_{ii}=\frac{1-{r}_{i}^{2}}{{r}_{i}^{2}}\), (\({r}_{i}^{2}\) is the reliability of DRP of individual i), and \({\sigma }_{{g}}^{2}\)and \({\sigma }_{{e}}^{2}\) are the additive genetic variance and the random residual variance, respectively.
In this study, GBLUP was carried out using DMU software [33]. The AI-REML method in the DMUAI procedure was used to estimate the variance components.
BayesB
In BayesB, the proportion of markers with no effect is assumed to be \(\pi\), and the proportion of markers with an effect is \(1-\pi\), and the prior distribution of SNP effect, \({\beta }_{k}\), was assumed to be t-distributed. The formula of BayesB can be written as follows:
where \(\varvec y\) represents the vector of standardized DRPs, \({\varvec{x}}_{\varvec{k}}\) is the vector of genotypes for the kth SNP, and \({\beta }_{k}\) is the effect of the kth SNP. The prior distribution of \({\beta }_{k}\) is as follows:
in which \(v\) is the degree of freedom, \({S}_{\beta }^{2}\) is the scale parameter. In the present study, for the BayesB method, we set the proportion of no-effect SNPs (\(\pi\)) to be 0.95.
BayesCπ
In BayesCπ, the marker effects are sampled from a mixture of null and normal distributions. The expression for BayesCπ aligns with that of BayesB except for the prior distribution of \({\upbeta }_{k}\), which is as follows:
where \({\sigma }_{\beta}^{2}\) is the variance of SNP effect. Additionally, in BayesCπ, the value of \(\pi\) is treated as an unknown with uniform (0,1) prior and is estimated through sampling [34].
For both BayesB and BayesCπ methods, the MCMC chain was run for 50,000 iterations, the first 20,000 iterations were discarded as burn-in, and every 50 samples of the remaining 30,000 iterations were saved to estimate SNP effects and variance components. The analysis was performed using the Julia package JWAS [35].
Random forest
Random forest is a ML algorithm that employs voting or averaging the outcomes of multiple decision trees to determine the classification or predicted values of new instances [36]. Essentially, RF is a collection of decision trees, with each tree exhibiting slight differences from the others. RF reduces the risk of overfitting by averaging the predictions of numerous decision trees [7]. The RF regression can be expressed as follows:
where \({y}\) represents the predicted value from the RF regression, \({t}_{m}\left({\psi }_{m}\left({y}:{\varvec X}\right)\right)\) represents an individual regression tree, and \(M\) represents the number of decision trees in the forest. Predictions were obtained by propagating predictor variables through the flowchart of each tree, with the estimated values at the terminal nodes serving as the predictions. The final predictions for unobserved data were determined by averaging the predictions across all trees in the RF. To optimize the model, a grid search approach was employed to identify the most suitable hyperparameter \(M\) and the maximum tree depth, with an inner five-fold cross-validation (CV) being conducted to tune these hyperparameters.
Datasets
In this study, phenotypic and genomic data were collected from Chinese Holstein cattle. The population and phenotype information are shown in Table 1. The phenotypic data included three milk production traits: milk yield (MY), fat yield (FY) and protein yield (PY); three type traits: conformation (CONF), feet and leg (FL) and mammary system (MS); and one health trait: somatic cell score (SCS). A total of 6,558, 6,210 and 5,962 individuals were genotyped for milk production traits, type traits and SCS, respectively. DRPs derived from the official estimated breeding values (EBV) provided by the Dairy Association of China following the method proposed by Jairath et al. [37] were used as pseudo-phenotypes for genomic predictions. The DRP reliability for each animal was estimated as \({r}_{DRP}^{2}=\frac{{ERC}_{i}}{{ERC}_{i}+\lambda }\), with \(\lambda =\frac{1-{h}^{2}}{{h}^{2}}\), in which \({ERC}_{i}\) refers to the effective record contribution and \({h}^{2}\) refers to the heritability of the trait. Note that \({ERC}_{i}=\lambda \times\frac{{REL}_{i}}{1-{REL}_{i}}\), where \({REL}_{i}\) is the reliability of EBV for individual i. All individuals were genotyped using the BovineSNP50 chip containing 54,609 SNPs from Illumina (Illumina, San Diego, CA, USA). Missing genotypes were imputed using Beagle 5.4 [38]. After imputation, SNPs with minor allele frequency (MAF) less than 0.01 and significantly deviating from Hardy-Weinberg equilibrium (P < 1.0E-6) were removed using PLINK software [39]. After genotype quality control, 45,944 autosomal SNPs remained for further analyses.
Cross-validation and genomic prediction accuracy
Prediction accuracy, mean square error (MSE) and dispersion were used to assess the prediction performance of different methods. A 5 × 5 CV (five-fold CV repeated five times, totaling 25 tests) process was carried out. The prediction accuracy was assessed with the Pearson correlation coefficient between standardized DRP (sDRP) and predicted values (PV) of the validation population divided by the mean accuracy \(\stackrel{-}{r}\) (square root of reliability) of DRP in validation data:
Besides, following the study by Legarra and Reverter [40], the slope of the regression of sDRP on PV was calculated to assess the dispersion of the prediction, although some studies used the regression coefficient as a measure of bias and referred to it as unbiasedness [30, 41]. In addition, MSE was also used as a measure for the performance of different methods, which considered both prediction bias and variability. In each prediction scenario, the reference and validation populations for all methods were the same in each replicate of the five-fold CV, and the final results of accuracy, dispersion and MSE are the averages of five repetitions. Furthermore, multiple t-tests were conducted based on the outcomes of five replicates, with P-values adjusted using the Bonferroni method, to compare the prediction accuracy of different methods.
Estimating phenotypic variance explained in the BANNs framework
Given that the BANNs framework offers posterior estimates for all weights in neural networks, it also enables the estimation of phenotypic variance explained (PVE). Here, PVE was defined as the total proportion of phenotypic variation explained by sparse genetic effects (both additive and non-additive effects) [42]. Within the BANNs framework, such estimation can be conducted at both the SNP and SNP-set levels as follows [27]:
where \(\text{V}\left(\bullet \right)\) denotes the variance function, \({\varvec{\beta }}_{\theta }\) and \({\varvec{\beta }}_{w}\) represent the vectors of the marginal posterior means for the input and outer layer weights, respectively. \(\varvec{H}\left({\varvec{\beta }}_{\theta }\right)=\left[h\left({\varvec{X}}_{1}{\varvec{\beta }}_{\theta 1}+{b}_{1}^{\left(1\right)}\right),\dots , h({\varvec{X}}_{G}{\varvec{\beta }}_{\theta G}+{b}_{G}^{\left(1\right)})\right]\) represents the matrix of deterministic nonlinear neurons in the hidden layer given \({\varvec{\beta }}_{\theta }\). The estimates of variance hyperparameters \({\tau }_{\theta }^{2}\) and \({\tau }_{w}^{2}\) in the variational EM algorithm were used to approximate the residual variance observed during the two-layer training process [27]. In fact, the formula is similar to the traditional form used for estimating PVE, with the distinction that the contribution of non-additive genetic effects is also taken into account through the nonlinear Leaky ReLU activation function \(h\left(\bullet \right)\). In other words, the PVE estimated at the SNP level considers only additive effects, while the PVE estimated at the SNP-set level takes into account both additive and non-additive genetic effects.
Results
Annotation summary
The distribution of the number of SNPs in each SNP-set under the two partitioning schemes is shown in Fig. 1. With regards to BANN_gene, of a total of 16,857 SNP-sets, 9,413 contained one SNP (including intergenic regions), while the remaining SNP-sets had varying numbers of SNPs, ranging from 2 to 108. For BANN_100kb, among the 22,626 SNP-sets, 21,466 sets had no more than 3 SNPs (7,152, 8,848 and 5,466 SNP-sets containing 1, 2 and 3 SNPs, respectively), and none of the SNP-sets had more than 6 SNPs. Therefore, it was evident that the distribution of SNPs within BANN_100kb SNP-sets was more uniform than in BANN_gene.
Genomic prediction accuracy
Comparison of prediction performance among BANN_gene, GBLUP, RF and Bayesian methods
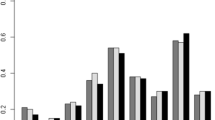
Figure 2 shows the accuracy, dispersion and MSE of genomic predictions for seven dairy cattle traits using six methods (Table S1 reports the underling values of Fig. 2). In terms of accuracy, BANN_gene performed best compared to GBLUP, RF and Bayesian methods. The average improvement of BANN_gene over GBLUP, RF, BayesB and BayesCπ were 3.75%, 2.86%, 2.73% and 0.85%, respectively, across all seven traits. For milk production traits, BANN_gene demonstrated better performance compared to GBLUP, RF, BayesB or BayesCπ, especially for MY. For example, the accuracy of BANN_gene for MY was 0.491, which resulted in a 7.68% significant improvement compared to GBLUP. The accuracy of BANN_gene for milk production traits, compared to GBLUP, RF, BayesB and BayesCπ, improved by an average of 3.93%, 3.25%, 1.90% and 1.53%, respectively. In case of type traits, BANN_gene significantly outperformed GBLUP, RF and BayesB, while BayesCπ performed similarly with BANN_gene. The improvement of BANN_gene over GBLUP, RF and BayesB was 3.52%, 2.33% and 3.84% on average, respectively.
Accuracy (a), mean squared error (MSE) (b) and dispersion (c) of genomic prediction on seven traits of dairy cattle using five-fold cross-validation with five replications. CONF Conformation, FL Feet and leg, MS Mammary system, FY Fat yield, MY Milk yield, PY Protein yield, SCS Somatic cell score. The error bar represents the standard error
Compared to GBLUP, RF and Bayesian methods, BANN_gene yielded the lowest or the second lowest MSE. It yielded the smallest MSE for FL, FY, MY and SCS traits, while for other traits, BANN_gene showed the second smallest MSE. However, in terms of overall dispersion, BayesCπ achieved the most appropriate dispersion (i.e., slopes closer to 1), followed by BANN_gene.
In addition, for the comparison of the two Bayesian methods, we found that BayesCπ obtained better results than BayesB across all metrics of accuracy, dispersion, and MSE; besides, as indicated by the estimated standard errors of marker effects (as shown in Table 2), BayesCπ produced smaller standard errors for marker effects across all traits.
Comparison of prediction performance among BANN_100kb, GBLUP, RF and Bayesian methods
BANN_100kb achieved the highest accuracy in all scenarios when compared to the conventional GBLUP and Bayesian methods, where the accuracy of BANN_100kb was improved by an average of 4.86%, 3.95%, 3.84% and 1.92% compared to GBLUP, RF, BayesB and BayesCπ, ranging from 2.12% to 7.46%, 2.63% to 5.38%, 1.87% to 6.93% and 1.25% to 3.23%, respectively. For milk production traits, BANN_100kb consistently achieved the highest accuracy, particularly for FY and MY traits, where BANN_100kb exhibited significant improvements of 5.42% and 7.46%, respectively, compared to GBLUP. Compared to GBLUP, BayesB and BayesCπ, BANN_100kb displayed average improvements in accuracy of 4.48%, 2.45% and 2.08%, respectively. For type traits, BANN_100kb also obtained the highest accuracy, with average improvements over GBLUP, RF, BayesB and BayesCπ of 5.36%, 4.14%, 5.68% and 1.71%, respectively. These results suggest that BANN_100kb captured some intrinsic nonlinear features within the dairy cattle data, whereas GBLUP and Bayesian methods did not. Regarding MSE, BANN_100kb showed the lowest value for all traits. As for dispersion, the dispersions of the four methods were roughly as follows: bBayesCπ < bBANN_100kb < bGBLUP < bBayesB < bRF.
Comparison of prediction performance between BANN_gene and BANN_100kb
Comparison of the BANNs methods used for differently partitioned SNP subsets (BANN_gene vs. BANN_100kb) showed that BANN_100kb consistently demonstrated superior accuracy with an average improvement of 1.80%, 1.79%, 1.73% and 1.82% over BANN_gene for CONF, FL, MS and FY traits, respectively. However, for MY and SCS traits, the accuracy of BANN_100kb closely resembled that of BANN_gene, with accuracies of 0.49 and 0.491 for MY and 0.351 and 0.352 for SCS. Overall, BANN_100kb resulted in an average improvement of 1.07% compared to BANN_gene across all traits (1.77% for type traits; 0.54% for milk production traits), although the improvements were not significant for most traits.
Concerning MSE, BANN_100kb consistently produced lower MSE than BANN_gene in almost all scenarios. Specially, BANN_100kb had an average MSE that was 0.007 lower than that of BANN_gene for milk production traits and an average MSE that was lower than BANN_gene by 0.0013 for type traits. In terms of dispersion, BANN_100kb achieved a generally more appropriate dispersion compared to BANN_gene for both milk production and type traits.
Posterior inclusion probabilities in the BANNs framework
Table 3 summarizes the average, maximum and minimum values of PIPs across all variants on SNPs and SNP-sets from the BANNs framework. Since BANN_gene and BANN_100kb shared the same SNP layer, both methods yielded identical PIP results at the SNP level. However, at the SNP-set level, BANN_100kb obtained a lower standard error in PIP across all seven traits compared to BANN_gene, as evidenced by the smaller range between the maximum and minimum PIP values obtained by BANN_100kb. In addition, for both BANN_gene and BANN_100kb methods, the maximum PIP values obtained at the SNP-set level were significantly higher than those at the SNP level for all traits.
Estimating phenotypic variance explained in the BANNs framework
Figure 3 presents the average PVE for the seven traits in five replicates of five-fold CV. For all traits, the PVE estimates obtained at the SNP-set level were substantially greater than those at the SNP level, regardless of whether they were derived from BANN_gene or BANN_100kb. In addition, as BANN_gene and BANN_100kb shared the same SNP layer, they yielded identical PVE estimates at the SNP level, while at the SNP-set level, BANN_100kb obtained larger PVE estimates. The average PVE estimated at the SNP level for both BANN_gene and BANN_100kb was 0.303, while the average PVE estimated at the SNP-set level was 0.738 and 0.754 respectively. Moreover, we observed that at the SNP-set level, the PVE for type traits (i.e., CONF, FL and MS) was generally greater than that for milk production traits (i.e., MY, FY, PY and SCS). For example, BANN_gene and BANN_100kb had average PVEs of 0.732 and 0.746 respectively for milk production traits, while for type traits, their average PVEs were 0.746 and 0.764, respectively. This might partly explain why type traits achieved higher accuracy compared to milk production traits.
Computation time
The average computation time to complete each fold of five-fold CV for all genomic prediction methods is shown in Table S2. The running time of the methods was measured in minutes on an HP server (CentOS Linux 7.9.2009, 2.5 GHz Intel Xeon processor and 515 GB total memory). Among all methods, GBLUP was the fastest algorithm across all traits, with each fold of CV taking an average of 41.76 min to complete the analysis. The computational efficiency of BayesB, with an average of 132.08 min, was comparable to that of BayesCπ, which averaged 148.91 min. As the BANNs framework involves the construction of neural networks, the computation time for BANN_gene (average 275.79 min) and BANN_100kb (average 284.49 min) was longer, roughly twice that of BayesB or BayesCπ. Additionally, we found that the computational efficiency of RF (average 274.10 min) to be close to that of BANN_gene and BANN_100kb. This may be due to RF being an ensemble algorithm, involving the construction of several hundred decision trees, along with data sampling and feature selection for each tree, leading to its computationally intensive process.
Discussion
The BANNs framework was extended and applied to genomic prediction of dairy cattle for the first time in this study. In addition, two SNP-set partitioning strategies (based on gene annotations and 100 kb windows) under the BANNs framework were also explored. The superiority of the BANNs methodology was demonstrated by using dairy cattle datasets and comparing them to GBLUP, RF and Bayesian methods (BayesB, BayesCπ). BANN_100kb, which partitioned SNP-sets based on 100 kb intervals, outperformed GBLUP, RF, BayesB and BayesCπ methods in terms of prediction accuracy and MSE across all investigated scenarios.
Non-additive effects often play an important role in the phenotypic variation of complex traits [43]. This is also evident from the PVE results in this study, where the PVE at the SNP-set level, considering both additive and non-additive genetic effects, was substantially higher than the PVE at the SNP level, which accounts only for additive effects (Fig. 3). By incorporating nonlinear Leaky ReLU activation functions within the hidden layer, BANN_100kb effectively captured interactions among input variables, enabling the BANNs framework to model sparse genetic effects that encompass both additive and non-additive effects. In contrast, GBLUP and Bayesian methods focus on additive genetics, overlooking potential complex nonlinear relationships between markers and phenotypes (e.g., dominance, epistasis, genotype by environment interactions) [9]. Additionally, in the BANNs approach, the bias term \({b}_{g}^{\left(1\right)}\) for SNP-sets enables each node in the hidden layer to alter the slope for different genotypic combinations, offering a more flexible estimation of generalized heritability. Theoretically, as more nodes and hidden layers are added to the network architecture, BANNs models will possess an increased capacity to account for non-additive genetic effects, akin to classical Gaussian process regression methods [27]. Consequently, BANNs may exhibit greater advantages when applied to high density SNP markers or whole-genome sequencing (WGS) data, as the use of WGS data has not improved the accuracy of genomic prediction compared to using high-density SNP panels [44, 45]. The BANNs framework could potentially provide a promising direction in this context. This is worth investigating in further studies.
It was found that Bayesian methods generally outperformed GBLUP. Bayesian models’ prediction accuracy is affected by the consistency between the underlying assumptions of the model and the true distribution of marker effects. Bayesian models improved prediction accuracy by shrinking the effects of noisy markers to zero. However, the performance of Bayesian methods over GBLUP mainly depends on the presence of QTLs with large effects on the analyzed trait [46]. As milk production traits (e.g., FY, MY, PY and SCS) were characterized by major effect QTLs [47], both BayesB and BayesCπ outperformed the GBLUP method, which assumed all SNP effects follow the same normal distribution. In addition, GWAS on dairy cattle [48] and beef cattle [49] have found that only a few SNPs were significant for type traits, suggesting that most genetic variants have similar medium or small effects on the traits. This might be the reason for the similar performance of BayesB and GBLUP in type traits (e.g., CONF, FL and MS). Additionally, it was observed that BayesB yielded more over/under dispersion compared to other methods. Despite BayesCπ producing less over/under dispersion, its prediction accuracy and MSE values across all traits still remained inferior to those of BANN_100kb.
In this study, we observed that the predictive performance of BANN_gene was not as strong as that of BANN_100kb. As shown in Fig. 3, the PVE values obtained by BANN_100kb at the SNP-set level were greater than that obtained by BANN_gene at the same level for all traits. This indicates a higher proportion of phenotypic variance explained by genetic effects in the BANN_100kb method, which may partially account for its higher accuracy. In addition, as evidenced by the distribution of SNPs (Fig. 1), the 100 kb interval partitioning method resulted in a more uniform SNP distribution and formed a larger number of SNP-sets (a total of 22,626 SNP-sets). In contrast, with the gene-based partitioning approach, the distribution of SNPs in the SNP-sets was highly uneven (the number of SNPs in each set ranged from 1 to 108) and many SNP-sets contained only one SNP. In fact, BANNs are likely to rank SNP-set enrichments that are driven by just a single SNP as less reliable than enrichments driven by multiple SNPs with nonzero effects [27]. Besides, SNP-sets containing only one SNP struggle to capture interactions or combinatorial effects among multiple loci. When the phenotype is affected by multiple variants within a gene region, a SNP-set containing only one SNP may not represent the total genetic contribution of that region, potentially leading to the model overlooking some biological information and thereby affecting its predictive ability. However, retaining these SNPs might still be beneficial compared to removing them, as BANNs are able to prioritize trait relevant SNPs and SNP-sets [27], and some of these single-SNP sets may contain SNPs that are associated with the traits of interest. In addition, in neural networks, the uneven connectivity between hidden and input layer neurons might also affect the predictive ability of the model, primarily for the following reasons: (I) Uneven connectivity might resulted in an imbalanced weight distribution, causing the network to be unable to capture different aspects of the input data in a balanced manner. This might result in biased feature extraction from the input data, ultimately affecting the model’s generalization ability. (II) Uneven connectivity might lead to unstable gradient updates, resulting in issues such as slow convergence, local optima, gradient explosion, or vanishing gradients during the training process. (III) Due to the uneven connectivity between hidden and input layer neurons, the network might struggle to capture complex relationships and features within the input data. This limitation could have constrained the expressiveness of the network and negatively affected its predictive ability. Consequently, the more uniform distribution of SNPs in BANN_100kb facilitated the network in capturing complex relationships and features within the input data; moreover, the larger number of SNP-sets in BANN_100kb potentially aided the network in extracting more meaningful information. These factors above potentially contributed to the greater advantage of BANN_100kb over BANN_gene. However, when based on high-density SNP panel or WGS data, the number of SNPs within each gene region will significantly increase, enhancing the reliability of SNP-set enrichment rankings [27]. Therefore, BANN_gene may outperform BANN_100kb under these conditions.
Although BANN_100kb has achieved superior predictive performance in this study, there remain several potential extensions to the BANNs framework. (I) It would be beneficial to explore different prior assumptions and consider alternative (more scalable) approaches for approximate Bayesian inference [50]. (II) Employing deep learning techniques by incorporating additional hidden layers in the neural network. (III) Consider environmental covariates (as well as potential genotype by environment interactions) in the model [27]. (IV) Evidence suggested that modeling multiple phenotypes into analytical models often results in a substantial improvement of statistical power [51]; therefore, extending the BANNs framework to accommodate multiple phenotypes and exploiting phenotype correlations to identify pleiotropic epistatic effects might be beneficial. Moreover, investigating the performance of more SNP partitioning strategies through future experiments would be interesting. For example, (i) LD-based partitioning: since the uneven distribution of LD along the genome (i.e., the LD heterogeneity of LD among regions) has a great impact on genomic prediction [52], dividing SNP-sets according to LD structure allows SNPs with higher LD to be grouped together, which may improve the ability to explain genetic variation, thus better reflecting the effects of genomic selection; (ii) function-annotation-based partitioning: the genetic variance explained by different functional regions varies across the entire genome [53, 54], so dividing SNPs based on gene functional regions could make the resulting SNP-sets more biologically meaningful, such as coding region SNPs, non-coding region SNPs, intronic SNPs, etc. Finally, given that BANNs require more computation time compared to conventional methods (as shown in Table S2), further optimization of the BANNs framework code to reduce computation time remains a worthwhile endeavor.
Conclusions
This study applied the BANNs framework to the field of genomic prediction in dairy cattle, and compared it with GBLUP, RF and Bayesian methods. Our results demonstrated that the BANNs framework holds greater potential for enhancing genomic prediction accuracy than traditional GBLUP, RF and Bayesian methods by modelling interactions between markers, emerging as a novel choice for dairy cattle genomic prediction. Further research might explore the performance of BANNs framework when applied to high density SNP markers and WGS data, together with function-annotation-based partitioning of SNP-sets.
Availability of data and materials
The datasets used or analyzed during the present study are available from the corresponding author on reasonable request.
Abbreviations
- BANNs:
-
Biologically annotated neural networks
- BN:
-
Bayesian network
- CONF:
-
Conformation
- CV:
-
Cross-validation
- DRP:
-
De-regressed proofs
- EBV:
-
Estimated breeding values
- EM:
-
Expectation-maximization
- FL:
-
Feet & leg
- FY:
-
Fat yield
- GBLUP:
-
Genomic best linear unbiased prediction
- GEBVs:
-
Genomic estimated breeding values
- G matrix:
-
Genomic relationship matrix
- LD:
-
Linkage disequilibrium
- MAF:
-
Minor allele frequency
- MCMC:
-
Markov chain Monte Carlo
- ML:
-
Machine learning
- MME:
-
Mixed model equations
- MS:
-
Mammary system
- MSE:
-
Mean squared error
- MY:
-
Milk yield
- PIPs:
-
Posterior inclusion probabilities
- PV:
-
Predicted values
- PVE:
-
Phenotypic variance explained
- PY:
-
Protein yield
- RF:
-
Random forest
- SCS:
-
Somatic cell score
- SGD:
-
Stochastic gradient descent
- WGS:
-
Whole-genome sequencing
References
Meuwissen THE, Hayes BJ, Goddard ME. Prediction of total genetic value using genome-wide dense marker maps. Genetics. 2001;157:1819–29.
García-Ruiz A, Cole JB, VanRaden PM, Wiggans GR, Ruiz-López FJ, Van Tassell CP. Changes in genetic selection differentials and generation intervals in US Holstein dairy cattle as a result of genomic selection. Proc Natl Acad Sci USA. 2016;113:E3995-4004.
Meuwissen T, Hayes B, Goddard M. Genomic selection: a paradigm shift in animal breeding. Anim Front. 2016;6:6–14.
Doublet AC, Croiseau P, Fritz S, Michenet A, Hoze C, Danchin-Burge C, et al. The impact of genomic selection on genetic diversity and genetic gain in three French dairy cattle breeds. Genet Sel Evol. 2019;51:52.
Schaeffer LR. Strategy for applying genome-wide selection in dairy cattle. J Anim Breed Genet. 2006;123:218–23.
An B, Liang M, Chang T, Duan X, Du L, Xu L, et al. KCRR: a nonlinear machine learning with a modified genomic similarity matrix improved the genomic prediction efficiency. Brief Bioinform. 2021;22:bbab132.
Gonzalez-Camacho JM, Ornella L, Perez-Rodriguez P, Gianola D, Dreisigacker S, Crossa J. Applications of machine learning methods to genomic selection in breeding wheat for rust resistance. Plant Genome. 2018;11:170104.
Montesinos-Lopez OA, Martin-Vallejo J, Crossa J, Gianola D, Hernandez-Suarez CM, Montesinos-Lopez A, et al. A benchmarking between deep learning, support vector machine and bayesian threshold best linear unbiased prediction for predicting ordinal traits in plant breeding. G3-Genes. Genom Genet. 2019;9:601–18.
Wang X, Shi SL, Wang GJ, Luo WX, Wei X, Qiu A, et al. Using machine learning to improve the accuracy of genomic prediction of reproduction traits in pigs. J Anim Sci Biotechnol. 2022;13:60.
Weissbrod O, Geiger D, Rosset S. Multikernel linear mixed models for complex phenotype prediction. Genome Res. 2016;26:969–79.
Wallen SE, Prestlokken E, Meuwissen THE, McParland S, Berry DP. Milk mid-infrared spectral data as a tool to predict feed intake in lactating Norwegian red dairy cows. J Dairy Sci. 2018;101:6232–43.
Ehret A, Hochstuhl D, Gianola D, Thaller G. Application of neural networks with back-propagation to genome-enabled prediction of complex traits in Holstein-Friesian and German Fleckvieh cattle. Genet Sel Evol. 2015;47:22.
Ebrahimie E, Ebrahimi F, Ebrahimi M, Tomlinson S, Petrovski KR. Hierarchical pattern recognition in milking parameters predicts mastitis prevalence. Comput Electron Agr. 2018;147:6–11.
Zheng H, Wang H, Yan T. Modelling enteric methane emissions from milking dairy cows with Bayesian networks. In: Proceedings of the 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). Shenzhen, China. 2016. p. 1635–1640. https://doi.org/10.1109/BIBM.2016.7822764.
Abdollahi-Arpanahi R, Gianola D, Peñagaricano F. Deep learning versus parametric and ensemble methods for genomic prediction of complex phenotypes. Genet Sel Evol. 2020;52:12.
Ogutu JO, Piepho H-P, Schulz-Streeck T. A comparison of random forests, boosting and support vector machines for genomic selection. BMC Proc. 2011;5:S11.
Zhao T, Zeng J, Cheng H. Extend mixed models to multilayer neural networks for genomic prediction including intermediate omics data. Genetics. 2022;221:iyac034.
Pook T, Freudenthal J, Korte A, Simianer H. Using local convolutional neural networks for genomic prediction. Front Genet. 2020;11:561497.
Brito Lopes F, Magnabosco CU, Passafaro TL, Brunes LC, Costa MFO, Eifert EC, et al. Improving genomic prediction accuracy for meat tenderness in Nellore cattle using artificial neural networks. J Anim Breed Genet. 2020;137:438–48.
Liu S, Gao Y, Canela-Xandri O, Wang S, Yu Y, Cai W, et al. A multi-tissue atlas of regulatory variants in cattle. Nat Genet. 2022;54:1438–47.
Liu S, Yu Y, Zhang S, Cole JB, Tenesa A, Wang T, et al. Epigenomics and genotype-phenotype association analyses reveal conserved genetic architecture of complex traits in cattle and human. BMC Biol. 2020;18:80.
Yao Y, Liu S, Xia C, Gao Y, Pan Z, Canela-Xandri O, et al. Comparative transcriptome in large-scale human and cattle populations. Genome Biol. 2022;23:176.
MacLeod IM, Bowman PJ, Vander Jagt CJ, Haile-Mariam M, Kemper KE, Chamberlain AJ, et al. Exploiting biological priors and sequence variants enhances QTL discovery and genomic prediction of complex traits. BMC Genom. 2016;17:144.
Brondum RF, Su G, Lund MS, Bowman PJ, Goddard ME, Hayes BJ. Genome position specific priors for genomic prediction. BMC Genom. 2012;13:543.
Patxot M, Banos DT, Kousathanas A, Orliac EJ, Ojavee SE, Moser G, et al. Probabilistic inference of the genetic architecture underlying functional enrichment of complex traits. Nat Commun. 2021;12:6972.
Zhao T, Fernando R, Cheng H. Interpretable artificial neural networks incorporating bayesian alphabet models for genome-wide prediction and association studies. G3 (Bethesda). 2021;11:jkab228.
Demetci P, Cheng W, Darnell G, Zhou X, Ramachandran S, Crawford L. Multi-scale inference of genetic trait architecture using biologically annotated neural networks. PLoS Genet. 2021;17:e1009754.
Blei DM, Kucukelbir A, McAuliffe JD. Variational inference: a review for statisticians. J Am Stat Assoc. 2017;112:859–77.
Dos Santos JPR, Fernandes SB, McCoy S, Lozano R, Brown PJ, Leakey ADB, et al. Novel bayesian networks for genomic prediction of developmental traits in biomass sorghum. G3 (Bethesda). 2020;10:769–81.
Moser G, Lee SH, Hayes BJ, Goddard ME, Wray NR, Visscher PM. Simultaneous discovery, estimation and prediction analysis of complex traits using a bayesian mixture model. PLoS Genet. 2015;11:e1004969.
Zhang Y, Qi G, Park JH, Chatterjee N. Estimation of complex effect-size distributions using summary-level statistics from genome-wide association studies across 32 complex traits. Nat Genet. 2018;50:1318–26.
Lloyd-Jones LR, Zeng J, Sidorenko J, Yengo L, Moser G, Kemper KE, et al. Improved polygenic prediction by bayesian multiple regression on summary statistics. Nat Commun. 2019;10:5086.
Madsen P, Milkevych V, Gao H, Christensen OF, Jensen J. DMU - A Package for Analyzing Multivariate Mixed Models in Quantitative Genetics and Genomics. Poster session presented at ICAR Conference and World Congress on Genetics Applied to Livestock Production 2018, Auckland, New Zealand.
Habier D, Fernando RL, Kizilkaya K, Garrick DJ. Extension of the bayesian alphabet for genomic selection. BMC Bioinform. 2011;12:186.
Cheng H, Fernando R, Garrick D. JWAS: Julia implementation of Whole-genome Analyses Software. In: Proceedings of the World Congress on Genetics Applied to Livestock Production. vol. Methods and Tools - Software. 2018. p. 859.
Breiman L. Random forests. Mach Learn. 2001;45:5–32.
Jairath L, Dekkers JC, Schaeffer LR, Liu Z, Burnside EB, Kolstad B. Genetic evaluation for herd life in Canada. J Dairy Sci. 1998;81:550–62.
Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84:210–23.
Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7.
Legarra A, Reverter A. Semi-parametric estimates of population accuracy and bias of predictions of breeding values and future phenotypes using the LR method. Genet Sel Evol. 2018;50:53.
Song H, Zhang Q, Ding X. The superiority of multi-trait models with genotype-by-environment interactions in a limited number of environments for genomic prediction in pigs. J Anim Sci Biotechnol. 2020;11:88.
Zhou X, Carbonetto P, Stephens M. Polygenic modeling with bayesian sparse linear mixed models. PLoS Genet. 2013;9:e1003264.
Momen M, Morota G. Quantifying genomic connectedness and prediction accuracy from additive and non-additive gene actions. Genet Sel Evol. 2018;50:45.
van Binsbergen R, Calus MPL, Bink MCAM, van Eeuwijk FA, Schrooten C, Veerkamp RF. Genomic prediction using imputed whole-genome sequence data in Holstein Friesian cattle. Genet Sel Evol. 2015;47:71.
Zhang C, Kemp RA, Stothard P, Wang Z, Boddicker N, Krivushin K, et al. Genomic evaluation of feed efficiency component traits in Duroc pigs using 80K, 650K and whole-genome sequence variants. Genet Sel Evol. 2018;50:14.
Shi S, Li X, Fang L, Liu A, Su G, Zhang Y, et al. Genomic prediction using bayesian regression models with global-local prior. Front Genet. 2021;12:628205.
Wang D, Ning C, Liu JF, Zhang Q, Jiang L. Short communication: replication of genome-wide association studies for milk production traits in Chinese holstein by an efficient rotated linear mixed model. J Dairy Sci. 2019;102:2378–83.
Wu X, Fang M, Liu L, Wang S, Liu J, Ding X, et al. Genome wide association studies for body conformation traits in the Chinese holstein cattle population. BMC Genom. 2013;14:897.
Vallée A, Daures J, van Arendonk JAM, Bovenhuis H. Genome-wide association study for behavior, type traits, and muscular development in Charolais beef cattle. J Anim Sci. 2016;94:2307–16.
Zhang C, Shahbaba B, Zhao HK. Variational Hamiltonian Monte Carlo via score matching. Bayesian Anal. 2018;13:485–506.
Runcie DE, Qu J, Cheng H, Crawford L. MegaLMM: mega-scale linear mixed models for genomic predictions with thousands of traits. Genome Biol. 2021;22:213.
Ren D, Cai X, Lin Q, Ye H, Teng J, Li J, et al. Impact of linkage disequilibrium heterogeneity along the genome on genomic prediction and heritability estimation. Genet Sel Evol. 2022;54:47.
Finucane HK, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, Loh PR, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47:1228–35.
Fang LZ, Cai WT, Liu SL, Canela-Xandri O, Gao YH, Jiang JC, et al. Comprehensive analyses of 723 transcriptomes enhance genetic and biological interpretations for complex traits in cattle. Genome Res. 2020;30:790–801.
Acknowledgements
The authors gratefully acknowledge the constructive comments from reviewers. We thank Professor Stuart Barker (School of Environmental and Rural Science, University of New England) for his English editing to this paper.
Funding
This study was supported by the National Key Research and Development Program of China (2022YFD1302204), the earmarked fund CARS36, and Ningxia Key Research and Development Program of China (2023BCF01004; 2019NYYZ09).
Author information
Authors and Affiliations
Contributions
YZ and ZZ designed and supervised the study. XW wrote the program, analyzed the data, and wrote the manuscript. SLS and MYAK provided help on language polishing. SLS, YZ and ZZ revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Animal samples used in this study were approved by the Animal Care and Use Committee of China Agricultural University. There was no use of human participants, data or tissues.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflict of interest.
Supplementary Information
Additional file 1: Table S1
Accuracy, dispersion, and mean squared error (MSE) of genomic prediction on seven traits of dairy cattle using five-fold cross-validation with five replications; Table S2 The average computation time to complete each fold of five-fold CV for all genomic prediction methods.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wang, X., Shi, S., Ali Khan, M. et al. Improving the accuracy of genomic prediction in dairy cattle using the biologically annotated neural networks framework. J Animal Sci Biotechnol 15, 87 (2024). https://doi.org/10.1186/s40104-024-01044-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40104-024-01044-1