Abstract
Mangroves are salt-tolerant forest ecosystems of tropical and subtropical intertidal regions. They are among most productive, diverse, biologically important ecosystem and inclined toward threatened system. Identification of mangrove species is of critical importance in conserving and utilizing biodiversity, which apparently hindered by a lack of taxonomic expertise. In recent years, DNA barcoding using plastid markers rbcL and matK has been suggested as an effective method to enrich traditional taxonomic expertise for rapid species identification and biodiversity inventories. In the present study, we performed assessment of available 14 mangrove species of Goa, west coast India based on core DNA barcode markers, rbcL and matK. PCR amplification success rate, intra- and inter-specific genetic distance variation and the correct identification percentage were taken into account to assess candidate barcode regions. PCR and sequence success rate were high in rbcL (97.7 %) and matK (95.5 %) region. The two candidate chloroplast barcoding regions (rbcL, matK) yielded barcode gaps. Our results clearly demonstrated that matK locus assigned highest correct identification rates (72.09 %) based on TaxonDNA Best Match criteria. The concatenated rbcL + matK loci were able to adequately discriminate all mangrove genera and species to some extent except those in Rhizophora, Sonneratia and Avicennia. Our study provides the first endorsement of the species resolution among mangroves using plastid genes with few exceptions. Our future work will be focused on evaluation of other barcode markers to delineate complete resolution of mangrove species and identification of putative hybrids.
Similar content being viewed by others
Background
Mangroves are unique ecosystem exist along the sheltered inter-tidal coastline, in the margin between the land and sea in tropical and subtropical areas. This ecosystem endowed with productive wetland having flora and fauna adapted to local environment such as fluctuated water level, salinity and anoxic condition (Tomlinson 1986; Hutchings and Saenger 1987). They are most productive and biologically important ecosystems of the world which provide goods and services to human society in coastal and marine systems (FAO 2007). They have unique features such as aerial breathing roots, extensive supporting roots, buttresses, salt-excreting leaves and viviparous propagules (Duke 1992; Shi et al. 2006). The term ‘mangroves’ are referred to either individual plant or intertidal ecosystem or both, as ‘Mangrove plants’ and ‘Mangrove ecosystem’ (MacNae 1968). However, in this context we used mangrove term as a mangrove plants. Anthropogenic activity and climate are responsible for destruction of coastal mangroves vegetation. Globally among 11 of the 70 mangrove species were listed threatened species by International Union for Conservation of Nature (IUCN) (Polidoro et al. 2010).
Mangrove species diversity and distribution reported existence of 34 major and 20 minor mangrove species belonging to 20 genera and 11 families across the world (Tomlinson 1986). Ricklefs and Latham (1993) reported the existence of 19 genera with 54 mangrove species including few hybrids. According to world atlas of mangroves database, 73 mangrove species along with few recognized hybrids are distributed in 123 countries with territorial coverage of 150,000 km2 area globally (Spalding et al. 2010). Indian mangrove vegetation represents fourth largest in the world, distributed along the coastline and occupies 8 % of the total world mangrove covering 6749 km2 areas (Naskar and Mandal 1999). The entire mangrove habitats in India are situated in three zones: east coast (4700 km2), west coast (850 km2) and Andaman & Nicobar Islands (1190 km2). East coast zone ranges from Sundarban forest of West Bengal to Cauvery estuary of Tamil Nadu and comprises 70 % mangrove (Untawale and Jagtap 1992; Jagtap et al. 1993; Sanyal et al. 1998). West coast region stretches from Bhavnagar estuary of Gujarat to Cochin estuary of Kerala and constitute 15 % mangrove (Mandal and Naskar 2008). Mangrove flora of India constitutes about 60 species belonging to 41 genera and 29 families (Untawale 1985). Along the west coast of India, 34 species of mangroves belonging to 25 genera and 21 families have been reported. There are about 11, 20, 14 and 10 species of mangroves reported along the coast of Gujarat, Maharashtra, Goa and Karnataka respectively in western India. Goa state is located in western coast of India and mangrove vegetation in Goa occupies 500 ha of area (Government of India, 1997). The Cumbarjua canal (15 km) links the two river channels of Mandovi and Zuari, forming an estuarine complex which supports a substantial mangrove extent. D’Souza and Rodrigues (2013) reported the presence of 17 mangrove species in Goa that include 14 true and 3 associated mangrove species.
DNA barcoding is currently used effective tool that enables rapid and accurate identification of plant (Li et al. 2015). The Consortium for the Barcode of Life (CBOL) recommended rbcL + matK as the core barcode. However, these core barcode further combined with the psbA-trnH intergenic non-coding spacer region which improved discrimination power of core barcode. The non-coding intergenic region psbA-trnH exhibits high rates of insertion/deletion and sequence divergence (Kress and Erickson 2007). These features make trnH-psbA highly suitable candidate plant barcode for species resolution. Later on, the nuclear ribosomal internal transcribed spacer (ITS) region considered as supplementary barcode, though China Plant Barcode of Life claimed ITS region had higher discriminatory power than plastid core barcodes (CBOL Plant Working Group 2009; Hollingsworth et al. 2011; China Plant BOL Group 2011). Hollingsworth et al. (2011) observed ITS region has some limitations which prevent it from being a core barcode such as incomplete concerted evolution, fungal contamination and difficulties of amplification and sequencing. Plastid gene large subunit of the ribulose-bisphosphate carboxylase gene (rbcL) is of 1350 bp in length and choice for DNA barcoding (Chase 1993).The maturase gene matK is about 1500 bp long and located within the trnK gene encoding the tRNALys (UUU). Substitution rate of the matK gene is highest among the plastid genes (Hilu et al. 2003). Plastid gene matK can discriminate more than 90 % of species in the Orchidaceae but less than 49 % in the nutmeg family (Kress and Erickson 2007; Newmaster et al. 2008). In another case, identification of 92 species from 32 genera using the matK barcode could achieve a success rate of 56 % (Fazekas et al. 2008). However, a recent study of the flora of Canada revealed 93 % success in species identification with rbcL and matK, while the addition of the trnH-psbA intergenic spacer achieved discrimination up to 95 % (Burgess et al. 2011). Gonzalez et al. (2009) reported that species discrimination was lower (<50 %) for rbcL + matK combination in the study of tropical tree species in French Guiana. Lower discrimination were reported in closest and complex taxa of Lysimachia, Ficus, Holcoglossum and Curcuma using rbcL and matK (Xiang et al. 2011; Zhang et al. 2012; Li et al. 2012; Chen et al. 2015). The lowest discriminatory power was observed in closely related groups of Lysimachia with rbcL (26.5–38.1 %), followed by matK (55.9–60.8 %) and combinations of core barcodes (rbcL + matK) had discrimination of 47.1–60.8 % (Zhang et al. 2012).
Delineating mangrove species from putative hybrids using morphological characters are always questionable. Putative hybrids were reported within the major genera of Rhizophora, Sonneratia and Lumnitzera and recently in Bruguiera (Tomlinson 1986; Duke and Ge 2011). In the present study, we assessed mangrove species using plastid coding loci viz. rbcL and matK. Mangroves from Goa are rich in diversity and accounted 14 species belonging to four order and five families. This is our first step towards DNA barcoding of mangroves based on plastid genes. Our study might be helpful in identification as well as developing various strategies towards mangrove conservation.
Methods
Sample collection
In the present study, leaf samples of 14 mangrove species were collected from Goa, located on the west coast of India with geographical latitude of 15.5256°N and longitude of 73.8753°E. Mangrove species identification was performed based on morphological characteristics using a comparative guide to the Asian mangroves and mangroves of Goa (Yong and Sheue 2014; Dhargalkar et al. 2014; Setyawan et al. 2014). Herbarium of these specimens was deposited at Botanical Survey of India, western regional centre, Pune, India. The morphology based identification keys used to authenticate the taxon identities of 14 mangroves species from Goa were listed in supplementary information (Additional file 1: Table S1). The well identified voucher specimens along with their taxonomic information and collection details are listed (Table 1) with their photographs in supplementary information (Additional file 1: Fig. S1). The sequences obtained using barcode markers: rbcL and matK were submitted to the NCBI GenBank (Accession numbers indicated in Table 1), and publicly accessible through the dataset of project DNA Barcoding of Indian Mangroves (Project code: IMDB) in Barcode of Life Data systems (BOLD) (doi:10.5883/DS-IMDBNG) (Ratnasingham and Hebert 2007).
DNA extraction
High content of mucilage, latex, phenolics, secondary metabolites and polysaccharides in these plants make it a difficult system for protein and nucleic acid isolation from mangrove plants. Cetyl-trimethyl ammonium bromide (CTAB) protocol for DNA extraction from mangroves (Parani et al. 1997a) was modified. Leaf tissue was pulverized in liquid nitrogen and pulverized leaf sample (0.2 g) were mixed with CTAB buffer (20 mM EDTA; 1.4 M NaCl; 2 % PVP-30; 1 % β-mercaptoethanol; 10 % SDS and 10 mg/ml proteinase K). The suspension was incubated at 60 °C for 60 min with gentle mixing and centrifuged at 14,000 rpm for 10 min at room temperature with equal volume of chloroform: isoamyl alcohol (24:1). The aqueous phase was transferred to a new tube and DNA was precipitated with 0.6 volume of cold isopropanol (−20 °C) and chilled 7.5 M ammonium acetate followed by storing at −20 °C for 1 h. The precipitated DNA was centrifuged at 14,000 rpm for 10 min at 4 °C followed by washing with 70 % ethanol. DNA was finally dissolved in TE buffer (10 mM Tris–HCl, 1 mM Na2EDTA, pH 8.0) and its quantity and quality was confirmed by agarose gel electrophoresis and nanodrop (Thermo Scientific, USA).
PCR and sequencing
Amplification of plastid genes (rbcL and matK) was carried out in 50-μl reaction mixture containing 10–20 ng of template DNA, 200 μM of dNTPs, 0.1 μM of each primers and 1 unit of Taq DNA polymerase (Thermo Scientific, USA). The reaction mixture was amplified in Bio-Rad (T100 model) thermal cycler with temperature profile for rbcL (94 °C for 4 min; 35 cycles of 94 °C for 30 s, 55 °C for 30 s, 72 °C for 1 min; repeated for 35 cycles, final extension 72 °C for 10 min) and for matK (94 °C for 1 min; 35 cycles of 94 °C for 30 s, 50 °C for 40 s, 72 °C for 40 s; repeated for 37 cycles, final extension 72 °C for 5 min). The amplified products were separated by agarose gel (1.2 %) electrophoresis and stained with ethidium bromide (Sambrook et al. 1989). Two pair of universal primers rbcL (rbcLa_F and rbcLa_R) and matK_390f and matK_1326r were used for the amplification purpose (Kress and Erickson 2007; Vinitha et al. 2014; Chen et al. 2015). To amplify R. apiculata matK locus, we designed matK_RA reverse primer as follows: 5′-AAAGTTCGTTTGTGCCAATGA-3′. PCR products were purified according to manufacturer’s instruction (Chromous Biotech) and further sequencing reactions were carried out using the Big Dye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) and analyzed on ABI 3500xL Genetic Analyzer (Applied Biosystems).
Data analysis
Sequence alignment and assembly was achieved in Codon code Aligner v.3.0.1 (Codon Code Corporation) and MEGA 6 (Tamura et al. 2013). The NCBI BLAST was performed to confirm identity of specimens (Altschul et al. 1990). All known mangroves sequences were searched with our sequenced samples using ‘BLASTn’ tool against NCBI database and highest-scoring hit from each query is taken as the mangrove identification. Intraspecific, interspecific and barcode gap analysis was performed at Barcode of Life Data systems web portal. Further, rbcL and matK sequences were concatenated using DNASP v5.10 and analyzed in MEGA 6 for their resolution inference (Rozas, 2009). The effectiveness of the analysed barcodes in rbcL, matK and rbcL + matK was evaluated using TaxonDNA v1.6.2, Species Identifier 1.8 (Meier et al. 2006) and BLASTClust (http://toolkit.tuebingen.mpg.de/blastclust). Neighbor-joining (NJ) trees were constructed using MEGA 6.0 and K2P genetic distance model, and node support was assessed based on 1000 bootstrap replicates. Species with multiple individuals forming a monophyletic clade in phylogenetic trees with a bootstrap value above 60 % were considered as successful identification.
Results
DNA barcode and sequence analysis
Mangroves belonging to 14 species, 9 genera and 5 families were collected. We acquired high quality DNA barcodes for 45 specimens belonging to 14 species, which were sequenced for rbcL and matK. The sequencing result of rbcL produced an average of 510 bp without any insertion, deletion and stop codon, whereas matK sequencing produced 712 bp with few insertion and deletions in the form of gaps without stop codon. Overall GC content observed in rbcL was 43.29 % (SE = 0.09), while in matK it was 33.18 % (SE = 0.18). The mean GC content of codon at positions 1-3 in rbcL was 54.66 % (SE = 0.1), 45.77 % (SE = 0.09) and 29.44 % (SE = 0.21), and in matK, it was 33.15 % (SE = 0.18), 30.92 % (SE = 0.36), 29.91 % (SE = 0.25) respectively. The specimen data, collection site details and sequences were submitted to BOLD database in form of project IMDB (doi:10.5883/DS-IMDBNG) (For details, Table 1). The specimens were verified from sequenced data by performing NCBI BLAST. This is performed for preliminary verification for all mangroves at species level but downside in our case study is limited reference data for comparison. The rbcL and matK correctly identified genera up to 100 %, while species identification with rbcL and matK leads to 64 and 85 % identification respectively.
Intraspecific and interspecific relationship
Barcoding of mangrove exhibited absolute average interspecific differentiation of 0.35 % and 0.9 % in rbcL and matK respectively, while for species average intraspecific variability was 0.24 % in rbcL and 0.20 % in matK (Table 2) with low species resolution in few taxa. The intraspecific and interspecific analysis for rbcL revealed largest average pairwise distance of 0.68, while in matK it was 2.05 and 2.32 respectively. The highest range of congeneric differentiation in Bruguiera and Avicennia were observed in rbcL from 0 to 0.68, whereas for matK, it ranged from 1.29 to 2.31 in Avicennia, further suggesting significant genetic divergence within Avicennia.
Barcode gap analysis
The barcode gap analysis revealed highest intraspecific distance (>2 %) in 9 specimens of rbcL and 6 specimens of matK, while low intraspecific distance (<2 %) in 11 specimens of rbcL and 9 specimens of matK. Here, low intraspecific distance (<2 %) suggests low species resolution, thus leading to species overlap.
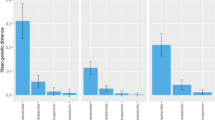
With rbcL the largest nearest neighboring distance of 8.43 was observed in Avicennia alba with mean intraspecific distance of 0.11 (Fig. 1a). The maximum intraspecific distance of 0.68 was observed within three individuals of Kandelia candel, Bruguiera gymnorrhiza, A. officinalis and Sonneratia caseolaris (Fig. 1b). With matK, maximum intraspecific distance of 2.05 was observed in Excoecaria agallocha with three individuals per species (Fig. 1d), while largest distance to the nearest neighbor of 24.65 was observed in A. officinalis with mean intraspecific distance of 0.12 (Fig. 1c). Overall average nearest neighboring divergence observed among mangroves using rbcL was 1.39 % (S.E = 0.17) and matK was 4.07 % (S.E = 0.5) (Fig. 1a).
Scatterplots confirming the existence and magnitude of the barcode gap. a For rbcL mean intra-specific versus Nearest Neighbor (NN). b For rbcL individuals per species vs. max intra-specific. c For matK mean intra-specific versus Nearest Neighbour. d For matK individuals per species versus max intra-specific
Species identification and assignment
The species were assigned to their taxa based on three methods, similarity based method using TaxonDNA, BLAST score based single linkage (BLASTClust) and tree based method (NJ). To assess the species assignment of single region and multi regions, we used the ‘Best Match’ (BM) and ‘Best Closest Match’ (BCM) criteria from TaxonDNA. For TaxonDNA analysis, we need to set threshold (T) below which 95 % of all intraspecific distances were found. All the results above the threshold (T) were treated as ‘incorrect’. Similarly, if all matches of the query sequence were below threshold (T), the barcode assignment was considered to be correct identification. The matches of the query sequence were equally good, but correspond to a mixture of species, then test was treated as ambiguous identification. For the single barcode region, matK had the highest rate of correct identification using BM (72.09 %) and BCM (39.53 %) than rbcL with (BM 47.72 %), BCM (31.81 %) (Table 3). The concatenated regions (rbcL + matK) demonstrated to resolve species at the level of 66.6 % using BM and BCM criteria (Table 3). The species specific clustering using match and mismatch criteria was evaluated in TaxonDNA and BLASTClust, where sequences with highest similarity and identity were considered as successfully identified. Those species with an identical barcode sequence to an individual of other species were considered as ambiguous, and sequences matching with different species names were treated as failure identifications. Species having single sample and unique sequence were considered as potentially distinguishable. The BLASTClust analysis revealed slightly different results than that of TaxonDNA, where the rate of species resolution and cluster formation was low as that of TaxonDNA (Table 4). Species with multiple individuals forming a monophyletic clade in NJ trees with a bootstrap value above 60 % were considered as successful identifications (Kress et al. 2010). The matK and rbcL + matK discriminated mangrove species in NJ model test method, while rbcL alone failed to identify those species (Fig. 2a–c). Further analysis revealed similar rates of species resolution using both methods for matK as well as rbcL (Table 5). Rhizophora, Sonneratia and Avicennia genera were failed to discriminate their species using plastid markers rbcL, matK and rbcL + matK.
Discussion
To the best of our knowledge, current study is the first attempt of performing DNA barcoding based assessment of mangroves from Goa using plastid core markers rbcL and matK. Some countable reports based on molecular taxonomy and phylogeny of Indian mangroves are available using nuclear, mitochondrial and plastid markers (ITS, rbcL, RFLP, RAPD, PCR-RAPD and AFLP) (Parani et al. 1997a, b; Lakshmi et al. 1997, 2000; Setoguchi et al. 1999; Schwarzbach and Ricklefs 2000). Besides this there are many reports of mangroves identification based on morphological characters (Untawale 1985; Tomlinson 1986; Untawale and Jagtap 1992). Present study revealed discrimination of mangroves based on DNA barcoding at species level excluding some taxa (Rhizophora, Sonneratia and Avicennia). Highest rate of PCR amplification and sequencing was observed in rbcL (97.7 %), while amplification as well as sequencing rate of matK was 95.5 %. Similarly, highest success rate of identification was observed with matK (80.5 %) in local temperate flora of Canada and in combination rbcL + matK identified 93 % flora (Burgess et al. 2011). Species identification success rate using rbcL seems to be higher, whereas rbcL recovery ranged from 90 to 100 % (Little and Stevenson 2007; Ross et al. 2008; CBOL Plant Working Group 2009). matK showed difficulties in PCR amplification and sequencing. Fazekas et al. (2008) showed that matK markers provide possibility of 88 % sequencing success, with the use of 10 primer pair combinations. Similarly, a lower amplification and sequencing success of matK has been reported in several other studies and amplification ranges from 42 to 70 % (Ford et al. 2009; Gonzalez et al. 2009; Kress et al. 2010; Hollingsworth et al. 2011). In contrast, CBOL reported that single pair of matK primer was successfully amplified and sequenced 84 % angiosperm species (CBOL Plant Working Group, 2009). We faced many hindrances in amplification and sequencing of Rhizophora genera species R. apiculata using universal matK primers. R. apiculata was amplified and sequenced using universal rbcL marker but for matK amplificaiton, we designed a reverse primer. The possible explanation for the trouble could be due to secondary metabolite might hindered amplification of target genes or failure of primers to amplify genes.
Initially, species identification was performed by NCBI BLAST using rbcL and matK sequence data, the BLAST could yield accurate identifications results (Hollingsworth et al. 2009; Kress et al. 2010; Kuzmina et al. 2012). On a similar note BLAST was performed revealing its least efficacy in species identification. It has been used for verification purpose in recent years and comparisons based on test datasets (Ford et al. 2009). Parmentier et al. (2013) reported that species assignment using BLAST method was reliable for genus identification of African rainforest tree (95–100 % success), but less for species identification (71–88 %). Sometimes it gave erroneous identifications, most often due to the limited number of available reference sequences. In the present study, BLAST result with default parameter, for rbcL successfully identified genera (100 %) and species identification rate was 64.28 % for 14 mangroves species. matK was able to identify genera (100 %) and species identification up to 85.71 % successfully. The possible reason for this erroneous assignment in some taxa at species level due to availability of limited sequences in the BOLD or GenBank database (Parmentier et al. 2013). Our result underscored the importance of BLAST method to assigned correct mangroves genera identification (with rbcL and matK). Both Sonneratia alba and Avicennia marina were incorrectly identified at species level using rbcL and matK. Some mangrove species viz. R. apiculata, B. cylindrica and A. alba were misidentified at species level using rbcL.
The genetic divergence analysis exhibited highest divergence in Avicennia species, while barcode gap and nearest neighbor analysis revealed low species resolution and barcode gap with nearest neighboring distance (<2 %), further confirming species overlap in Avicennia (A. officinalis (rbcL:0; matK: 0–1.71) and A. marina (rbcL: 0–0.34; matK: 0), Bruguiera (B. gymnorrhiza (rbcL: 0; matK: 0.61) and B. cylindrica (rbcL: 0–1.71; matK: 0.61), Rhizophora (R. mucronata (rbcL: 0; matK: 0.14) and R. apiculata (rbcL: 0; matK: 0.14), Sonneratia (S. caseolaris (rbcL: 0; matK: 0) and S. alba (rbcL: 0; matK: 0). Low genetic distances between species was largely due to the presence of species-rich genera with low sequence variation for the plastid genome (Burgess et al. 2011).
The species identification and taxon assignment was evaluated using TaxonDNA and BLASTClust for rbcL, matK and rbcL + matK. Overall matK marker showed good performance at species and genus level (Tables 3, 4). In contrast to matK; rbcL alone showed poor performance at species level identification. Combined, rbcL + matK markers showed better performance at species and genus level identification (Tables 3, 4, 5). Accordingly, plant CBOL group (2009) reported only 72 % species level resolution using combined rbcL and matK. Similar result was observed after combined rbcL and matK at species level resolution (Chen et al. 2015). Lowest resolution was recorded in closely related groups of Lysimachia with combination of rbcL and matK universal markers (Zhang et al. 2012). However, the identification rates based on TaxonDNA and phylogenetic tree methods (Tables 3, 5) were significant with matK as compared to rbcL. Low resolution using DNA barcoding regions has been documented in many other plants such as the genus Araucaria (32 %), Solidago (17 %) and Quercus (0 %) (Little and Stevenson 2007; Leon-Romero et al. 2012). In TaxonDNA analysis, for rbcL threshold (T) was observed 0 %, similar result was recorded for rbcL in the Zingiberaceae family (Chen et al. 2015). However, threshold (T) for Indian Zingiberaceae family members were recorded as 0.20 % for rbcL and 0 % for rpoB and accD (Vinitha et al. 2014). In BLASTClust, the rbcL and matK regions showed similar identification rates, while concatenation of both these regions increased the efficiency of species resolution as well as cluster formation (Gonzalez et al. 2009; Blaalid et al. 2013). In case of closest taxa of mangroves viz. Avicennia, Rhizophora and Sonneratia species, there is a need to explore new DNA barcode markers, which may leads to species level resolution.
Conclusions
DNA barcoding can be a very effective tool to identify mangroves. Here, we tested DNA barcodes of plant plastid DNA, rbcL and matK to resolve available mangrove species. For the single barcode region, matK had the highest rate of correct identification using BM and BCM than rbcL. When both regions were concatenated (rbcL + matK) their efficiency to resolve species was 66.6 % using BM and BCM criteria. In the present work, we lay the foundation towards DNA barcoding applications for mangroves plant genera viz. Acanthus, Kandelia, Ceriops, Bruguiera, Aegiceras and Excoecaria. matK is proposed to be a suitable candidate DNA barcode marker for mangrove species identification. Compiled mangroves barcoding result had some limitations, most of which are due to imperfect discrimination ability of the markers, natural hybridization and homoplasy. Further need to explore with additional markers which may improve mangrove species identification for practical conservation.
Abbreviations
- RbcL :
-
ribulose bisphosphate carboxylase large subunit
- matK :
-
maturase K
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Blaalid R, Kumar S, Nilsson HR, Abarenkov K, Kirk MP, Kauserud H (2013) ITS1 versus ITS2 as DNA metabarcodes for fungi. Mol Ecol Resour 13:218–224
Burgess KS, Fazekas AJ, Kesanakurti PR, Graham SW, Husband BC, Newmaster SG, Percy DM, Hajibabaei M, Barrett SCH (2011) Discriminating plants pecies in a local temperate flora using the rbcL + matK DNA barcode. Method Ecol Evol 2:333–340
CBOL Plant Working Group (2009) A DNA barcode for land plants. PNAS USA 106:12794–12797
Chase MW (1993) Phylogenetics of seed plants: an analysis of nucleotide sequences from the plastid gene rbcL. Ann Missouri Bot Gar 80:528–580
Chen J, Zhao J, Erickson DL, Xia N, Kress WJ (2015) Testing DNA barcodes in closely related species of Curcuma (Zingiberaceae) from Myanmar and China. Mol Ecol Resour 15:337–348
China Plant BOL Group (2011) Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. PNAS USA 108:19641–19646
D’Souza J, Rodrigues BF (2013) Biodiversity of arbuscular mycorrhizal (AM) fungi in mangroves of Goa in West India. J For Res 24:515–523
Dhargalkar VK, D’Souza R, Kavlekar DP, Untawale AG (2014) Mangroves of Goa. Forest department, Government of Goa and Mangroves society of India, Goa, India, pp 12–31
Duke NC (1992)Tropical mangrove ecosystems (Coastal and Estuarine studies). In: Robertson AI, Alongi DM (eds) American geophysical union, Washington, pp 63–100
Duke NC, Ge XJ (2011) Bruguiera (Rhizophoraceae) in the Indo-West Pacific: a morphometric assessment of hybridization within single flowered taxa. Blumea Biodiver Evol Biogeogr Plants 56:36–48
Fazekas AJ, Burgess KS, Kesanakurti PR, Graham SW, Newmaster SG, Husband BC, Percy DM, Hajibabaei M, Barrett SCH (2008) Multiple multilocus DNA barcodes from the plastid genome discriminate plant species equally well. PLoS ONE 3:e2802
Food and Agriculture Organization (2007) The world’s mangroves 1980–2005. United Nations, Rome, pp 1–6
Ford CS, Ayres KL, Toomey N, Haider N, Van Alphen Stahl J et al (2009) Selection of candidate coding DNA barcoding regions for use on land plants. Bot J Linean Soc 159:1–11
Gonzalez MA, Baraloto C, Engel J, Mori SA, Pétronelli P et al (2009) Identification of Amazonian Trees with DNA Barcodes. PLoS ONE 4:e7483
Government of India (1997) The State of Forest Report. Forest Survey of India, Ministry of Environment & Forests. New Delhi, vol 38, pp 5–6
Hilu KW, Borsch T, Muller K et al (2003) Angiosperm phylogeny based on matK sequence information. Am J Bot 90:1758–1776
Hollingsworth ML, Clark A, Forrest LL et al (2009) Selecting barcoding loci for plants: evaluation of seven candidate loci with species level sampling in three divergent groups of land plants. Mol Ecol Resour 9:439–457
Hollingsworth PM, Graham SW, Little DP (2011) Choosing and using a plant DNA barcode. PLoS ONE 6:e19254
Hutchings PA, Saenger P (1987) Ecology of mangroves. Australian Ecology Series University of Queensland Press, St Lucia, pp 14–44
Jagtap TG, Chavan VS, Untawale AG (1993) Mangrove ecosystems of India: a need for protection. Ambio 22:252–254
Kress J, Erickson DL (2007) A two locus global DNA barcode for land plants: the Coding rbcL Gene complements the non-coding trnH-psbA spacer region. PLoS ONE 2:e508
Kress WJ, Erickson DL, Swenson NG et al (2010) Advances in the use of DNA barcodes to build a community phylogeny for tropical trees in a Puerto Rican forest dynamics plot. PLoS ONE 5:e15409
Kuzmina ML, Johnson KL, Barron HR, Hebert PDN (2012) Identification of the vascular plants of Churchill, Manitoba, using a DNA barcode library. BMC Ecol 12:25
Lakshmi M, Rajalakshmi S, Parani M, Anuratha CS, Parida A (1997) Molecular phylogeny of mangroves I. Use of molecular markers in assessing the intra-specific gene variability in the mangrove species, Acanthus ilicifolius Linn. (Acanthaceae). Theor Appl Genet 94:1121–1127
Lakshmi M, Parani M, Ram N, Parida A (2000) Molecular phylogeny of mangroves IV. Intraspecific genetic variation in mangrove species Excoecaria agallocha L. (Euphorbiaceae). Genome 43:110–115
Leon-Romero Y, Mejıa O, Soto-Galera E (2012) DNA barcoding reveals taxonomic conflicts in the Herichthys bartoni species group (Pisces: Cichlidae). Mol Ecol Resour 12:1021–1026
Li HQ, Chen JY, Wang S, Xiong SZ (2012) Evaluation of six candidate DNA barcoding loci in Ficus (Moraceae)of China. Mol Ecol Resour 12:783–790
Li X, Yang Y, Henry RJ, Rossetto M, Wang Y, Chen S (2015) Plant DNA barcoding: from gene to genome. Biol Rev 90:157–166
Little DP, Stevenson DW (2007) A comparison of algorithms for the identification of specimens using DNA barcodes: examples from gymnosperms. Cladistics 23:1–21
MacNae M (1968) A general account of the fauna and flora of mangrove swamps and forest in the Indo-west pacific region. Adv mar biol 6:73–270
Mandal RN, Naskar KR (2008) Diversity and classification of Indian mangroves: a review. Trop Ecol 49:131–146
Meier R, Kwong S, Vaidya G, Ng PKL (2006) DNA barcoding and taxonomy in diptera: a tale of high intraspecific variability and low identification success. Syst Biol 55:715–728
Naskar KR, Mandal NR (1999) Ecology and biodiversity of Indian mangroves. Daya Publishing House, New Delhi, pp 386–388
Newmaster SG, Fazekas AJ, Steeves RAD, Janovec J (2008) Testing candidate plant barcode regions in the Myristicaceae. Mol Ecol Resour 8:480–490
Parani M, Lakshmi M, Elango S, Ram N, Anuratha CS, Parida A (1997a) Molecular phylogeny of mangroves II. Intra and inter-specific variation in Avicennia revealed by RAPD and RFLP markers. Genome 40:487–495
Parani M, Rao CS, Mathan M, Anuratha CS, Narayanan KK, Parida A (1997b) Molecular phylogeny of mangroves III. Parentage analysis of Rhizophora hybrid using random amplified polymorphic DNA (RAPD) and restriction fragment length polymorphism (RFLP) markers. Aquat Bot 58:165–172
Parmentier I, Duminil J, Kuzmina M, Philippe M, Thomas DW et al (2013) How effective are DNA barcodes in the Identification of African Rainforest Trees? PLoS ONE 8:e54921
Polidoro BA, Carpenter KE, Collins L, Duke NC, Ellison AM et al (2010) The loss of species: mangrove extinction risk and geographic areas of global concern. PLoS ONE 5:e10095
Ratnasingham S, Hebert PDN (2007) BOLD: the barcode of life data system (www.barcodinglife.org). Mol Ecol Notes 7:355–364
Ricklefs RE, Latham RE (1993) Global patterns of diversity in mangrove floras. In: Ricklefs RE, Schluter D (eds) Species diversity in ecological communities: historical and geographical perspectives. University of Chicago Press, Chicago, pp 215–230
Ross HA, Murugan S, Li WLS (2008) Testing the reliability of genetic methods of species identification via simulation. Syst Biol 57:216–230
Rozas J (2009) Polymorphism Analysis using DnaSP. In: Posada D (ed) Bioinformatics for DNA sequence analysis; methods in molecular biology series. Humana Press, New Jersey, vol 537, pp 337–350
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning, a laboratory manual, vol I, Cold Spring Harbor Laboratory Press, New York, pp 5.1–5.17
Sanyal P, Mandal RN, Ghosh D, Naskar KR (1998) Studies on the mangrove patch at Subarnarekha river mouth of Orissa state. J Interacad 2:140–149
Schwarzbach AE, Ricklefs RE (2000) Systematic affinities of Rhizophoraceae and Anisophylleaceae and intergeneric relationships within Rhizophoraceae, based on chloroplast DNA, nuclear ribosomal DNA, and morphology. Am J Bot 87:547–564
Setoguchi H, Kosuge K, Tobe H (1999) Molecular phylogeny of Rhizophoraceae based on rbcL gene sequences. J Plant Res 112:443–455
Setyawan AD, Ulumuddin YI, Ragavan P (2014) Review: Mangrove hybrid of Rhizophora and its parental species in Indo-Malayan region. Nusantara Biosci 6:69–81
Shi S, Huang Y, Zeng K, Tan F, He H, Huang J, Fu Y (2006) Molecular phylogenetic analysis of mangroves: independent evolutionary origins of vivipary and salt secretion. Mol Phylogenet Evol 40:298–304
Spalding M, Kainuma M, Collins L (2010) World atlas of mangroves. Earthscan eBook, pp 1–11
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Tomlinson PB (1986) The botany of mangroves. Cambridge University Press, Cambridge, pp 1–22
Untawale AG (1985) Mangroves of India: present status and multiple use practices, UNDP/UNESCO Regional Mangrove Project, pp 67
Untawale AG, Jagtap TG (1992) Floristic composition of the deltaic regions of India. Memoirs Geol Soc India 22:243–263
Vinitha RM, Kumar SU, Aishwarya K, Sabu M, Thomas G (2014) Prospects for discriminating Zingiberaceae species in India using DNA barcodes. J Integr Plant Biol 56:760–773
Xiang XG, Hu H, Wang W, Jin XH (2011) DNA barcoding of the recently evolved genus Holcoglossum (Orchidaceae: Aeridinae): a test of DNA barcode candidates. Mol Ecol Resour 11:1012–1021
Yong WHJ, Sheue CR (2014) A comparative guide to the asian mangroves. doi:10.13140/2.1.3765.1843
Zhang CY, Wang FY, Yan HF, Hao G, Hu CM, Ge XJ (2012) Testing DNA barcoding in closely related groups of Lysimachia L. (Myrsinaceae). Mol Ecol Resour 12:98–108
Authors’ contributions
AAS collected the samples, performed the experiments and drafted the manuscript. RAJ helped in data analysis. KK conceived of the study, participated in its design, coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Acknowledgements
AAS acknowledge the Senior Research Fellowship provided by University Grant Commission, India. Authors thank Mr. Vinod Naik, Department of Forest, Goa for his kind help in mangrove sample collection. This work was supported by research initiation grant from the Institute.
Competing interests
The authors declare that there is no competing interests.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Saddhe, A.A., Jamdade, R.A. & Kumar, K. Assessment of mangroves from Goa, west coast India using DNA barcode. SpringerPlus 5, 1554 (2016). https://doi.org/10.1186/s40064-016-3191-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40064-016-3191-4