Abstract
Multidrug resistant organisms (MDROs) constitute a major public health threat globally. Clinical isolates of Pseudomonas aeruginosa remains one of the most studied MDROs however there is paucity of information regarding the susceptibility of its animal and plants isolates to antipseudomonas drug in Nigeria. From a total of 252 samples consisting of plants, animals and clinical samples, 54, 24 and 22 P. aeruginosa were isolated from vegetables, animals and clinical sources respectively. All the isolates were identified by standard biochemical methods. Antimicrobial susceptibility testing (AST) of the 100 P. aeruginosa isolates against 7 antipseudomonal drugs was carried out by disk diffusion method, the phenotypic detection of ESBL was done by double disk synergy test (DDST) while plasmid extraction on 20 selected isolates based on their resistance to 2 or more classes of antibiotics was carried out by alkaline lysis method and analysed with Lambda DNA/Hind lll marker respectively. The AST results revealed highest resistance of 91 and 55 % to ceftazidime and carbenicillin respectively while highest susceptibilities of 99 % for piperacillin–tazobactam and imipenem were recorded in overall assay. Fifteen out of 100 isolates specifically (10) from vegetables, (3) clinical and (2) poultry isolates showed synergy towards the beta-lactamase inhibitor indicating production of ESBL by DDST method. Detection of plasmids was among vegetable (n = 4), poultry (n = 4), cow (n = 3) and clinical isolates (n = 1). Plasmid profile for the selected isolates revealed 6 of the strains had one plasmids each while 5 strains possessed 2–4 plasmids and 1 strain had 5 plasmids. The sizes of the plasmid range from <1 to ≥23kbp. Detection of ESBL and Plasmids among the investigated isolates is suggestive of multiple interplay of resistance mechanism among the isolates. Plants and animal isolates of P. aeruginosa harbouring multiple mechanisms of resistance is of concern due to the danger it poses on the public health.
Similar content being viewed by others
Background
Antimicrobial resistance (AMR) among bacterial species is now global public threat. AMR in bacteria is commonly associated with different resistant genes, transfer mechanisms, and antibiotic resistance reservoirs among different species. Indiscriminate use of antibiotics and prescriptions has been linked together as one of the instrumental factors leading to the emergence of multidrug resistant organisms (MDROs) in the community/hospital (Bronzwaer et al. 2002; Tarai et al. 2013). Unabated spread and proliferation of AMR genes among pathogenic bacteria has regrettably been the cause of high morbidity and mortality among critically ill patients, and has been found to contribute substantially to rising cost of healthcare due to prolong admission and expensive drugs for treatments annually (Carmeli et al. 2002; Roberts et al. 2009).
The use of antibiotics in farming and livestock management has also contributed to the spread of antibiotic resistance genes in the environment (Kümmerer 2004). In Agriculture, management of livestock and crop production often involve the use antibiotics in form of consumables that are usually excreted as biologically active metabolites in the environment. This can increase selective pressure and favours the growth of antibiotic resistant strains of bacteria (Kümmerer 2004). Studies on bacteria isolated from raw vegetables sold in different markets and eateries, that are resistant to commercially available antibiotics have been documented previously (Bhutani et al. 2015; Nipa et al. 2011).
Other studies have also focused on incidence of pet, food processing and farm animals as reservoirs for antibiotic resistant bacteria (Barber et al. 2003; Guardabassi et al. 2004; Faldynova et al. 2008). Worthy of mention are documented reports of high incidence of AMR bacteria from poultry, cattle as well other food processing animals, which are frequently consumed (Faldynova et al. 2008; Ayeni et al. 2016). It is also believed that health workers and families may serve as a conduit for the entry of resistance genes into the community and hospital environment where further spread into pathogen is possible (Molbak et al. 1999; Voss et al. 2005).
Pseudomonas aeruginosa is a Gram-negative, rod shaped; motile bacterium found abundant in various habitats and has been implicated in several human infections. Most P. aeruginosa infections are difficult to treat due to its ability to resist many structurally unrelated antibiotics, which is because of the presence of intrinsic and acquired antibiotic resistance mechanisms (Lister et al. 2009). Antimicrobial drug resistance mechanisms and its acquisition in P. aeruginosa are remarkable, to the favour of the pathogen. Several mechanisms of resistance to antimicrobial agents ranging from efflux pump to mobile genetic elements and hydrolysing enzymes have been described in clinical isolates of P. aeruginosa (Mesaros et al. 2007; Odumosu et al. 2013, 2015a, b). Notably among these mechanisms is the production of various classes of β-lactamases that mediates their resistance to beta-lactam drugs. Over the last decades, enzymes in this category have been detected in P. aeruginosa, especially extended-spectrum beta-lactamase (ESBL) such as OXA, VEB, PER, SHV and TEM types of ESBL (Odumosu et al. 2015; Mirsalehian et al. 2010).
Pseudomonas aeruginosa has been reported to account for up to 10 % of all human infections and is one of the important bacterial pathogens commonly isolated from various clinical and environmental samples (Rajat et al. 2012). It is also one of the major cause of diseases such as otitis, mastitis, endometritis, hemorrhagic pneumonia and urinary tract infections in both livestock and companion animals (Kidd et al. 2011; Poonsuk and Chuanchuen 2012; Salomonsen et al. 2013). However, in comparison with reported cases of hospital acquired infections, community associated infection due to P. aeruginosa is rarely reported (Huhulescu et al. 2011). Its ubiquity in the environment is with a high possible risk of plant-animal-human transfer of antimicrobial genes. However, due to the paucity of it’s the susceptibility from animals and plant origin to antispseudomonas drugs in Nigeria, a justification for this study was established. The objective of this study was to provide a snapshot of their resistance to antipseudomonas drug and production of ESBL. This will provide us a better understanding of how AMR are disperse in our environment and subsequently guide against it. For a good comparison, clinical isolates of P. aeruginosa from various samples were also included.
Methods
Between 2014 and 2015, from 252 samples comprising of 112 clinical samples, 82 vegetables, 41 cow dungs and 17 poultry dungs, 100 consecutive and non-duplicated P. aeruginosa were randomly isolated of which 22 were obtained from clinical isolates, 54 from vegetables, 7 from cows and 17 from poultry. These samples were collected at different farms, hospitals at different geographical locations across south-western Nigeria states making these isolates to have different epidemiological relationships. All samples were obtained by informed consent of the patients in this study, proper ethical clearance approval was obtained from Babcock University Health Research Ethics committee NHREC/17/12/2013.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed and interpreted according to the Clinical Laboratory Standard Guidelines 2010 (CLSI 2010) on Mueller–Hinton agar (Oxoid UK) for 7 antibiotics: piperacillin–tazobactam (100/10 µg), cefepime (30 µg) imipenem (10 µg), amikacin (30 µg), ciprofloxacin (5 µg), carbenicillin (100 µg) and ceftazidime (30 µg) (OXOID UK). P. aeruginosa ATCC 27853 and E. coli ATCC 25922 were used as quality control.
Phenotypic detection of ESBL by double-disk synergy test method (DDST)
A modified method for DDST (Jarlier et al. 1988), was performed for all the beta-lactam-resistant and intermediate resistant strains as a standard disk diffusion assay on Mueller–Hinton agar. A 0.5 Mac Farland standard suspension of the test bacteria were evenly inoculated on the Mueller–Hinton agar by gentle swabbing using sterile swabbing sticks. Disks containing 30 μg of aztreonam, ceftazidime, and cefepime, were placed 20 mm apart (centre to centre) consecutively from piperacillin/tazobactam (110 μg) and incubated for 18–24 h at 37 °C. Enhancement of the inhibition zone towards the piperacillin–tazobactam disc, indicating synergy between piperacillin–tazobactam and any one of test antibiotics, was regarded as presumptive ESBL production. Klebsiella pneumoniae ATCC 700603 and Escherichia coli ATCC 25922 were used for positive and negative controls respectively.
Plasmid isolation and profiling
Plasmids DNA for selected ESBL positives and multidrug resistance strains totaling 20 was performed as described Kado and Liu (Kado and Liu 1981). Briefly, bacterial cultures from Mueller–Hinton broth were suspended in microcentrifuge tubes with lysis buffer, the suspension was heated for 15 min at 70 °C and mixed with an equal volume of phenol:chorophorm:isoamyl alcohol (25:24:1) for plasmid extraction. The supernatants were loaded on a 1 % agarose gel in Tris–acetate–EDTA buffer and run for an hour at 80 V. The representative plasmids were digested with EcoRI while Lambda DNA/HindIII marker was used for molecular size estimation by comparing their motilities on agarose gel with the marker. The DNA bands were visualized and photographed by using Gel Documentation system after staining with gel red and with UV transillumination.
Statistical analysis
The Chi square test was used to compare the difference in the prevalence of isolates recovered from the various categories of samples at significant level, α = 5 %.
Result
Pseudomonas aeruginosa distribution among the samples collected is as follows; poultry 100 %, vegetables 54/82 (66 %), clinical isolates 22/112 (19.6 %) and cow dungs 7/41 (17 %), The results of the antimicrobial susceptibility testing for P. aeruginosa shows a high-level resistance (100 %) to ceftazidime among clinical and animal isolates, while vegetable isolates showed 45 (83.3 %) resistance (Tables 1, 2, 3, 4, 5). The overall susceptibility studies show 55 and 91 % isolates were resistant to carbenicillin and ceftazidime respectively. The susceptibilities of the isolates to imipenem, piperacillin–tazobactam, cefepime, ciprofloxacin and amikacin were 99, 99, 98, 96 and 88 % respectively. Phenotypic detection of ESBL among the P. aeruginosa isolates was confirmed in 15 of 100. Accordingly, a total percentage of 43 positive ESBL was detected in overall, this comprised of 18 % (10/54) from vegetables, 13 % (3/22) from clinical and 12 % (2/17) from poultry isolates. ESBL was detected at the 25 mm distance synergy towards cefepime and aztreonam in all the isolates. Plasmid profiles for the selected 20 isolates are represented in Table 6. Twelve isolates harboured multiple plasmids of copies ranging from 1 to 5 and estimated size ranges of < 1 to ≥ 23 (kbp). Highest number of plasmids 4 (33.33 %) were detected from vegetables and poultry isolates, 3 (25 %) was detected in cow while only 1 (8.33 %) was detected in clinical isolates. All the isolates harbouring plasmids were resistant to carbenicillin and ceftazidime but shows complete susceptibilities to cefepime, imipenem, and piperacillin–tazobactam antibiotics. Statistical analysis shows no significant difference in the prevalence of isolate recovered from the various categories at α = 5 % significant level.
Discussion
The result of the investigation in this study has revealed the various distribution of P. aeruginosa across the samples indicative of the pathogen ubiquity especially in the environment. A high count of P. aeruginosa was found in ready to eat vegetables than those isolated from animal and human sources. Similar isolation of high prevalence of P. aeruginosa from vegetables has been previously reported (Oluyege et al. 2015; Sabry et al. 2011). Presence of P. aeruginosa in animal faeces that is intentionally used as fertilizers or unintentionally in form of droppings from free-range animals in the soil suggests a possible source of contamination of the raw vegetables as well as the hands of sellers or water used in washing the food substances. Clinical isolates of P. aeruginosa recovered in the samples were lower in comparison to the number of samples collected. Even though P. aeruginosa is ubiquitous, the prevalence of its isolation among animal isolates in this study was lower compared to other general such as Enterococci and Escherichia which are commonly encountered and as previously reported from other studies (Ayeni et al. 2016; Ojo et al. 2013).
Comparing the sensitivity patterns of the isolates recovered from the three sources, it was observed that isolates from poultry showed highest number of resistance to ceftazidime and carbenicillin, the two most resisted antipseudomonas drug in this study. Amikacin resistance and intermediate susceptibility among poultry isolates were also highest (29.4 %) compared to all other sources. The observed high resistance by poultry isolates in this study suggest the possibility of selective pressure developed as a result of antibiotic use in management of poultry. It has been previously reported that food processing animals especially poultry serves as reservoirs for antibiotic resistance genes due to antibiotic use in poultry management (Diarrassouba et al. 2007; Mena et al. 2008). Such use of drugs is less common in the management of cattle and vegetable propagation because cattle rearing in Nigeria is largely carried out by local herdsman in a nomadic fashion with little or no education on the use of growth promotion in livestock business (Author’s personal observation).
In the present study, 100 % susceptibilities to ciprofloxacin, cefepime, imipenem and piperacillin/tazobactam was observed among the plants and animal P. aeruginosa isolates. This is in agreement with a recent study from France where a high proportion of the P. aeruginosa isolates from cattle and horse were susceptible to similar antibiotics as reported in this study (Marisa et al. 2015). However among the clinical isolates, there were pockets of resistance to cefepime, imipenem and piperacillin/tazobactam at 4.5 % each, while 18.2 % of the isolates showed resistance to ciprofloxacin suggesting the frequent use of these antibiotics clinically may play a significant role in its increasing resistance.
The phenotypic detection of ESBL among the isolates in this study was carried out with a modified method by including a fourth generation cephalosporin (cefepime) and tazobactam as the beta-lactamase inhibitor. Cefepime is resistant to the production of AmpC beta-lactamase commonly produced among P. aeruginosa that always mask the detection of ESBL phenotypically (Tzelepi et al. 2000). Previous studies have documented tazobactam inhibitory activity against ESBL and AmpC beta-lactamase to be almost tenfold greater than clavulanic acid (Bush et al. 1993; Phillippon et al. 2002).
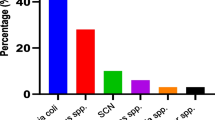
Out of 100 isolates investigated for ESBL phenotypically, 15 % of the isolates were positive with synergies towards cefepime. This result is higher than observed frequency of 4.0 and 8.1 % reported from Turkey and Iran respectively (Gençer et al. 2002; Tavajjohi et al. 2011) but is in contrast with two previous Nigerian studies that reported higher frequencies of ESBL in P. aeruginosa (Aibinu et al. 2007; Akinjogunla et al. 2010) although percentages sometimes is a reflection of the population of the study sample hence in essence may not reflect higher or lower proportion perse (Figs. 1, 2).
In the present study, high resistance to third generation cephalosporin (ceftazidime) and carboxypenicillins (carbenicillin) both of beta-lactam class of antibiotics is worrisome. Ninety-one percent (91 %) of the P. aeruginosa investigated from clinical, animal and plant sources were resistant to ceftazidime in this study while 55 % were resistant to carbenicillin. According to previous studies, resistance to ceftazidime and carbenicillin by P. aeruginosa is mostly due to the presence of extended-spectrum beta-lactamases such as OXA types and PER that are commonly detected in P. aeruginosa and are usually encoded on mobile genetic elements such as plasmids and integrons (Odumosu et al. 2013, 2015; Alikhani et al. 2014). Interestingly the P. aeruginosa investigated in this study for the presence of plasmids all had diverse plasmids profile with copies ranging from 1 to 5 and estimated size ranges of <1 to ≥23 (kbp) (Table 6). Plasmids are mobile genetic elements and can also facilitate the dispersal of resistance genes among the bacterial population and can also serve as vehicle for other resistance mechanisms. Although the present study did not investigate the association of plasmids with the observed resistance, previous studies have shown plasmids with capacity for multiple resistance to antimicrobial agents. Incidence of these genes in animal and plants for human consumption is dangerous because of the risk of transmission of resistant strains from animal origin to humans. Cattles and poultry are frequently consume in Nigeria as part of normal diet and some people eat roasted chicken and beef without proper boiling to kill all the available pathogen. This may lead to serious public health hazard if such animal or plant sources are consumed.
Conclusion
The present study provides a view into the antibiotic susceptibility profile of P. aeruginosa isolated from plants animals and humans to antipseudomonas drugs. The multiplicity of antibiotic resistance, detection of ESBL and plasmids among the isolates in this study is disturbing considering the fact that it establishes their potential abilities to the spread of antimicrobial resistance. The abundance MDROs from animal and plants in this study suggests that risk of human susceptibility to AMR bacteria from these sources is plausible. Although this study did not provide a direct evidence of their link and to what extend these sources contribute to AMR in comparison to clinical sources and hospital environments. Urgent measures are necessary to restrict the continuous abuse of antibiotics especially in livestock production. Further studies will be carried out in continuation, to fill the pending gaps in this study such as recognizing the relationship between the AMR bacteria from the animals and humans.
References
Aibinu I, Nwanneka T, Odugbemi T (2007) Occurrence of ESBL and MBL in clinical isolates of Pseudomonas aeruginosa from Lagos. Nigeria. J Amer Sci 3:81–85
Akinjogunla OJ, Odeyemi AT, Olasehinde GI (2010) Epidemiological studies of urinary tract infection (UTI) among post-menopausal women in Uyo Metropolis South-South Nigeria. J Am Sci 6:1674–1681
Alikhani MY, Tabar ZK, Mihani F, Kalantar E, Karami P, Sadeghi M, Khosroshahi SA, Farajnia S (2014) Antimicrobial resistance patterns and prevalence of blaPER-1 and blaVEB-1 genes among ESBL-producing Pseudomonas aeruginosa isolates in West of Iran. Jundishapur J Microbiol 7:1–5
Ayeni FA, Odumosu BT, Oluseyi AE, Ruppitsch W (2016) Identification and prevalence of tetracycline resistance in enterococci isolated from poultry in Ilishan, Ogun State, Nigeria. J Pharm Bioall Sci 8:69–73
Barber DA, Miller GY, McNamara PE (2003) Models of antimicrobial resistance and foodborne illness: examining assumptions and practical applications. J Food Protect 66:700–709
Bhutani N, Muraleedharan C, Talreja D, Rana SW, Walia S, Kumar A, Walia SK (2015) Occurrence of multidrug resistant extended spectrum beta-lactamase-producing bacteria on iceberg lettuce retailed for human consumption. BioMed Res Int. doi:10.1155/2015/547547
Bronzwaer S, Cars O, Buchholz U, Molstad S, Goettsch W, Veldhuijzen IK et al (2002) A European study on the relationship between antimicrobial use and antimicrobial resistance. Emerg Infect Dis 8:278–282
Bush K, Macalintal C, Rasmussen BA, Lee VJ, Yang Y (1993) Kinetic interaction of tazobactam with beta-lactamases from all major structural classes. Antimicrob Agents Chemother 37:851–858
Carmeli Y, Eliopoulos G, Mozaffari E, Samore M (2002) Health and economic outcomes of vancomycin-resistant enterococci. Arch Intern Med 162:2223–2228
CLSI (2010) Performance standards for antimicrobial susceptibility testing; 24th informational supplement. M100-S24. Clinical and Laboratory Standards Institute, Wayne
Diarrassouba F, Diarra MS, Bach S, Delaquis P, Pritchard J, Topp E et al (2007) Antibiotic resistance and virulence genes in commensal Escherichia coli and Salmonella isolates from commercial broiler chicken farms. J Food Prot 70:1316–1327
Faldynova M, Videnska P, Havlickova H, Sisak F, Juricova H, Babak V et al (2008) Prevalence of antibiotic resistance genes in faecal samples from cattle, pigs and poultry. Vet Med Czech 58:298–304
Gençer S, Öznur AK, Benzonana N, Batırel A, Özer S (2002) Susceptibility patterns and cross resistances of antibiotics against Pseudomonas aeruginosa in a teaching hospital of Turkey. Ann Clin Microbiol Antimicrob 1:1–4
Guardabassi L, Schwarz S, Lloyd DH (2004) Pet animals as reservoirs of antimicrobial-resistant bacteria. J Antimicrob Chemother 54:321–332
Huhulescu S, Simon M, Lubnow M, Kaase M, Wewalka G, Pietzka AT (2011) Fatal Pseudomonas aeruginosa pneumonia in a previously healthy woman was most likely associated with a contaminated hot tub. Infection 39:265–269
Jarlier V, Nicolas M, Fournier G, Philippon A (1988) Extended spectrum β-lactamase conferring transferable resistance to newer β-lactam agent in Enterobacteriaceae: hospital prevalence and suscpeitbility patterns. Rev Infect Dis 10:867–878
Kado CI, Liu ST (1981) Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol 145:1365–1373
Kidd TJ, Gibson JS, Moss S, Greer RM, Cobbold RN, Wright JD et al (2011) Clonal complex Pseudomonas aeruginosa in horses. Vet Microbiol 149:508–512
Kümmerer K (2004) Resistance in the environment. J Antimicrob Chemother 54(2):311–320
Lister PD, Wolter DJ, Hanson ND (2009) Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 22:582–610
Marisa H, Didier H, Cecile P, Pascal C, Christophe G, Jean-Yves M, Xavier B (2015) Population structure and antimicrobial susceptibility of Pseudomonas aeruginosa from animal infections in France. BMC Vet Res 11:9
Mena C, Rodrigues D, Silva J, Gibbs P, Teixeira P (2008) Occurrence, identification, and characterization of Campylobacter species isolated from Portuguese poultry samples collected from retail establishments. Poult Sci 87:187–190
Mesaros N, Nordmann P, Plésiat P (2007) Pseudomonas aeruginosa: resistance and therapeutic options at the turn of the new millennium. Clin Microbiol Infect 6:560–578
Mirsalehian A, Feizabadi M, Nakhjavani FA, Jabalameli F, Goli H, Kalantari N (2010) Detection of VEB-1, OXA-10 and PER-1 genotypes in extended-spectrum-lactamase-producing Pseudomonas aeruginosa strains isolated from burn patients. Burns 36:70–74
Molbak K, Baggesen DL, Aarestrup FM, Ebbesen JM, Engberg J, Frydendahl K et al (1999) An outbreak of multidrug-resistant, quinolone-resistant Salmonella enterica serotype Typhimurium DT104. New Eng J Med 341:1420–1425
Nipa MN, Mazumdar RM, Hasan MM, Fakruddin MD, Islam S, Bhuiyan HR, Iqbal A (2011) Prevalence of multi drug resistant bacteria on raw salad vegetables sold in major markets of Chittagong City, Bangladesh. Middle-East J Sci Res 10:70–77
Odumosu BT, Adeniyi BA, Chandra R (2013) Analysis of integrons and associated gene cassettes in clinical isolates of multidrug resistant Pseudomonas aeruginosa from Southwest Nigeria. Ann Clin Microbiol Antimicrob 21(12):1
Odumosu BT, Adeniyi BA, Chandra R (2015a) First detection of OXA-10 extended-spectrum beta-lactamases and the occurrence of mexR and nfxB in clinical isolates of Pseudomonas aeruginosa from Nigeria. Chemotherapy 61:87–92
Odumosu BT, Adeniyi BA, Chandra R (2015b) Occurrence of aminoglycoside-modifying enzymes genes (aac (6′)-I and ant (2′′)-I) in clinical isolates of Pseudomonas aeruginosa from Southwest Nigeria. Afri Health Sci 15:1277–1281
Ojo OE, Bello AO, Ogunjobi OO, Ajadi RA (2013) Antimicrobial resistance of gram-negative aerobic bacteria isolates from the faeces of diarrhoeic and non-diarrhoeic dogs in Abeokuta, Nigeria. Trop Vet 31:55–64
Oluyege JO, Oluwaniyi TT, Ijasan OC (2015) Composition of antibiotic resistant bacteria from irrigated vegetable farmland. J Microbiol Res 5:161–168
Phillippon A, Arlet G, Jacoby GA (2002) Plasmid-determined AmpC type β-lactamases. Antimicrob Agents Chemother 46:1–11
Poonsuk K, Chuanchuen R (2012) Contribution of the MexXY multidrug efflux pump and other chromosomal mechanisms on aminoglycoside resistance in Pseudomonas aeruginosa isolates from canine and feline infections. J Vet Med Sci 74:1575–1582
Rajat RM, Ninama GL, Mistry K, Parma R, Patel K, Vegad MM (2012) Resistance pattern in Pseudomonas aeruginosa species isolated at a tertiary care hospital, Amabadad. Natl J Med Res 2(2):156–159
Roberts R, Hota B, Ahmad I, Scott RD, Foster SD, Abbasi F, Schabowski S, Kampe LM, Ciavarella GG, Supino M, Naples J, Cordell R, Levy SB, Weinstein RA (2009) Hospital and societal costs of antimicrobial-resistant infections in a Chicago teaching hospital: implications for antibiotic stewardship. Clin Infect Dis 49:1175–1184
Sabry AH, Abdullah DA, Youssuf AG, Bahig AE (2011) Bacterial load of fresh vegetables and their resistance to the currently used antibiotics in Saudi Arabia. Foodborne Path Dis 8:1011–1018
Salomonsen CM, Themudo GE, Jelsbak L, Molin S, Hoiby N, Hammer AS (2013) Typing of Pseudomonas aeruginosa from hemorrhagic pneumonia in mink (Neovison vison). Vet Microbiol 163:103–109
Tarai B, Das P, Kumar D (2013) Recurrent challenges for clinicians: emergence of methicillin-resistant Staphylococcus aureus, vancomycin resistance, and current treatment options. J Lab Phys 5:71
Tavajjohi Z, Rezvan M, Ahmad K (2011) Detection and characterization of multidrug resistance and extended-spectrum-beta-lactamase-producing (ESBLS) Pseudomonas aeruginosa isolates in teaching hospital Zahra. Afr J Microbiol Res. 5:3223–3228
Tzelepi E, Giakkoupi A, Sofianou D, Loukova V, Kemeroglou A, Tsakris A (2000) Detection of extended-spectrum beta-lactamases in clinical isolates of Enterobacter cloacae and Enterobacter aerogenes. J Clin Microbiol 38:542–546
Voss A, Loeffen F, Bakker J, Klaassen C, Wulf M (2005) Methicillin resistant Staphylococcus aureus in pig farming. Emerg Infect Dis 11:1965–1966
Authors’ contributions
BTO conceived and planned the study. OA carried out the experiment under the supervision of BTO and co-supervised by IO. HDA provided clinical isolates. BTO wrote the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Odumosu, B.T., Ajetunmobi, O., Dada-Adegbola, H. et al. Antibiotic susceptibility pattern and analysis of plasmid profiles of Pseudomonas aeruginosa from human, animal and plant sources. SpringerPlus 5, 1381 (2016). https://doi.org/10.1186/s40064-016-3073-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40064-016-3073-9