Abstract
Bacterial bloodstream infections are a major public health problem, which leads to high morbidity and mortality of patients. On time diagnosis and appropriate medication will be the best way to save the lives of affected ones. The aim of the present study was to determine the bacterial profile of bloodstream infections and their antibiotic susceptibility pattern in Mekelle Hospital. Cross sectional study method was carried out in 514 (269 females and 245 males) febrile patients in Mekelle hospital from March to October 2014. Standard bacteriological methods were used for blood collection, bacterial isolation and antimicrobial susceptibility pattern. Out of the total 514 febrile patients, 144 (28%) culture positive were isolated. Staphylococcus aureus 54 (37.5%), Coagulase-negative staphylococci 44 (30.6%), Escherichia coli 16 (3.1%), Citrobacter spp. 9 (1.7%) and Salmonella typhi 8 (1.6%) were the most dominant isolates, collectively accounting for >90% of the isolates. Antimicrobial resistance pattern for gram positive and gram negative bacteria was 0–83.3% and 0–100%, respectively. High resistance was seen to Trimethoprim-sulphamethoxazole 101 (70.1%), Oxacillin 65 (62.5%), Ceftriaxone 79 (58.9%) and Doxycycline 71 (49.3%). Fifty-nine percent of the isolated bacteria in this study were multi drug resistant. Most bacterial isolates were sensitive to Gentamicin, Ciprofloxacin and Amoxicillin clavulanic acid. All gram positive isolates in this current study were sensitive to vancomycin. Prevalence of bacterial isolates in blood was high. It also reveals isolated bacteria species developed multi drug resistance to most of the antibiotics tested, which highlights for periodic surveillance of etiologic agent, antibiotic susceptibility to prevent further emergence and spread of resistant bacterial pathogens.
Similar content being viewed by others
Background
Bloodstream infection (BSI) remains one of the most important causes of morbidity and mortality globally (Zenebe et al. 2011). Though the problem is still common in developed nations (Diekma et al. 2002), the burden is high in sub Saharan countries with 53% children mortality rate (Aiker et al. 2011).
Many bacteria have been reported which cause bacteraemia with variation in distribution from place to place (Daniel et al. 2006; Asrat and Amanuel 2001; James et al. 2004; Gohel et al. 2014; Rina et al. 2007). Infection by these organisms causes prolonged patient hospitalization, increased healthcare costs and mortality rate of patients (Tziamabos and Kasper 2005).
Treatment of bacteraemia is usually done by timely administration of appropriate antibiotics. However, since many bacterial pathogens have developed resistance to most of the antibiotics, it has become a serious health problem with many economic and social inferences all over the world (JoAnn 2009). Researches in Ethiopia revealed that there is high bacterial drug resistance to commonly used antibiotics mainly due to the lack of national guideline for antibiotic use, absence of good laboratory facilities to do antimicrobial drug susceptibility test. As a result clinicians use empirical way to treat their patients, there is also high self treatment of humans, and animals without prescription of doctors. These all would lead to emergence and rapid dissemination of resistance (Zenebe et al. 2011).
Research findings have reported that inappropriate treatment of BSI aggravates to increased mortality of patients and emerging of drug resistance strains (JoAnn 2009; Zenebe et al. 2011; Ali and Kebede 2008). In Ethiopia there are only a few studies on organisms involved in bloodstream bacterial infection and their susceptibility pattern (Dagnew et al. 2013; Asrat and Amanuel 2001; Zenebe et al. 2011; Ali and Kebede 2008); however, we could not get any published data in the study area and in the region as a whole. Therefore, we conducted this study to determine the common bacterial agents associated with bacteraemia and their antimicrobial susceptibility patterns in febrile patients in Mekelle hospital in Tigray, Northern Ethiopia.
Methods
Study design, study area and sampling process
Prospective cross-sectional study design was used to conduct the study among febrile out patients from March to October 2014. Mekelle Hospital, which is located 783 km North of Addis Ababa, is the largest hospital in the region giving services since the last 58 years. Sample size was calculated considering 24.2% prevalence (Ali and Kebede 2008), 5% precision, and 95% confidence level; with 15% contingency, thus a total of 514 febrile patients were included. Patients who were on antibiotic within the last 2 weeks of visiting the hospital were excluded from this study.
Data collection and laboratory procedures
About 5 ml of venous blood for adults and 2–3 ml for children was collected aseptically using 70% alcohol and 2% tincture of iodine and transferred into a bottle containing 45 ml of tryptic soy broth sterile culture medium (BBL™ USA). Blood culture broths were then transported within 30 min to Ayder referral hospital microbiology laboratory and incubated at 37°C.
Bacterial identification
Blood culture broths were checked for sign of bacterial growth (turbidity, haemolysis, clot formation) daily up to 7 days. Bottles which showed signs of growth were further processed by gram stain and sub-culture was made onto blood agar, MacConkey agar, Manitol salt agar (all Oxoid Ltd, UK) and incubated at 37°C for 24 h. Blood culture broths with no bacterial growth after 7 days were sub-cultured before being reported as a negative result. Bacterial isolates were identified by colony morphology, gram staining reaction, biochemical tests using Catalase test, Coagulase test, Triple Sugar Iron agar (TSI) (OXOID, UK),citrate utilization test (BBL™),Urease test (BBL™) and Motility Indole Lysine (MIL) (BBL™) test using the standard procedure for bacterial identification.
Antimicrobial susceptibility test
Following identification of the bacterial isolates, disc diffusion method was done according to Clinical Laboratory Standards Institute (CLSI) guidelines (M100 S24, 2014). The antibiotics discs and their concentrations were: Amoxicillin-clavunilic acid; (30 µg) (OXOID, UK), Ceftriaxone, (30 µg) (BBL™),Vancomycin (30 µg) (BBL™), Oxacillin (1 µg) (BBL™), Ciprofloxacin (5 µg) (BBL™), Gentamicin (120 µg) (BBL™), Norfloxacin (10 µg) (OXOID), Doxycycline (30 µg) (OXOID UK), Erythromycin (15 µg) (BBL™), Nitrofurantonin (300 µg) (BBL™) and Trimethoprim-sulphamethoxazole (25 µg) (BBL™ USA). We selected these antimicrobial agents as they are available and frequently prescribed for the management of bacterial infections in the hospital and in Ethiopia as whole.
S. aureus and CoNS isolates which were found to be vancomycin resistant by disk diffusion methods were sub-cultured in skim milk agar and stored at −60°C for further Vancomycin minimum inhibitory concentration (MIC) determination by broth micro dilutions (BMD) methods (CLSI, M100 S24, 2014).
Quality control
Reference strains E. coli (ATCC 25922) and S. Aureus (ATCC 25923) were used as a control reference strains for identifications and drug susceptibility testing. Negative control was done by randomly taking the prepared culture media and incubating over nigh to see for any growth. In this study multi-drug resistance was defined as simultaneous resistance to more than two antimicrobial agents (Magiorakos et al. 2012).
Data analysis
SPSS version 20 software was used for statistical analysis. Chi square test (χ2) was used to determine relationship between dependent and independent variable. P value <0.05 was used to indicate significant association.
Results
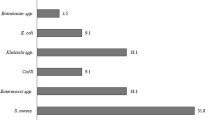
From the total 514 febrile patients, 269 (52.3%) were females and 245 (47.7%) males. Their age ranges from 1–80 years [mean 28.912 ± 1.46 (SD)]. Two hundred seventy-two (52.9%) of the participants were in the age range of 15–29 years. A total of 144 (28%) bacterial strains were isolated. Predominant isolates were S. aureus 54 (10.3%), Coagulase negative staphylococci (CoNS) 44 (8.5%), E. coli 16 (3.1%), Citrobacter spp. 9 (1.7%) and S. typhi 8 (1.6%) (Table 1).
More bacteria were isolated form females than males, though it was not statistically significant (P = 0.06, χ2 = 3.47, df = 1). The spectrum of bacteria varies with the age of patients, where 48.6% of the bacteria isolates were found in the age group of 15–29 years but not statistically significant associated (P = 0.54, χ2 = 46.4, df = 48).
In vitro antibiotic susceptibility of the bacterial isolates (Table 2) showed resistance for gram positive bacteria from 0 to 83%. Thirty-six (66.7%) of S. aureus isolates were resistant to Trimethoprim-sulphamethoxazole, 31 (57.4%) to Ceftriaxone, 29 (53.7%) to Doxycycline and 38 (70.4%) to Oxacillin. Thirty-six (81.8%) and 27 (61.4%) resistance was seen by CoNS to Trimethoprim-sulphamethoxazole and Doxycycline, respectively. All S.aureus and CoNS isolates were sensitive to vancomycin. Over all, high resistance was seen by gram positive isolates to Trimethoprim-sulphamethoxazole (74%) and Doxycycline (57.7%). Amoxicillin clavulanic acid was an effective antibiotic for gram positive bacteria next to Vancomycin in our study.
Antimicrobial resistance level of gram negative bacteria in this study was from 0 to 100%. E. coli was resistant to Ceftriaxone and Nitrofurantonin (60% each), Acinetobacter spp. were 80% resistant to Ceftriaxone and 60% to Trimethoprim-sulphamethoxazole. S. typhi isolates were resistant to Doxycycline, Trimethoprim-sulphamethoxazole and Nitrofurantonin (75% each). Over all, high resistance was seen by gram negative isolates to Nitrofurantonin 53.3% and Ceftriaxone 46.7%. Amoxicillin clavulanic acid and Ciprofloxacin were, however, effective for gram negative bacteria.
Antibiogram pattern of the isolates in this study showed that, S. aureus 34 (63%), CoNS 27 (61.2%), S. pyogenes 3 (50%), Citrobacter spp. 5 (62.5%), E. coli 8 (53.3%) and Acinetobacter spp. 4 (80%) showed multiple drug resistance (MDR). In general, 59% of the isolates in our study developed multidrug resistance to different antibiotics (Table 3).
MIC of Vancomycin by BMD to disc diffusion resistant CoNS and S. aureus showed that 80% of tested isolates had Vancomycin MIC results of ≤1 µg/ml. Two (13.3%) S. aureus and 1 (16.7.4%) CoNS isolate showed MIC results of ≥1.5 µg/ml. Only one (6.7%) S. aureus isolates alone showed MIC results of ≥2 µg/ml which is intermediate resistant, where as the rest S. aureus and all CoNS and were sensitive to vancomycin as per the 2014 guideline of CLSI.
Discussion
Bacteria isolation rate in this study 28%, was comparable with the result done in Gonder, Ethiopia, 24.2% (Ali and Kebede 2008), but higher than Addis Ababa, Ethiopia 21.4% (Asrat and Amanuel 2001), Jimma, Ethiopia (8.8%) (Zenebe et al. 2011), Gonder, Ethiopia 18.2% (Dagnew et al. 2013), Nigeria 18.2% (Nwadioha et al. 2010), Malawi 17.6% (Archibald et al. 2000), Nepal 23.1% (Amatya et al. 2007), India 9.94% (Manjula et al. 2005) and Nepal 10% (Usha and Pushpa 2007). It was, however; lower than reports from Zimbabwe 37.1% (Obi and Mazarura 1996), Gambia 34% (Philip et al. 2007) and India 40% (Neuma and Chitnis 1996). Possible explanation for the difference in prevalence of BSI across countries could be due to the difference in blood culture system, the study design, geographical location, nature of patient population, epidemiological difference of the etiological agents, seasonal variations, difference in infection control policies between nations (Gohel et al. 2014; Zenebe et al. 2011; Dagnew et al. 2013).
In our present study, 72.2% of infections were caused by gram-positive and 27.8% by gram-negative bacteria. Similar finding was seen from studies in Gonder, Ethiopia (69 and 31%) (Dagnew et al. 2013), Jimma, Ethiopia, (60.9 and 39.1%) (Zenebe et al. 2011) Gondar, Ethiopia (70.2 and 29.8%) (Ali and Kebede 2008), Zimbabwe (71.9 and 28.1%) (Obi and Mazarura 1996), Addis Ababa, Ethiopia (62.6 and 37.4%) (Shitaye et al. 2010).
On the contrary, gram-negative bacteria have been reported as the commonest cause of bacteraemia among febrile patients in Nigeria (69.3 and 30.7%) (Nwadioha et al. 2010), Saudi Arabia (62.2 and 33.8%) (Elbashier et al. 1998), Tanzania (69.7, 30.3%) (Meremo et al. 2012) and Nepal (89.19 and 10.81%) (David et al. 2004). Possible explanation for the difference could be due to epidemiological difference of the etiological agents.
S. aureus, CoNS, E. coli, S. typhi, Citrobacter spp., S. pyogenes and Acinetobacter spp. were the most common bacterial pathogens causing bacteraemia in this study. More or less similar observations have been seen in cases of bacteraemia in different countries, though, the proportion and prevalence of the bacterial agents varied (Dagnew et al. 2013; Asrat and Amanuel 2001; Zenebe et al. 2011; Ali and Kebede 2008; Obi and Mazarura 1996; Elbashier et al. 1998; Wisplinghoff et al. 2004).
S. aureus (37.5%) were the most commonly isolated bacteria in contrary to the other studies (Dagnew et al. 2013; Asrat and Amanuel 2001; Rina et al. 2007; Ali and Kebede 2008; Obi and Mazarura 1996) where coagulase negative staphylococci was the most isolated and in Tanzania, Salmonella spp., were reported as the dominant bacteria (Meremo et al. 2012). In our study, the second most bacterial isolate was CoNS (30.6%).This prevalence was comparable to the study done in other parts of Ethiopia 26.1% (Ali and Kebede 2008), 33.3% (Dagnew et al. 2013), but lower than reports from Addis Ababa Ethiopia (43.3%) (Shitaye et al. 2010) and Zimbabwe (42.9%) (Obi and Mazarura 1996). Though CoNS were mainly recognized as a contaminant until the 1970s, studies have reported an increasing incidence of infections due to these bacteria (Dagnew et al. 2013; Boisson et al. 2002).
In this study, E. coli (10.4%) was the predominant gram negative bacteria followed by S. typhi and Citrobacter spp. (5.6% each). However, results from other parts of Ethiopia (Dagnew et al. 2013; Asrat and Amanuel 2001) reported that Klebsiella spp. and E. coli as the dominant isolates. In contrary to other studies (Asrat and Amanuel 2001; Zenebe et al. 2011; Ali and Kebede 2008; Usha and Pushpa 2007), we have not isolated H. influenzae in our study which in lines with the finding from Gonder (Dagnew et al. 2013). In our current study we found all cases of BSI with single microorganism which in lines with earlier reports (Dagnew et al. 2013; Ghanshyam et al. 2008; Angyo et al. 2001). Unlike to our study; however, septicaemia of poly-microbial aetiology were reported by other studies (Obi and Mazarura 1996; Ghanshyam et al. 2002).
In this study bloodstream infection was not age dependent which was in contrast to other previous reports that showed septicaemia was relatively higher in neonates (Dagnew et al. 2013; Komolafe and Adegoke 2008; Shitaye et al. 2010).This may be due to small numbers of children in our study participants.
Overall the resistance of gram positive bacteria was from 0 to 83%, and for gram negative from 0 to 100% which is similar to the result from Jimma which was 0–85.7% and 0–100% for gram negative and positive, respectively (Zenebe et al. 2011). This was however, different from the study result done in Addis Ababa, where the rate for gram positive bacteria ranged from 12 to 76%, and for gram negatives ranged from 8 to 46% (Asrat and Amanuel 2001) and Gonder 23.5–58.8% and 20–100% for gram positive and negative, respectively (Dagnew et al. 2013). The increased resistant blood isolates in this study may be a signal of indiscriminate and continuous use of sub-therapeutic doses of commonly available antimicrobials both in the veterinary and public health sectors (Zenebe et al. 2011). This could challenge the management of patients very difficult.
Amoxicillin clavulanic acid was found to be effective against both gram positive and gram negative isolates in this study. Other studies reported Ciprofloxacin as an effective (Dagnew et al. 2013; Asrat and Amanuel 2001; Zenebe et al. 2011). Ciprofloxacin was found to be effective against gram negative isolates comparable with findings from Ethiopia and other nations (Dagnew et al. 2013; Komolafe and Adegoke 2008; Shitaye et al. 2010).
Antibiograms resistance pattern of the isolates revealed 59% them showed multidrug resistance. This suggests a high resistance gene pool perhaps due to gross misuse and inappropriate usage of the antibacterial agents (Komolafe and Adegoke 2008).
Except one isolate of S. aureus which was intermediate resistant, all MICs tested CoNS and S. aureus were sensitive to Vancomycin as per the CLSI, 2014 guideline, which was similar to other studies done elsewhere (Sakoulas et al. 2004; Soriano et al. 2008). Possible explanation for the absence of vancomycin resistant bacteria in our study is due to the fact that use of vancomycin by doctors is restricted in the treatment of patients in the study area and in the nation as whole. Unlike to this; however, researchers have reported vancomycin resistant strains from France (Ploy et al. 1998), Korea (Kim et al. 1998), South Africa (Ferraz et al. 2000) and Nigeria (Moses et al. 2013) showing that it is becoming a global threat.
There are also other reports that indicated that there is an increasing vancomycin MICs over time at individual institutions. However, as most clinical laboratories use automated systems to carry out susceptibility testing, these systems do not provide an accurate vancomycin MICs. Hence it has been recommended that clinical laboratories to provide accurate and reliable vancomycin MICs report to clinicians to help select appropriate therapy for the management of patients (Hsu et al. 2008; Steinkraus et al. 2007).
In conclusion, prevalence of BSI in this current study was high. This study added to the knowledge of the epidemiology of the isolates with high rates of resistance to most used antibiotics. Therefore timely investigation of bacterial flora of the bloodstream infections and monitoring of their antibiotic susceptibility pattern is important to reduce the incidence of bloodstream infections and multi drug resistant strains.
References
Aiker AM, Mturi N, Niugana P (2011) Risk and cause of paediatrics hospital acquired bacteremia Klifi district hospital, Kenya: prospective cohort study. Lancet J 10(37):2012–2017
Ali J, Kebede Y (2008) Frequency of isolation and antimicrobial susceptibility pattern of bacterial isolation from blood culture in Gondar university hospital. Ethio Med J 46(2):155–161
Amatya NM, Sheresha B, Lekhak B (2007) Etiological agents of bacteremia and antibiotic susceptibility pattern in Kathmandu hospital. J Nepal Med Assoc 46(167):112–118
Angyo IA, Opkeh ES, Opajobi SO (2001) Predominant bacterial agents of childhood septicaemia in Jos. Niger J Med 6:75–77
Archibald LK, McDonald LC, Nwanyanwu O, Kazembe P, Dobbie H, Tokars J et al (2000) A hospital-based prevalence survey of bloodstream infections in febrile patients in Malawi: implications for diagnosis and therapy. JID 181:1414–1420
Asrat D, Amanuel Y (2001) Prevalence and antibiotic susceptibility pattern of bacterial isolates from blood culture in Tikur Anbessa hospital, Addis Ababa, Ethiopia. Ethiop Med J 39(Suppl 2):97–104
Boisson K, Thouverez M, Talon D, Bertrand X (2002) Characterization of coagulase-negative staphylococci isolated from blood infections: incidence, susceptibility to glycopeptides, and molecular epidemiology. Eur J Clin Microbiol Infect Dis 6(9):660–665
Clinical and Laboratory Standards Institute (CLSI) (2014) Performance standards for antimicrobial susceptibility testing; twenty-fourth informational supplement, CLSI document M100-S24. Wayne and Pennsylvania
Dagnew M, Yismaw G, Gizachew M, Gadisa A, Abebe T, Tadesse T et al (2013) Bacterial profile and antimicrobial susceptibility pattern in septicemia suspected patients attending Gondar University Hospital, Northwest Ethiopia. BMC Res Notes 6:283
Daniel RK, Scott AF, James MB, Sanjay S (2006) Brief Report: incidence, etiology, risk factors, and outcome of hospital acquired fever. J Gen Intern Med 21:1184–1187
David RM, Christopher WW, Mark DZ, Peter MD, Ram HB, Andrew JK et al (2004) The etiology of febrile illness in adults presenting to Patan Hospital in Kathmandu, Nepal. Am J Tro Med Hyg 70(6):670–675
Diekma DJ, Beekman SE, Chapin KC, Morel KA, Munson E, Deorn GV (2002) Epidemiology and outcome of nosocomial and community onset bloodstream infection. Eur J Clin Microbiol Infect Dis 21(Suppl 9):660–665
Elbashier AM, Malik AG, Knot AP (1998) Bloodstream infections: micro-organisms, risk factors and mortality rate in Qatif Central Hospital. Ann Saudi Med 18(2):176–180
Ferraz V, Duse AG, Kassel M, Black AD, Ito T, Hiramatsu K (2000) Vancomycin—resistant S. aureus occurs in South Africa. S Afr Med J 90:1113
Ghanshyam DK, Ramachandram VC, Piyush G (2002) Bacteriological analysis of blood culture isolates from neonates in a tertiary care hospital in India. J Health Popul Nutr 6(4):343–347
Ghanshyam DK, Ramachandram VC, Piyush G (2008) Bacteriological analysis of blood culture. Malays J Microbiol 4(2):51–61
Gohel K, Jojera A, Soni S, Gang S, Sabnis R, Desai M (2014) Bacteriological profile and drug resistance patterns of blood culture isolates in a tertiary care nephrourology teaching institute. Biomed Res Int 2014:153747
Hsu DI, Hidayat LK, Quist R, Hindler J, Karlsson A, Yusof A et al (2008) Comparison of method-specific vancomycin minimum inhibitory concentration values and their predictability for treatment outcome of meticillin-resistant Staphylococcus aureus (MRSA) infections. J Antimicrob Agents 32:378–385
James AK, Mark EJ, Deborah CD, Clyde T, Daniel FS, Gregory AV (2004) Prevalence and antimicrobial susceptibilities of bacteria isolated from blood cultures of hospitalized patients in the United States in 2002. Ann Clin Microbio Antimicrobi 3(7):1–8
JoAnn D (2009) Antibiotic resistance: the ongoing challenge for effective drug therapy. JAAPA 22(3):18–22
Kim M-N, Pai CH, Woo JH, Ryu JS, Hiramatsu K (1998) Vancomycin—intermediate S. aureus in Korea. J Clin Microbiol 38:3879–3881
Komolafe AO, Adegoke AA (2008) Incidence of bacterial Septicaemia in Ile-Ife Metropolis, Nigeria. Malays J Microbiol 4(2):51–61
Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG et al (2012) Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18(3):268–281
Manjula M, Pyria D, Varsha G (2005) Antimicrobial susceptibility pattern of blood isolates from a teaching Hospital in North India. Jpn J Infec Dis 58:174–176
Meremo A, Mshana SE, Kidenya BR, Kabangila R, Peck R, Kataraihya JB (2012) High prevalence of Non-typhoid salmonella bacteraemia among febrile HIV adult patients admitted at a tertiary Hospital, North-Western Tanzania. Int Arch Med 5:28
Moses A, Uchenna U, Nworie O (2013) Epidemiology of vancomycin resistant staphyloccus aureus among clinical isolates in a tertiary hospital in Abakaliki, Nigeria. AJEID 1(3):24–26
Neuma S, Chitnis DS (1996) Antibiogram study over bacterial isolated from cases of bacteremia. Indian J Med Sci 50:325–329
Nwadioha I, Nwokedi EOP, Kashibu E, Odimayo MS, Okwori EE (2010) A review of bacterial isolates in blood cultures of children with suspected septicemia in a Nigerian. Afr J Microbiol Res 4(4):222–225
Obi CL, Mazarura E (1996) Aerobic bacteria isolated from blood cultures of patients and their antibiotic susceptibilities in Harare, Zimbabwe. Cent Afr J Med 42(Suppl 12):332–336
Philip CH, Charles OO, Usman NAI, Ousman S, Samuel A, Naomi S et al (2007) Bacteraemia in patients admitted to an urban hospital in West Africa. BMC Infect Dis 7(2):1–8
Ploy MC, Grelaud C, Martin C, de Lumley L, Denis F (1998) First clinical isolate of vancomycin—intermediate Staphylococcus aureus in a French hospital. Lancet 251:1212
Rina K, Nadeem SR, Kee PN, Parasakthi N (2007) Etiology of blood culture isolates among patients in a multidisciplinary teaching hospital in Kuala Lumpur. J Microbiol Immunol Infect 40:432–437
Sakoulas GPA, Moise-Broder J, Schentag A, Forrest Moellering RC Jr, Eliopoulos GM (2004) Relationship of MIC and bactericidal activity to efficacy of vancomycin for treatment of methicillin-resistant Staphylococcus aureus bacteremia. J Clin Microbiol 42:2398–2402
Shitaye D, Asrat D, Woldeamanuel Y, Worku B (2010) Risk factors and etiology of neonatal sepsis in TikurAnbessa University Hospital, Ethiopia. Ethiop Med J 48(1):11–21
Soriano A, Marco F, Martinez JA, Pisos E, Almela M, Dimova VP et al (2008) Influence of vancomycin minimum inhibitory concentration on the treatment of methicillin-resistant S. aureus bacteremia. Clin Infect Dis 46:193–200
Steinkraus G, White R, Friedrich L (2007) Vancomycin MIC creeps in non-vancomycin-intermediate S. aureus (VISA), vancomycin susceptible clinical methicillin-resistant S. aureus (MRSA) blood isolates from 2001–05. J Antimicrob Chemother 60:788–794
Tziamabos AO, Kasper DL (2005) Principle and practice of infectious diseases. Frank Polizano J 26:2810–2816
Usha A, Pushpa D (2007) Bacterial profile of bloodstream infections and antibiotics resistance pattern of isolates. JK Sci 9(4):186–190
Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB (2004) Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis 39(3):309–317
Zenebe T, Kannan S, Yilma D, Beyene G (2011) Invasive bacterial pathogens and their antibiotic susceptibility patterns In Jimma University Specialized Hospital, Jimma, Southwest Ethiopia. Ethiop J Health Sci 21:1
Authors’ contributions
AGW was the primary researcher, conceived the study, designed, participated in data collection, laboratory work, conducted data analysis, drafted and finalized the manuscript for publication. LN, SA, TA, SM performed in data collection, analysis data, assisted in reviewed the initial and final drafts of the manuscript. TD and AL interpreted the results, and reviewed the initial and final drafts of the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We greatly appreciate Mekelle University, NORAD III project for the grant. We are also grateful to laboratory staff of Mekelle hospital and the study participants.
Compliance with ethical guidelines
Competing interests The authors declare that they have no competing interests.
Consent for publication The study was approved and ethically cleared by the Research and Ethical Review Committee of Mekelle University, College of Health Sciences (REC REF 2014-152). Written informed consent was obtained from each participants and parents or care takers. All patients’ information was kept confidential and secret using codes.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Wasihun, A.G., Wlekidan, L.N., Gebremariam, S.A. et al. Bacteriological profile and antimicrobial susceptibility patterns of blood culture isolates among febrile patients in Mekelle Hospital, Northern Ethiopia. SpringerPlus 4, 314 (2015). https://doi.org/10.1186/s40064-015-1056-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40064-015-1056-x