Abstract
Monoamine oxidases (MAOs) regulate neurotransmitters, and changes in their regulation lead to neurogenerative diseases (NDs). Therefore, MAO inhibitors are used to treat NDs. Ferulic acid, a phenolic compound found in various plant species, has been demonstrated to have a variety of biological functions, including anti-inflammatory, anticancer, and neuroprotective effects. In this study, ten ferulic acid hydrazide derivatives (FA1–FA10) were synthesized, and their ability to inhibit monoamine oxidase (MAO) enzymes was tested. Six candidates demonstrated a more pronounced pattern of inhibitory action against MAO-B than against MAO-A. FA3 had the highest inhibitory efficacy in MAO-B inhibition (IC50 value of 1.88 μM), followed by FA9 (2.08 μM). FA3 has a Ki of 1.92 ± 0.73 μM. A reversibility experiment of MAO-B inhibition by FA3 was conducted using dialysis, and the recovery pattern showed FA3 was a reversible MAO-B inhibitor with a similar recovery to safinamide, a reversible reference inhibitor. These results indicate that FA3 is an effective reversible MAO-B inhibitor. In molecular dynamics and docking, FA3 paired with pi-pi stacking helped stabilize the protein ligand in the active site of MAO-B. According to this study, lead compounds can be used as therapeutic agents to treat neurological conditions, such as Parkinson's disease (PD).
Similar content being viewed by others
Introduction
The FAD-dependent enzyme monoamine oxidase (MAO) is located in the outer mitochondrial membrane [1]. The enzyme has two isoforms: MAO-A and MAO-B [2]. These isoforms vary in their tissue distribution and substrate-inhibitor recognition sites despite having many sequence similarities [3]. MAOs are essential regulators of neurotransmitter levels. Changes in MAO levels have been linked to various neurological disorders [4]. They play a crucial role in the metabolism of neurotransmitters, such adrenaline, noradrenaline, dopamine, -phenylethylamine, and benzylamine by catalyzing the oxidative deamination of a variety of monoamines [5]. MAOs can cause emotional and behavioral alterations, and neurodegenerative diseases (NDs) by degrading these biogenic amines [6]. Alzheimer’s disease (AD) and Parkinson’s disease (PD) are examples of progressive, chronic, and incurable NDs that afflict millions of people worldwide [7,8,9,10]. Inhibitors of MAO are a class of medicines used to treat such disorders [11, 12].
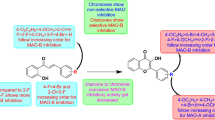
To develop new, reversible, competitive, and selective MAO-B inhibitors, the pharmacophoric groups of current MAO-B inhibitors, such as lazabemide, safinamide, and chalcones have been used [13, 14]. Recently, hydrazide derivatives have been reported to have MAO-B inhibition capabilities [15, 16]. The MAO-B inhibiting activity of the ferulic acid scaffold has been widely described in the literature [17]. Ferulic acid is a well-known naturally occurring phenolic compound found in various plant species. It has been demonstrated to have a variety of biological functions, including anti-inflammatory, anticancer, and neuroprotective effects. Among its compounds, ferulic acid hydrazide has drawn a lot of interest because of its potential as a multifunctional medication [18,19,20].
This article describes the design, synthesis, and MAO-B inhibitory activity of ferulic acid hydrazide derivatives. The design of ferulic acid hydrazide derivatives was based on the hypothesis that the incorporation of different aldehydes into the hydrazide moiety of ferulic acid could lead to compounds with improved bioactivity. Accordingly, a series of ferulic acid hydrazide derivatives were synthesized by reacting ferulic acid hydrazide with different aldehydes [21,22,23], using a single scaffold with an unsaturated ketone, carboxamide, and olefinic connection [24, 25].
Materials and methods
Synthesis
The compounds were synthesized as per the following scheme:
Step 1: General procedure for the synthesis of ferulic acid chloride.
The synthesis was performed as indicated in Scheme 1. For the preparation of ferulic acid chloride, thionyl chloride 1.6 mL (0.0220 mol) was added gradually to 1 g of ferulic acid (0.00514 mol) in a round bottom flask. After adding thionyl chloride, 10 mL of chloroform was added, and the mixture was stirred for 8 h. The reaction mixture was maintained in a water bath at a temperature of 60 ℃. After cooling the reaction mixture, excess SOCl2 was removed. The yield was found to be 76%.
Synthesis of ferulic acid hydrazones.
Hydrazine hydrate solution was added dropwise to dry ferulic acid chloride (0.0040 mol, 0.85 g). The reaction mixture was kept in an ice bath for 2 h followed by continuous stirring at room temperature for another 2 h. After completion of the reaction, as indicated in thin-layer chromatography (TLC), 15 mL of cold water was added, and the resulting hydrazide was filtered off and dried. The yield was found to be 80%.
A mixture of hydrazide (0.00067 mol, 0.14 g) and substituted aromatic aldehydes (0.00067 mol) was stirred in methanol (5 mL). After 10 min, a few drops of glacial acetic acid were added and refluxed for the prescribed time period mentioned in Additional file 1: Table S1. The completion of the reaction was monitored by TLC. The product obtained was filtered, dried, and recrystallized from ethanol.
Enzyme assays of MAO-A and MAO-B
The activities of MAO-A and MAO-B were assayed using kynuramine (0.06 mM) and benzylamine (0.30 mM), respectively, as substrates by measuring the absorbance change continuously at 316 nm and 250 nm, respectively [26].
Enzyme kinetics and inhibition studies
Enzyme kinetics for MAO-B were carried out at five different substrate concentrations (0.0375–0.6 mM, respectively) [26]. For inhibition study, residual activity was assayed after addition of 10 μM inhibitor as an initial screening step, and IC50 values were determined for potential compounds with residual activity of less than 80%, using GraphPad Prism software 5 [27]. The selectivity index (SI) value of MAO-B was calculated as IC50 of MAO-A/IC50 of MAO-B [28]. The inhibition type of the leading compound for MAO-B was determined at the five different substrate concentrations and at three inhibitor concentrations around its IC50 [29]. Toloxatone, clorgyline, safinamide, and pargyline were used as the reference inhibitors [30]. The enzyme kinetic pattern and Ki value were determined by comparing the Lineweaver–Burk plots and their secondary plots, respectively [30].
Reversibility studies
The reversibility of the leading compound for MAO-B was evaluated by comparing undialyzed and dialyzed residual activities at a concentration of approximately 2 times the IC50 after preincubation for 30 min prior to measurement, as previously described [31]. Two types of reference inhibitors were used for MAO-B: the reversible inhibitor, safinamide and the irreversible inhibitor, pargyline. The reversibility pattern was determined by comparing the activities of undialyzed (AU) and dialyzed (AD) samples, and by considering the reference results.
Molecular docking
The Schrodinger suite [32] was used to carry out the molecular docking investigation of FA3. The human MAO-B (2V5Z) structure was retrieved from the Protein Data Bank [3]. The crystal structures were improved and optimized using the protein preparation wizard included in the Schrodinger suite, which also performed energy minimization, hydrogen atom addition, protonation state correction, and protonation state addition. The LigPrep tool was used to build the ligand structure. The co-crystallized ligands served as the automated center of the grid box. For docking simulations, the force Field OPLS_2005 default settings and extra precision (XP) docking protocol default settings were used [33, 34].
Molecular dynamic simulation
Schrodinger LLC’s Desmond simulation program was used to run molecular dynamics (MD) simulations [32]. The protein–ligand combination was initially created for the Desmond system builder panel utilizing compound FA3 against MAO-B in the aqueous solvent system. For complete protein–ligand simulations and stability trajectory analysis (root mean square deviation [RMSD], RMSF, and protein–ligand contact), the simulated parameters were 100 ns at 300 K, 1.01325 bar pressure, and 1000 frames [35, 36].
Results and discussion
Spectral characterization
Compound information like the color of the compound, retention factor (Rf) value, TLC mobile phase, IR, 1H, 13C, and ESI–MS was described below, and their spectra were provided in Additional file 1: (Figure S1–S30) Table S1.1H-NMR spectra confirmed the formation of imines (FA1-FA10). In the 1H-NMR spectra of FA1, the peak at 11.69 ppm suggested the formation of the NH group of hydrazone. The doublet signals were observed at 7.43 and 6.54 ppm with a coupling constant of 15 Hz for acrylic CHB=CHA protons [37]. The higher J value suggested that the molecule appeared as a trans form. The peak at 8.19 belongs to the aldehydic hydrogen atom of FA1. The shielded protons at 3.86 suggested the appearance of methoxy group in the final molecule. The presence of CO-NH group at 147.7 ppm in 13C-NMR confirmed the formation of acylhydrazone.
FA1: (2E,14Z)-N′-(3-bromobenzylidene)-3-(4-hydroxy-3-methoxyphenyl)acrylohydrazide
Yield: 68%; White powder; Rf value 0.65 (chloroform/methanol = 4/1); IR (ZnSe): 3251 (-NH-), 2943 (CHAr), 1623 (-CO-NH-), 1583 (-CH=N-), 858 (C-Br); 1H NMR (500 MHz) DMSO, δppm: 11.69 (s, 1H, NH), 9.55 (s, 1H, OH), 8.19–6.82 (8H, Ar–H), 7.57–7.54 (d, J = 15 Hz, 1H, CH), 6.54–6.51 (d, J = 15 Hz, 1H, CH), 3.86–3.83 (s, 3H, OCH3); 13C NMR (500 MHz): 147.7, 144.1, 132.6, 130.8, 129.0, 500.9, 121.0, 116.4, 115.6, 110.9, 55.4, 39.5; ESI–MS (m/z): calculated 374.0266, found 397.1429 [M+Na]+.
FA2: (2E,14Z)-N′-(4-chlorobenzylidene)-3-(4-hydroxy-3-methoxyphenyl)acrylohydrazide
Yield: 72%; White powder; Rf value 0.48 (chloroform/methanol = 4/1); IR (ZnSe): 3253 (NH), 2945 (CHAr), 1595 (CONH), 1510 (CH=N), 813 (CCl); 1H NMR (500 MHz) CDCl3, δppm: 11.48 (br, 1H, NH), 9.34 (s, 1H, OH), 7.83–7.80 (d, J = 15 Hz, 1H, CH), 7.36–7.33 (d, J = 15 Hz,1 H, CH), 7.66–6.96 (8H, Ar-H), 3.97 (s, 3H, CH3); 13C NMR (500 MHz): 168.0, 151.3, 144.9, 144.0, 143.0, 136.6, 131.9, 130.6, 129.0, 128.8, 120.1, 118.9, 116.8, 112.0, 56.2; ESI–MS (m/z): calculated 330.0771, found 331.1479 [M+H]+.
FA3: (2E,14Z)-N′-(4-(benzyloxy)-3-methoxybenzylidene)-3-(4-hydroxy-3-methoxyphenyl)acrylohydrazide.
Yield: 64%; Yellow powder; Rf value 0.45 (chloroform/methanol = 4/1); IR (ZnSe): 3256 (-NH-), 2944 (CHAr), 1620 (-CO–NH-), 1593 (-CH=N-); 1H NMR (500 MHz) DMSO, δppm: 11.44 (br, 1H, NH), 9.51 (s, 1H, OH), 7.39–7.36 (d, J = 15 Hz, 1H, CH), 6.52–6.49 (d, J = 15 Hz, 1H, CH), 7.96–6.81 (11H, Ar-H), 3.87–3.83 (s, 6H, CH3), 8.14 (s, 1H, N=CH), 5.14 (s, 2H, O-CH2); 13C NMR (500 MHz) DMSO, δppm: 161.6, 149.5, 149.2, 148.7, 148.5, 147.7, 146.1, 140.6, 136.7, 128.3, 127.8, 127.7, 127.3, 126.1, 121.7, 121.4, 120.9, 116.8, 115.6, 113.8, 113.0, 110.8, 108.5; ESI–MS (m/z): calculated 432.1685, found 433.2729 [M+H]+.
FA4: (2E,14Z)-N′-(4-ethylbenzylidene)-3-(4-hydroxy-3-methoxyphenyl)acrylohydrazide.
Yield: 78%; Brown powder; Rf value 0.42 (chloroform/methanol = 4/1); IR (ZnSe): 3241 (-NH-), 2945 (CHAr), 1624 (-CO-NH-), 1585 (-CH=N-); 1H NMR (500 MHz) DMSO, δppm: 11.49 (br, 1H, NH), 9.52 (s, 1H, OH), 7.40–7.37 (d, J = 15 Hz, 1H, CH), 6.52–6.49 (d, J = 15 Hz,1 H, CH), 7.11–6.78 (7H, Ar-H), 2.65–2.64 (s, 3H, O-CH3), 3.86 (s, 2H, CH2-CH3), 8.10 (s, 1H, N=CH), 1.22–1.19 (s, 3H, CH3); 13C NMR (500 MHz) DMSO, δppm: 166.2, 161.7, 148.7, 147.7, 146.0, 145.8, 142.7, 142.5, 140.8, 131.8, 131.7, 128.1, 126.8, 126.3, 126.0, 121.9, 116.7, 115.6, 113.6, 112.0, 110.8; ESI–MS (m/z): calculated 324.1474, found 325.2022 [M+H]+.
FA5: (2E,14Z)-N′-(4-fluorobenzylidene)-3-(4-hydroxy-3-methoxyphenyl)acrylohydrazide.
Yield: 72%; Off white powder; Rf value 0.71 (chloroform/methanol = 4/1); IR (ZnSe): 3251 (-NH), 2941 (CHAr), 1633 (-CO-NH-), 1600 (-CH=N), 1095 (C-F); 1H NMR (500 MHz) DMSO, δppm: 11.37 (br, 1H, NH), 9.53 (s, 1H, OH), 7.41 (d, J = 15 Hz, 1H, CH), 6.53 (d, J = 15 Hz, 1H, CH), 7.11–6.78 (7H, Ar-H), 3.85 (s, 3H, CH3), 8.23 (s, 1H, N=CH); 13C NMR (500 MHz): 161.7, 148.6, 144.8, 142.6, 140.9, 129.0, 121.8, 116.6, 115.8, 113.5, 112.1, 110.8, 55.6, 39.7; ESI–MS (m/z): calculated 314.1067, found 337.1576 [M+Na]+.
FA6: (2E,14Z)-N′-benzylidene-3-(4-hydroxy-3-methoxyphenyl)acrylohydrazide
Yield: 71%; Light brown powder; Rf value 0.56 (chloroform/methanol = 4/1); IR (ZnSe): 3190 (-NH-), 2966 (CHAr), 1587 (-CO-NH), 1594 (-CH=N-); 1H NMR (500 MHz) DMSO, δppm: 11.37 (br, 1H, NH), 9.53 (s, 1H, OH), 6.53 (d, J = 15 Hz, 1H, CH), 7.23 (d, J = 15 Hz, 1H, CH), 3.86 (s, 3H, CH3), 7.77–6.82 (8H, Ar–H), 8.23 (s, 1H, N=CH); 13C NMR (500 MHz): 161.7, 148.8, 147.7, 145.9, 142.6, 140.9, 134.3, 129.7, 126.6, 126.9, 121.8, 116.6, 113.5, 55.6, 39.7; ESI–MS (m/z): calculated 296.1161, found 297.1717 [M+H]+.
FA7: (E)-3-(4-hydroxy-3-methoxyphenyl)-N′-((Z)-4-methylbenzylidene)acrylohydrazide
Yield: 58%; Yellow powder; Rf value 0.76 (chloroform/methanol = 4/1); IR (ZnSe): 3189 (NH), 2965 (CHAr), 1590 (CONH), 1594 (CH=N); 1H NMR (500 MHz) DMSO, δppm: 11.30 (br, 1H, NH), 9.52 (s, 1H, OH), 7.40 (d, J = 15 Hz, 1H, CH), 6.52 (d, J = 15 Hz, 1H, CH), 3.86 (s, 3H, O-CH3), 2.52 (s, 3H, CH3), 7.65–6.85 (7H, Ar-H), 8.18 (s, 1H, N=CH); 13C NMR (500 MHz): 168, 151.3, 144.9, 144.0, 143.0, 140.7, 130.8, 129.2, 129.1, 128.8, 120.1, 118.9, 116.8, 112.0, 56.2, 24.3; ESI–MS (m/z): calculated 310.1317, found 311.1969 [M+H]+.
FA8: (2E,14Z)-N′-(4-methoxybenzylidene)-3-(4-hydroxy-3-methoxyphenyl) acrylohydrazide
Yield: 71%; Yellow powder, Rf value 0.65 (chloroform/methanol = 4/1); IR (ZnSe): 3193 (NH), 2960 (CHAr), 1577 (CONH), 1595 (CH=N); 1H NMR (500 MHz) DMSO, δppm: 11.24 (br, 1H, NH), 9.52 (s, 1H, OH), 6.56 (d, J = 15 Hz, 1H, CH), 7.40 (d, J = 15 Hz, 1H, CH), 3.86–3.81 (s, 6H, O-CH3), 7.90–6.40 (7H, ArH), 8.16 (s, 1H, N=CH),; 13C NMR (500 MHz): 166.1, 161.5, 160.6, 120.5, 126.9, 126.1, 121.9, 116.8, 115.6, 114.1, 113.7, 112.0, 110.8, 55.7, 39.6; ESI–MS (m/z): calculated 326.1267, found 349.2530 [M+Na]+.
FA9: (2E,14Z)-N′-(4-(benzyloxy)benzylidene)-3-(4-hydroxy-3-methoxyphenyl)acrylohydrazide
Yield: 58%; Yellow powder, Rf value 0.78 (chloroform/methanol = 4/1); IR (ZnSe): 3188 (NH), 2964 (CHAr), 1596 (CONH), 1595 (CH=N); 1H NMR(500 MHz) DMSO, δppm: 9.28 (s, 1H, CH), 7.80 (d, J = 15 Hz, 1H, CH), 7.78–6.95 (12H, Ar-H), 5.12 (s, 2H, O-CH2), 3.96 (s, 3H, O-CH3); 13C NMR (500 MHz): 160.4, 147.7, 146.6, 143.9, 143.1, 136.5, 128.7, 127.4, 126.6, 122.6, 115.2, 113.9, 110.9, 110.4, 77.2, 70.1, 56.0; ESI–MS (m/z): calculated 402.158, found 425.3581 [M+Na]+.
FA10: (2E,14Z)-3-(4-hydroxy-3-methoxyphenyl)-N′-((thiophen-2-yl) methylene) acrylohydrazide
Yield: 81%; Brown powder; Rf value 0.59 (chloroform/methanol = 4/1); IR (ZnSe): 3190 (NH), 2964 (CHAr), 1591 (CONH), 1592 (CH=N); 1H NMR (500 MHz) DMSO, δppm: 11.37 (br, 1H, NH), 9.56 (s, 1H, OH), 6.53 (d, J = 15 Hz, 1H, CH), 7.59 (d, J = 15 Hz, 1H, CH), 7.60–6.81 (6H, ArH), 3.83 (s, 3H, CH3) 8.22 (s, 1H, N=CH),; 13C NMR (500 MHz): 165.9, 161.6, 148.8, 147.7, 142.4, 141.1, 140.9, 139.1, 137.5, 130.5, 129.8, 128.6, 126.0, 121.8, 116.5, 113.2; ESI–MS (m/z):calculated 302.0725, found 325.1899 [M+Na]+.
Inhibition studies of MAO-A and MAO-B
At a concentration of 10 μM, five compounds showed low residual activity of < 50% for MAO-B, while only one compound showed low residual activity of < 50% for MAO-A (Table 1). FA3 had an IC50 value of 1.88 μM, showing the best inhibitory ability against MAO-B, followed by compound FA9 (IC50 = 2.08 μM) (Table 1, Additional file 1: Fig. S31). Overall, it was observed that the compounds did not exhibit effective inhibitory ability against MAO-A. The best compound for MAO-A inhibition was FA1 with an IC50 of 8.05 μM.
In the comparison of FA3 and FA9, a methoxy group of FA3 at the meta-position of the benzyl ether group, which is a common substituent at the para-position of the benzyl group, slightly increased inhibitory activity, that is, IC50 values from 2.08 to 1.88 μM. Comparing the IC50 levels of FA8 and FA9, a benzyl ether-type substituent in FA9 showed a better MAO-B inhibitory effect than a methoxy group in FA8. The comparison of derivatives with halogen substituents (FA1 and FA2 or FA5), showed that compounds with halogen functional groups at the para-position showed better inhibition ability than at the meta-position, while halogen substituents showed similar inhibitory properties under the same chemical conditions (FA2 and FA5).
Enzyme kinetics
Enzyme and inhibition kinetics were analyzed at five substrate concentrations and three inhibitor concentrations of FA3. In the Lineweaver–Burk plot, FA3 appeared to be a competitive MAO-B inhibitor (Fig. 1A). In the secondary plots, the Ki value was determined to be 1.92 ± 0.73 μM (Fig. 1B). These results suggest that FA3 acts as a potent competitive MAO-B inhibitor.
Reversibility studies
The reversibility of MAO-B inhibition by FA3 was analyzed using dialysis after 30 min of preincubations at 2-times the FA3 IC50 value (4.22 μM). The recovery pattern was compared using undialyzed (AU) and dialyzed (AD) relative activities. Inhibition of MAO-B by FA3 was recovered from 31.89% to 102.03% (Fig. 2). The recovery value of the compound was similar to that of safinamide (reversible type, from 21.50% to 71.25%), and it was distinguished from pargyline (irreversible type, from 19.16% to 30.35%). These results indicate that FA3 is a reversible inhibitor of MAO-B.
Molecular docking
To learn more about the binding of the lead compounds, molecular docking studies were conducted on a lead molecule (FA3) and MAO-B (2V5Z) interactions. We re-docked the native ligands to validate the docking procedure [34]. The docking scores (XP mode) for the lead compounds (FA3) were approximately − 8.591 kcal/mol, while the scores (− 9.648 kcal/mol) for safinamide were comparable. The amide side chain of safinamide pointed in the direction of the FAD molecule, whereas the fluoro-benzyl group was positioned towards the opening of the cavity. Similar positioning was shown in lead compound FA3 (Fig. 3), where the variable ferulic moiety was pointed in the direction of FAD and the benzyloxy group was facing the cavity entrance. The lead inhibitor FA3 occupied the whole substrate cavity and entered the MAO-B binding pocket (Fig. 3). Associations with the residues Ser200, Thr201, Ile199, Ile198, Phe343, Tyr60, Tyr435, Leu171, Cys172, Phe168, Trp119, Pro104, and Pro102 are primarily hydrophobic, and Gln206 is in polar contact. The FA3-MAO-B protein complex is stable because of the interaction between the ferulic moiety, and Tyr398 and the phenyl moiety and Tyr326 through pi-pi stacking.
Molecular dynamic simulation
The FA3 binding mode in the MAO-B inhibitor binding cavity was observed using Desmond's MD simulations. RMSD analysis revealed that the protein C-alpha and its ligand were monitored within a reasonable range for a lengthy simulation (100 ns). In contrast to the protein RMSD, the RMSD of the ligand was stable after 30 ns. The protein RMSD had an average value of 2.11 Å and a range of 1.2 to 3.10 Å (Fig. 4A).The protein-specific RMSD during the simulation was found to remain constant, with the exception of a minor change, stabilizing at a high of 3.10 Å at 68 ns. Adaptability of the protein system was assessed by calculating the RMSF of each protein amino acid residue. The 480–498 residue of the MAO-B protein fluctuated more. During the binding process, the atoms in the benzoyl ring of the RMSF ligand (Fig. 4B) displayed minor fluctuations. The 26 amino acid residues the ligand interacted with were Tyr60 (0.45 Å), Leu88 (0.76 Å), His90 (0.67 Å), Phe99 (0.78 Å), Gly101 (0.95 Å), Pro102 (0.92 Å), Pro104 (0.83 Å), His115 (0.99 Å), Trp119 (0.83 Å), Leu164 (0.71 Å), Leu167 (0.63 Å), Phe168 (0.6 Å), Thr201 (0.55 Å), Ile199 (0.67 Å), Ile198 (0.59 Å), Cys172 (0.52 Å), Leu171 (0.52 Å), Phe343 (0.44 Å), Leu171 (0.638 Å), Gln206 (0.55 Å), Ile316 (0.54 Å), Tyr326 (0.50 Å), Leu345 (0.57 Å), Tyr398 (0.62 Å), Gly434 (0.56 Å), and Tyr435 (0.418 Å). In the interaction histogram of FA3 and MAO-B, hydrogen bonds, hydrophobic interactions, and a few water bridges were observed (Fig. 4C, D). The number of distinct interactions between amino acids and ligands was normalized during a trajectory of 100 ns. The interaction histogram of the MD study revealed that several critical amino acids, including Gly434 (hydrogen bond), Tyr435 (hydrophobic), Tyr398 (hydrogen bond), Cys172 (hydrogen bond), Ile199 (water bridge and hydrophobic), and Phe99 (hydrophobic), interacted with FA3. The hydrophobic and hydrogen bonding contacts at the active site of MAO-B were substantial, because the observed interaction fraction with Gly434, Tyr435, and Ile199 was > 0.8. The hydrogen bonds, water bridges, and hydrophobic stability of the ligand protein complexes are shown in Fig. 4C, D. Gly434 participates in an 84% hydrogen bond with the OH of the ferulic ring. Tyr398 makes a 76% contribution through a pi-pi staking arrangement with FA3 the ferulic ring. Ile199, Phe99, Ile316, and Tyr326 displayed hydrophobic contact with the benzyloxy ring. Overall, it is expected that the lead chemical FA3 will inhibit MAO-B based on trajectory analysis and comprehensive MD simulation.
Collectively, we synthesized ferulic hydrazide-based compounds and assessed how efficiently they inhibited MAOs. FA3 was found to be a competitive, reversible MAO-B inhibitor. The stability of the complex was additionally identified by a docking investigation of MAO-B and FA3, which was made possible by the pi-pi stacking of Tyr326 and Tyr398. In the dynamic analysis, the participation rates of Tyr398 and Gly434 residues in the interaction with the ligand were 76% and 84%, respectively. The main compounds FA3 may be useful therapeutic agents for the treatment of neurological illnesses, such as PD, according to the study's overall findings.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
References
Devenish SRA (2020) The current landscape in Alzheimer’s disease research and drug discovery. Drug Discov Today 25(6):943–945
Ferino G, Vilar S, Matos MJ, Uriarte E, Cadoni E (2012) Monoamine oxidase inhibitors: ten years of docking studies. Curr Top Med Chem 12(20):2145–2162
Binda C, Wang J, Pisani L, Caccia C, Carotti A, Salvati P, Edmondson DE, Mattevi A (2007) Structures of human monoamine oxidase B complexes with selective noncovalent inhibitors: safinamide and coumarin analogs. J Med Chem 50(23):5848–5852
Mathew B, Mathew GE, Suresh J, Ucar G, Sasidharan R, Anbazhagan S, Vilapurathu JK, Jayaprakash, (2016) Monoamine oxidase inhibitors: perspective design for the treatment of depression and neurological disorders. Curr Enzyme Inhib 12(2):115–122
Kumar B, Gupta VP, Kumar VA (2017) Perspective on monoamine oxidase enzyme as drug target: challenges and opportunities. Curr Drug Targets 18:87–97
Schedin-Weiss S, Inoue M, Hromadkova L, Teranishi Y, Yamamoto NG, Wiehager B, Bogdanovic N, Winblad B, Sandebring-Matton A, Frykman S, Tjernberg LO (2017) Monoamine oxidase B is elevated in Alzheimer disease neurons, is associated with γ-secretase and regulates neuronal amyloid β-peptide levels. Alzheimer’s Res Ther 9(1):57
Ramsay RR (2016) Molecular aspects of monoamine oxidase B. Prog Neuropsychopharmacol Biol Psychiatry 69:81–89
Tao D, Wang Y, Bao XQ, Yang BB, Gao F, Wang L, Zhang D, Li L (2019) Discovery of coumarin Mannich base derivatives as multifunctional agents against monoamine oxidase B and neuroinflammation for the treatment of Parkinson’s disease. Eur J Med Chem 173:203–212
Tzvetkov NT, Stammler H-G, Neumann B, Hristova S, Antonov L, Gastreich M (2017) Crystal structures, binding interactions, and ADME evaluation of brain penetrant N -substituted indazole-5-carboxamides as subnanomolar, selective monoamine oxidase B and dual MAO-A/B inhibitors. Eur J Med Chem 127:470–492
Mathew B, Parambi DGT, Mathew GE, Uddin MdS, Inasu ST, Kim H, Marathakam A, Unnikrishnan MK, Carradori S (2019) Emerging therapeutic potentials of dual-acting MAO and AChE inhibitors in Alzheimer’s and Parkinson’s diseases. Arch Pharm 352(11):1900177
Tzvetkov NT, Stammler HG, Georgieva MG, Russo D, Faraone I, Balacheva AA, Hristova S, Atanasov AG, Milella L, Antonov L, Gastreich M (2019) Carboxamides vs methanimines: crystal structures, binding interactions, photophysical studies, and biological evaluation of (indazole-5-yl)methanimines as monoamine oxidase B and acetylcholinesterase inhibitors. Eur J Med Chem 179:404–422
Tripathi AC, Upadhyay S, Paliwal S, Saraf SK (2018) Privileged scaffolds as MAO inhibitors: retrospect and prospects. Eur J Med Chem 145:445–497
Mellado M, González C, Mella J, Aguilar LF, Viña D, Uriarte E, Cuellar M, Matos MJ (2021) Combined 3D-QSAR and docking analysis for the design and synthesis of chalcones as potent and selective monoamine oxidase B inhibitors. Bioorg Chem 108:104689
Maliyakkal N, Eom BH, Heo JH, Almoyad MAA, Parambi DGT, Gambacorta N, Nicolotti O, Beeran AA, Kim H, Mathew B (2020) A new potent and selective monoamine oxidase-B inhibitor with extended conjugation in a chalcone framework: 1-[4-(Morpholin-4-yl)phenyl]-5-phenylpenta-2,4-dien-1-one. ChemMedChem 15:1629–1633
Turan-Zitouni G, Hussein W, Saglık BN, Tabbi A, Korkut B (2018) Design, synthesis and biological evaluation of novel N-pyridyl-hydrazone derivatives as potential monoamine oxidase (MAO) inhibitors. Molecules 23:113
Can NO, Osmaniye D, Levent S, Saglık BN, Inci B, Ilgın S, Ozkay Y, Kaplancıklı ZA (2017) Synthesis of new hydrazone derivatives for MAO enzymes inhibitory activity. Molecules 22:1381
Singh YP, Rai H, Singh G, Singh GK, Mishra S, Kumar S, Srikkishna S, Modi G (2021) A review on ferulic acid and analogs-based scaffolds for the management of Alzheimer’s disease. Eur J Med Chem 215:113278
Nabavi SF, Devi KP, Malar DS, Sureda A, Daglia M, NabaviSM, (2015) Ferulic acid and Alzheimer’s disease: promises and pitfalls. Mini Rev Med Chem 15:776–788
Benchekroun M, Romero A, Egea J, Leon R, Michalska P, Buendía I, Jimeno ML, Jun D, Janockova J, Sepsova V, Soukup O, Bautista-Aguilera OM, Refouvelet B, Ouari O, Marco-Contelles J, Ismaili L (2016) The antioxidant additive approach for Alzheimer’s disease therapy: new ferulic (lipoic) acid plus melatonin modified tacrines as cholinesterases inhibitors, direct antioxidants, and nuclear factor (erythroid-derived 2)-like 2 activators. J Med Chem 59:9967–9973
Kumar N, Pruthi V (2014) Potential applications of ferulic acid from natural sources. Biotechnol Rep 4:86–93
Liu H, Liu L, Gao X, Liu Y, Xu W, He W, Jiang H, Tang J, Fan H, Xia X (2017) Novel ferulic amide derivatives with tertiary amine side chain as acetylcholinesterase and butyrylcholinesterase inhibitors: the influence of carbon spacer length, alkylamine and aromatic group. Eur J Med Chem 126:810–822
Sgarbossa A, Giacomazza D, di Carlo M (2015) Ferulic acid: a hope for Alzheimer’s disease therapy from plants. Nutrients 7:5764–5782
Turkez H, Arslan ME, Barboza JN, Kahraman CY, de Sousa DP, Mardinoğlu A (2022) Therapeutic potential of ferulic acid in Alzheimer’s disease. Curr Drug Deliv 19(8):860–873
Wolszleger M, Stan CD, Apotrosoaei M, Vasincu I, Panzariu A, Profire L (2014) New hydrazones of ferulic acid: synthesis, characterization and biological activity. Rev Med Chir Soc Med Nat Iasi 118:1150–1156
Mancuso C, Santangelo R (2014) Ferulic acid: pharmacological and toxicological aspects. Food Chem Toxicol 65:185–195
Lee HW, Ryu HW, Kang MG, Park D, Lee H, Shin HM, Oh SR, Kim H (2017) Potent inhibition of monoamine oxidase A by decursin from Angelica gigas Nakai and by wogonin from Scutellaria baicalensis Georgi. Int J Biol Macromol 97:598–605
Oh JM, Kang Y, Hwang JH, Park JH, Shin WH, Mun SK, Lee JU, Yee ST, Kim H (2022) Synthesis of 4-substituted benzyl-2-triazole-linked-tryptamine-paeonol derivatives and evaluation of their selective inhibitions against butyrylcholinesterase and monoamine oxidase-B. Int J Biol Macromol 217:910–921
Baek SC, Park MH, Ryu HW, Lee JP, Kang MG, Park D, Park CM, Oh SR, Kim H (2019) Rhamnocitrin isolated from Prunus padus var. seoulensis: a potent and selective reversible inhibitor of human monoamine oxidase A. Bioorg Chem 83:317–325
Baek SC, Lee HW, Ryu HW, Kang MG, Park D, Kim SH, Cho ML, Oh SR, Kim H (2018) Selective inhibition of monoamine oxidase A by hispidol. Bioorg Med Chem Lett 28(4):584–588
Oh JM, Jang HJ, Kim WJ, Kang MG, Baek SC, Lee JP, Park D, Oh SR, Kim H (2020) Calycosin and 8-O-methylretusin isolated from Maackia amurensis as potent and selective reversible inhibitors of human monoamine oxidase-B. Int J Biol Macromol 151:441–448
Lee HW, Ryu HW, Kang MG, Park D, Oh SR, Kim H (2016) Potent selective monoamine oxidase B inhibition by maackiain, a pterocarpan from the roots of Sophora flavescens. Bioorg Med Chem Lett 26:4714–4719
Shaw DE (2021) Desmond molecular dynamics system. Maestro Desmond interoperability tools. Research, Schrodinger Release.
Mathew B, Haridas A, Uçar G, Baysal I, Joy M, Mathew GE, Lakshmanan B, Jayaprakash V (2016) Synthesis, biochemistry, and computational studies of brominated thienyl chalcones: a new class of reversible MAO-B inhibitors. ChemMedChem 11:1161–1171
Parambi DGT, Oh JM, Baek SC, Lee JP, Tondo AR, Nicolotti O, Kim H, Mathew B (2019) Design, synthesis and biological evaluation of oxygenated chalcones as potent and selective MAO-B inhibitors. Bioorg Chem 93:103335
Oh JM, Rangarajan TM, Chaudhary R, Gambacorta N, Nicolotti O, Kumar S, Mathew B, Kim H (2022) Aldoxime- and hydroxy- functionalized chalcones as highly potent and selective monoamine oxidase-B inhibitors. J Mol Struc 1250:131817
Rehuman NA, Oh JM, Abdelgawad MA, Beshr EAM, Abourehab MAS, Gambacorta N, Nicolotti O, Jat RK, Kim H, Mathew B (2022) Development of halogenated-chalcones bearing with dimethoxy phenyl head as monoamine oxidase-B inhibitors. Pharmaceuticals 15:1152
Tripathi A, Choubey PK, Sharma P, Seth A, Saraf P, Shrivastava SK (2020) Design, synthesis, and biological evaluation of ferulic acid-based 1,3,4-oxadiazole hybrids as multifunctional therapeutics for the treatment of Alzheimer’s disease. Bioorg Chem 95:103506
Acknowledgements
Not applicable.
Funding
This work was supported by a Research Promotion Program of SCNU.
Author information
Authors and Affiliations
Contributions
Conceptualization: HK, BM; synthesis: ATP, MAA, MMG, SD; biological assay: JL; docking analysis: SK, MES, SS; writing—original draft preparation: ATP, JL, SK; writing—review and editing: BM, SD, HK; supervision: HK. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
Figure S1–S31.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Peedikayil, A.T., Lee, J., Abdelgawad, M.A. et al. Inhibitions of monoamine oxidases by ferulic acid hydrazide derivatives: synthesis, biochemistry, and computational evaluation. Appl Biol Chem 66, 66 (2023). https://doi.org/10.1186/s13765-023-00823-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13765-023-00823-0