Abstract
Background
Circular RNA pyridoxal kinase (circPDXK; hsa_circ_0061893) is newly identified to be aberrantly expressed in ovarian cancer (OVCA); however, its functional role in OVCA cells remains to be expounded.
Methods
Real-time quantitative polymerase chain reaction, western blotting, and immunohistochemistry quantified RNA and protein expression levels. MiRNA binding site prediction tools predicted direct interaction between two RNAs, and dual-luciferase reporter and RNA immunoprecipitation assays further confirmed that prediction. Cell-counting kit-8, colony formation, and 5-ethynyl-2ʹ-deoxyuridine assays measured cell growth; nude mice xenograft tumor experiment detected tumor growth. Transwell and Annexin V-fluorescein isothiocyanate/propidium iodide staining assays evaluated cell motility and apoptosis. Glycolysis process was determined by glucose uptake, lactate, and ATP assay kits.
Results
CircPDXK is highly expressed in OVCA patients’ tumor tissues and cells, concomitant with microRNA (miR)-654-3p downregulation and hexokinase II (HK2) upregulation. RNA interference of circPDXK could restrain cell viability, colony formation, DNA synthesis, migration, invasion, and glycolysis of OVCA cells, but also retard xenograft tumor growth. Allied with those are higher apoptosis rate, elevated Bax and E-cadherin levels, and depressed ki67 and HK2 levels. Compared to circPDXK inhibition, restoration of miR-654-3p functions analogical effects in OVCA cells in vitro. Mechanistically, there are direct interactions between miR-654-3p and circPDXK or HK2; moreover, miR-654-3p inhibition could weaken the functional roles of circPDXK interference in OVCA cells, and either HK2 ectopic expression abrogates the effects of miR-654-3p overexpression.
Conclusion
CircPDXK/miR-654-3p/HK2 axis could be a novel molecular mechanism of OVCA progression and glycolysis, and targeting circPDXK might overcome OVCA.
Highlights
-
1
CircPDXK is an abnormally upregulated circRNA in OVCA tumors and cells.
-
2
CircPDXK siRNA suppresses cell growth, migration, and invasion of OVCA cells and glycolysis.
-
3
CircPDXK shRNA retards tumor growth of OVCA cells in nude mice.
-
4
CircPDXK directly interacts with miR-654-3p and regulates HK2 expression.
Similar content being viewed by others
Introduction
Ovarian cancer (OVCA)) is the deadliest gynecological malignancy [1], and the tumors are featured by heterogeneities; for example, the tumor origin could be from ovarian surface epithelium (the most prevalent type), fallopian tube, and ectopic endometrial tissue [2]. Epithelial OVCA consists of low-grade and high-grade serous, endometrioid, and clear-cell ovarian carcinomas, etc. [3]. Uncontrolled growth and metastasis throughout the peritoneal cavity are the leading causes of chemoresistance and death in OVCA patients [4]. The high glycolysis process is a defining hallmark of most malignant tumors [5]. Hexokinase II (HK2) is the catalyzing enzyme involved in the first committed and irreversible step of glycolysis and contributes to tumorigenesis and cancer cell survival [6], as well as chemoresistance [7]. Notably, HK2 can act as both facilitator and gatekeeper of malignancy in cancer [8], making it a promising primary target to eradicate highly glycolytic tumors including OVCA [6, 9, 10]. Nevertheless, the upstream regulatory mechanism of HK2 expression in OVCA is elusive.
Circular RNA (circRNA) is an entire class of abundant and ubiquitous non-coding RNAs (ncRNAs) with a back splice site between 5ʹ- and 3ʹ-end [11], which is the hallmark feature of circRNA and different from other linear ncRNAs, such as long ncRNA and microRNAs (miRNAs). Of great importance, circRNAs have been well-documented to be implicated in human cancer initiation, progression, metabolism, drug resistance, and tumor microenvironment [12,13,14], rendering circRNAs as better predictive biomarkers and targets [15]. Mechanistically, circRNAs functioning as “miRNA sponges” could efficiently control miRNAs’ activity of inhibiting target gene expression, and thus construct a circRNA-centered competitive endogenous RNA (ceRNA) regulatory network [16]. Hsa_circRNA (circ)_0061893 is derived from the linear transcript of human pyridoxal kinase (PDXK), the enzyme that generates the active vitamin B6 which is an essential cofactor in all organisms [17]. Therefore, this circRNA is hereinafter referred to as circPDXK, and it is one of the most aberrantly expressed circRNAs in OVCA patients’ tumor tissues according to a circRNA microarray [18]. However, whether circPDXK expression alters in OVCA samples remains to be further validated; if so, its action and action-of-mechanism in regulating OVCA cell activities should be determined either.
MiRNA (miR)-654-3p mapping to the 14q32 region is one of the panels of 4 common differently-expressed miRNAs in OVCA cells and their exosomes [19]. Moreover, miR-654-3p plays an important anti-oncogenic role in many types of malignant tumors [20,21,22] including OVCA [23]. Thus, in this study, we aimed to explore the dysregulation of circPDXK in human OVCA samples and to test its functional role and ceRNA regulatory mechanism in OVCA progression and glycolysis by regulating miR-654-3p and HK2.
Materials and methods
Human tissues and cells
A sum of 52 OVCA patients was collected from Beijing Tongren Hospital, Capital Medical University and no patient received any antitumor therapy. Prior consent from the patients and approval from the Ethics Committees of this hospital were obtained before the collection of paired tumor tissue and adjacent non-tumorous tissue. All participants were treated in accordance with the Declaration of Helsinki (as revised in 2013).
One normal ovarian surface epithelial cell line HOSEpiC (#7310; Science Cell, Carlsbad, CA, USA), and four OVCA cell lines SKOV3 (HTB-77; ATCC, Manassas, VA, USA), OVCAR-3 (HTB-161; ATCC), A2780 (#93112519; ECACC, Salisbury, UK) and ES-2 (CRL-1978; ATCC) were employed in this study. HOSEpiC was cultured in the Ovarian Epithelial Cell Medium (Science Cell), SKOV3 and ES-2 were cultured in McCoy’s 5A Medium (Procell, Wuhan, China), and OVCAR-3 and A2780 were in RPMI-1640 medium (Science Cell); all cells were incubated in 2 mM Glutamine and 10% Fetal Bovine Serum at 37 °C with 5% carbon dioxide.
Actinomycin D treatment and cell transfection
0.2 μM of Actinomycin D (GlpBio, Montclair, CA, USA) was added to A2780 and SKOV3 cells at 80% confluence, and treated cells were collected at 0 h, 4 h, 8 h, 12 h, and 24 h for RNA expression detection of circPDXK and linear PDXK.
CircPDXK expression was silenced using RNA interference including small interfering (si)RNA in vitro and short hairpin (sh)RNA in vivo. MiR-654-3p expression was overexpressed and downregulated using mimic (sense RNA of miR-654-3p) and inhibitor (antisense RNA of miR-654-3p), respectively. HK2 expression was upregulated using pcDNA overexpression vector (pcDNA3.1( +); YouBio, Changsha, China). All transfections were performed using Neon™ Transfection System (Invitrogen, Carlsbad, CA, USA) in line with the protocol, and transfected cells were harvested at 48 h for further analysis. The sequence of si-circPDXK was AAAACUCGGCCUCAAACUGGUTT (#1) and UCAAAUCCCAAAACUCGGCCUTT (#2); PGPU6/GFP/Neo plasmid (Genepharma, Shanghai, China) and PGPU6/GFP/Neo siRNA expression vector kit (Genepharma) were used to construct sh-circPDXK with hairpin structure AGGCCGAGUUUUGGGAUUUGACAAGAGAUCAAAUCCCAAAACUCGGCCUUU.
Real-time quantitative polymerase chain reaction (qPCR) and RNase R assay
Total RNA was extracted from tissue and cell using TRIzol reagent (Sigma-Aldrich, St. Louis, MO, USA) based on the manufacturer’s protocols. Isolated total RNA was subjected to cDNA synthesis and qPCR analysis using the EXPRESS One-Step SYBR™ GreenER™ Kit with premixed ROX (Invitrogen) and special primer pairs (Table 1). β-actin and U6 served as the internal control to adjust for sample loading.
Isolated RNAs (5 μg) from SKOV3 and A2780 cells were added with RNase R working solution containing 0.5 μL RNase R (20 U/μL Lucigen, Middleton, WI, USA) and 9.5 μL 10 × RNase R Reaction Buffer for 30 min, followed by qPCR analysis for RNA expression detection of circPDXK and linear PDXK. The mock solution treatment without RNase R was the control group.
Cell-counting kit-8 (CCK8), colony formation, and 5-ethynyl-2’-deoxyuridine (EdU) assays
Transfected cells at 48 h were passaged in new cell plates with 3-paralleled wells at a certain cell density for cell growth assays. For cell viability assay, 2,500 cells/well/100 µL were used and further cultured for 0 h, 24 h, 48 h, and 72 h in 96-well plates. At each time point, 10 μL CCK8 reagent (Yeasen, Guangzhou, China) was added to each well for 4 h, and optical density (OD) values at 450 nm were measured on a SpectraMax microplate reader (Molecular Devices, Shanghai, China). For colony formation, 200 cells/well were used in 12-well plates, and single cells were further cultured to form colonies within 15 days. Colonies were visualized using the 0.5% crystal violet staining method, and pictures were taken under a microscope (Tokyo, Japan). For DNA synthesis ability, 5,000 cells/well/100 µL were used in 96-well plates, and cells were fixed for EdU detection after 2 day-incubation according to the procedures recommended by the manufacturer. In brief, new synthesized DNA was stained by EdU which was pre-labeled with Yefluor 488Azide (Component B) via click reaction, and total DNA was dyed by Hoechst 33342 (Component F). EdU positive cell rate among all Hoechst positive cells was measured per field under a microscope at 100 × .
Transwell assays
QCM ECMatrix Cell Invasion Assay with 8 µm inserts (Sigma-Aldrich) and QCM Chemotaxis Cell Migration Assay with 8 µm inserts (Sigma-Aldrich) was used to measure cell migration and invasion abilities in 24-well plates. 1 × 105 cells re-suspended in serum-free medium were included in the Transwell apparatuses, and medium containing 10% serum was in the lower chamber as an attractant to the upper cells. Transwell apparatuses were incubated in normal cell culture conditions for 48 h, and cells that migrated or invaded across the inserts were dyed with the Cell Stain reagent, and eventually counted under a microscope at 100 × .
Apoptosis assay and flow cytometry (FCM)
After different transfection for 48 h, 1 × 105 cells per group were harvested for Annexin V-fluorescein isothiocyanate (FITC) Apoptosis Kit (APExBio, Shanghai, China). Briefly, cells were re-suspended in 1 × Binding buffer and then added with Annexin V-FITC and propidium iodide (PI) for 5 min in the dark. Then, FITC signal detector (FL1) and PI signal (FL2) were captured and analyzed on a CytoFLEX S flow cytometer (Beckman Coulter, Brea, CA, USA). Apoptosis rate was determined by the percentage of the blots distributed in the upper right (UR) and left–right (LR) quadrants on the matched CytExpert software.
Western blotting
Total protein in tissue and cell was extracted from the lysates in 1 × RIPA Lysis buffer (Millipore, Billerica, MA, USA) supplemented with Protease Inhibitor Cocktail (Sigma-Aldrich), followed by concentration quantification using the BCA method. 30 μg proteins were utilized for western blotting procedures. Non-special antigens were blocked by 3% bovine serum albumin (BSA), and special antigens were incubated with corresponding antibodies of HK2 (CSB-PA132121; 1:5,000, CusaBio, Houston, TX, USA), ki67 (#13180-T48; 1:1,000, Sino Biological, Beijing, China), Bax (CSB-PA221042; 1:2,000, CusaBio), E-cadherin (CSB-PA218378; 1:2,500, CusaBio) and β-actin (CSB-PA244513; 1:5,000, CusaBio). After secondary antibody incubation, chromogen was developed with enhanced chemiluminescence and detected using gel imaging system (Bio-Rad, Hercules, CA, USA). The densitometry of the protein bands was analyzed using Quantity One software (Bio-Rad). β-actin served as the internal control to adjust for sample loading.
Glycolysis analysis
Glucose Uptake Assay Kit (KA4086; Abnova, Birmingham, UK) and Lactate Assay Kit (KA0833; Abnova) were used to measure glucose uptake and L( +)-lactate production in cells with the colorimetric detection method by reading OD) value at 570 nm on SpectraMax microplate reader (Molecular Devices); intracellular ATP level was detected using ADP/ATP Ratio Assay Kit (KA 16 7 3; Abnova) with luminescence measurement method by measuring relative light unit of oxyluciferin on a luminometer (Molecular Devices).
Dual-luciferase reporter assay
CircInteractome, starBase and circBank prediction tools were used to predict the potential target miRNAs in circPDXK sequence, and starBase tool predicted potential miR-654-3p binding sites in HK2. in The wild type (WT) of circPDXK and HK2 3ʹuntranslated region (3ʹUTR) harboring the predicted miR-654-3p target sites were cloned into luciferase reporter vector pmirGLO (Promega, Madison, WI, USA); besides, these predicted sites in circPDXK and HK2 3’UTR WT vectors were mutated using QuickChange Lightning multisite-directed mutagenesis kit (Stratagene, Cedar Creek, TX, USA), constructing the corresponding mutated type (MUT) vectors. SKOV3 and A2780 cells were co-transfected with recombinant report vectors and miR-654-3p mimic or miR-NC mimic, and luciferase activities of Firefly and the Renilla were analyzed via Dual-Glo® Luciferase Assay System (Promega) after 48 h. Relative luciferase activity was the ratio of Firefly to Renilla (the internal control to adjust for transfection efficiency).
RNA Immunoprecipitation (RIP)
Immunoprecipitation Kit (Sino Biological) was used for nucleic acid and protein complexes. Cell lysates of SKOV3 and A2780 were collected using NP40 Cell Lysis Buffer; 50 μL Immunomagnetic Beads Protein A/G was incubated with Ago2 antibody (anti-Ago2) or anti-IgG for 2 h at room temperature. The nucleic acid and protein complex was harvested in the 20 μL Elution Buffer, and treated with Proteinase K (APExBIO, Houston, TX, USA) followed by RNA isolation using TRIzol reagent (Sigma-Aldrich).
Immunohistochemistry (IHC) and xenograft tumor experiment
Paraffin-coated tissues were for IHC analysis as previously reported [24]. In brief, paraffin tissues (4-µm thick) were deparaffinized and rehydrated with xylene and a graded ethanol series; antigen retrieval was performed in 10 mM citrate buffer and blocked by 5% BSA prior to the incubation of HK2 antibody (CSB-PA132121; 1:5,000, CusaBio) or ki67 antibody (#12202; 1:500, Cell Signaling Technology, Shanghai, China). After secondary antibody incubation, chromogen was developed with 3,3′‐diaminobenzidine. Sections were counterstained with hematoxylin and eventually captured by microscope.
A2780 cells (3 × 106 cells/mice) stably expressed sh-NC or sh-circPDXK were re-suspended in normal saline and subcutaneously inoculated into the posterior flank of BALB/c nude mice (5–6 week-old; female, Vital River Laboratory, Beijing, China). Five mice were for each group, and all purchased mice were fed for 1 week before this experiment. The health and behavior of mice were monitored every day, and the length (l) and width (w) of formed tumors were measured every 7 days. After 5 weeks, mice were euthanized by cervical dislocation under anesthesia, and immediately tumors were excised, weighed, and photographed. Tumor volume was 0.5 × l × w2. This animal experiment was pre-approved by the Animal Care and Use Committee at Beijing Tongren Hospital, Capital Medical University, and performed following the guidelines for laboratory animal welfare (GBT 35892–2018). After picture capture, tumor tissues were either lysed for total RNA/protein isolation or formalin-fixed and paraffin-coated for in situ expression detection.
Statistical analysis
The experiments were conducted in triplicate, and each experiment was repeated three times. Data were shown as mean ± standard deviation and analyzed by Student’s t-test or analysis of variance followed by Tukey’s post hoc test. Correlation analysis was calculated by Pearson correlation coefficient (r). P < 0.05, 0.01, and 0.001 were considered significant (*), very significant, and extremely significant. GraphPad Prism 7.0 (GraphPad, La Jolla, CA, USA) was used to analyze data and image processing.
Results
CircPDXK is an upregulated circRNA in human OVCA samples
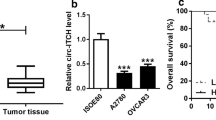
CircPDXK was generated from the exon 2, exon 3, exon 4, exon 5, and exon 6 of the PDXK gene, and the end of exon 2 and exon 6 were back-spliced to form the circular structure (Fig. 1A). Then, to verify the existence of circPDXK in OVCA cells, we designed divergent primers to amplify circPDXK and convergent primers to amplify β-actin mRNA. As shown in Fig. 1B, circPDXK was only detectable in cDNA but not genomic DNA (gDNA) from SKOV3 cells by RT-PCR with divergent primers, whereas β-actin could be amplified in both cDNA and gDNA using convergent primers. Next, RNase R and actinomycin D were applied to test the stability of circPDXK. Data suggested that circPDXK expression in OVCA cell lines (SKOV3 and A2780) was resistant and its host gene linear PDXK expression was sensitive to RNase R digestion (Fig. 1C and D). In addition, the half-life of circPDXK was much longer than 24 h after actinomycin D treatment, whereas that of linear PDXK was no more than 8 h (Fig. 1E and F). In human OVCA samples, circPDXK was aberrantly expressed and to be upregulated in tumor tissues and tumor cell lines from patients (Fig. 1G and H). These data showed that circPDXK was an upregulated circRNA in human OVCA with favorable structure stability.
The formation and expression of circPDXK. A Schematic diagram showed the back-splicing of exons 2–6 of PDXK. B The existence of circPDXK was detected in the SKOV3 cell line by RT-PCR with convergent or divergent primers and verified using Gel electrophoresis. C–F QPCR estimated relative RNA expression of circPDXK and linear PDXK in C, D RNase R treatment group and mock group, and E, F actinomycin D treatment groups for different durations in SKOV3 and A2780 cells. G, H QPCR estimated relative circPDXK expression in G tumor tissues and adjacent tissues from OVCA cancer patients (n = 52), and H OVCA cell lines (ES-2, OVCAR-3, SKOV3, and A2780) and normal control cell line HOSEpiC. **P < 0.01; ***P < 0.001
Inhibiting circPDXK via siRNA restrains cell growth and motility of OVCA cells and glycolysis
Then, circPDXK expression was manipulated to be inhibited in OVCA cells using siRNA transfection, and si-circPDXK#2 displayed a better silencing efficiency than si-circPDXK#1 in SKOV3 and A2780 cells (Fig. 2A); therefore, si-circPDXK#2-transfected cells were harvested for further functional assays. First of all, cell growth was measured. As consequently, cell viabilities during 3 days analyzed by CCK8 assay were overall lowered in si-circPDXK#2 group (Fig. 2B), allied with that was the decrease of colonies formed in 15 days (Fig. 2C); moreover, DNA synthesis capacity examined by EdU assay was consistently reduced in the condition of circPDXK silencing (Fig. 2D). Then, cell motility was measured using Transwell assays, and numbers of migrated cells and invaded cells were synchronously lessened in si-circPDXK#2-expressed cells than in negative control cells (Fig. 2E and F). data displayed a promotion of apoptosis rate due to si-circPDXK#2 addition (Fig. 2G). Molecularly, the expression of proliferation marker ki67was depressed, and apoptosis marker Bax and epithelial marker E-cadherin were highly expressed in SKOV3 and A2780 cells with circPDXK interference (Fig. 2H–J). Furthermore, tumor-related glycolysis was restrained by RNA-interfering circPDXK, as evidenced by the loss of glucose uptake, lactate production, and intracellular ATP level (Fig. 2K–M). Taken together, cell growth and motility of OVCA and glycolysis process were suppressed by inhibiting circPDXK.
The biological role of circPDXK dysregulation in OVCA cells. A QPCR estimated relative circPDXK expression in SKOV3 and A2780 cells transfected with si-NC, si-circPDXK#1 or si-circPDXK#2. (B-M) SKOV3 and A2780 cells were harvested after transfections of si-NC and si-circPDXK#2. B Cell viability was detected using CCK8 assay by measuring OD values (λ = 450 nm) at different culture durations. C Colony formation was determined using colony formation assay by counting the number of colonies. D DNA synthesis was examined using EdU assay by calculating EdU positive cell rate (%). Hoechst was total DNA dye, EdU was new DNA stain. E, F Transwell assays evaluated migration and invasion abilities by microscopically counting numbers of migrated cells and invaded cells in per field (100 ×). G FCM method and Annexin V-FITC/PI staining showed apoptosis rate (%). H–J Western blotting detected relative protein expression of ki67, Bax and E-cadherin with normalization to β-actin. K–M Glucose uptake, lactate and ATP assay kits separately examined relative glucose uptake, lactate production and intracellular ATP level. **P < 0.01; ***P < 0.001
CircPDXK directly interacts with miR-654-3p
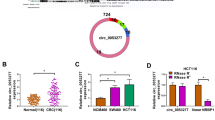
Functional mechanism of circPDXK was further identified via interaction with miRNAs. By retrieving miRNA binding site prediction tools, miR-582-3p and miR-654-3p were putative target miRNAs for circPDXK (Fig. 3A). In response to circPDXK silencing, miR-654-3p abundance was elevated with greater intensity than miR-582-3p (Fig. 3B and C). Based on the computational results, miR-654-3p target sites in sequence of circPDXK were mutated to construct the mutant luciferase report vector (Fig. 3D). In response to ectopic expression of miR-654-3p via mimic transfection (Fig. 3E), luciferase activity of circPDXK WT report vector was sensitive to be diminished in SKOV3 and A2780 cells, whereas that of circPDXK MUT was unresponsive (Fig. 3F and G). Moreover, circPDXK and miR-654-3p were spatiotemporally co-distributed in Ago2-mediated immunoprecipitation in SKOV3 and A2780 cells (Fig. 3H and I). These results indicated a direct interaction between circPDXK and miR-654-3p. In terms of expression status, miR-654-3p was downregulated in human OVCA tumor tissues and cell lines (Fig. 3J and K), and its tissue expression level was mildly linearly correlated with circPDXK in these 52 samples according to Pearson correlation analysis (Fig. 3L). These data hinted at a potential contribution of miR-654-3p in the circPDXK silencing role.
The functional mechanism of circPDXK via directly interacting with miRNA. A Venn diagram showed the potential target miRNAs according to CircInteractome, starBase, and circBank prediction tools. B, C QPCR estimated the relative expression of miR-582-3p and miR-654-3p in SKOV3 and A2780 cells after transfections of si-NC and si-circPDXK#2. D Schematic plot showed the predicted miR-654-3p target sites in circPDXK WT and their mutations in circPDXK MUT. E QPCR estimated relative miR-654-3p expression in SKOV3 and A2780 cells after transfections of miR-654-3p mimic and miR-NC mimic. F, G Dual-luciferase reporter assay determined relative luciferase activity of circPDXK WT and circPDXK MUT report vectors in condition of miR-654-3p or miR-NC overexpression. H, I RIP and qPCR assays confirmed relative RNA enrichment of circPDXK and miR-654-3p in the input and precipitations mediated by anti-Ago2 and anti-IgG. J, K QPCR estimated the relative miR-654-3p expression in (J) tumor tissues and adjacent tissues from OVCA cancer patients (n = 52), and (K) HOSEpiC, SKOV3 and A2780 cells. L Linear correlation coefficient (r) between circPDXK and miR-654-3p levels in 52 OVCA tumors was determined by Pearson correlation analysis. *P < 0.05; **P < 0.01; ***P < 0.001
Inhibition of miR-654-3p weakens the functional roles of circPDXK silencing
An inhibitor of miR-654-3p was introduced to exhaust miR-654-3p levels in SKOV3 and A2780 cells and in circPDXK-silenced cells (Fig. 4A and B). In function, in-miR-654-3p significantly improved cell viability, colony formation number and EdU positive rate of si-circPDXK#2-transfected cells (Fig. 4C–F), accompanied by promoted ki67 level (Fig. 4J–L). Migration and invasion abilities of OVCA cells with circPDXK interference were distinctively raised by further inhibiting miR-654-3p (Fig. 4G and H), concomitant with suppressed expression of E-cadherin (Fig. 4J–L). Additional inhibition of miR-654-3p abated the promoting effects of circPDXK siRNA on apoptosis rate and Bax expression (Fig. 4I–L). Either, circPDXK silencing-mediated glycolysis inhibition was partially canceled by silencing miR-654-3p (Fig. 4M–O). These results revealed that miR-654-3p inhibition showed a weakening effect on circPDXK silencing role in OVCA cells.
The weakening effect of miR-654-3p on circPDXK role. A–O SKOV3 and A2780 cells were harvested after single transfections of si-NC, si-circPDXK#2, in-miR-NC, and in-miR-654-3p, and co-transfections of si-circPDXK#2 along with in-miR-NC or in-miR-654-3p. A, B QPCR estimated the relative expression of miR-654-3p. C, D CCK8 assay measured OD values (λ = 450 nm) at different culture durations. E Colony formation assay determined the number of colonies. F EdU assay examined EdU-positive cell rate (%). G, H Transwell assays evaluated numbers of migrated cells and invaded cells per field (100 ×) under a microscope. I FCM method and Annexin V-FITC/PI staining showed apoptosis rate (%). J–L Western blotting detected relative protein expression of ki67, Bax, and E-cadherin with normalization to β-actin. M–O Glucose uptake, lactate, and ATP assay kits separately examined relative glucose uptake, lactate production, and intracellular ATP level. *P < 0.05; **P < 0.01; ***P < 0.001
Overexpression of miR-654-3p suppresses the progression of OVCA cells and glycolysis by target inhibiting HK2
As one rate-limiting enzyme in glycolysis, HK2 was the putative target gene for miR-654-3p, and the predicted target sites in the 3’UTR were mutated for luciferase assay (Fig. 5A). As a result, the luciferase activity of the HK2 3’UTR WT report vector instead of the MUT vector was attenuated by miR-654-3p mimic (Fig. 5B and C). HK2 expression at mRNA and protein levels was higher in OVCA patients’ tumor tissues than in adjacent normal tissues (Fig. 5D and F), and in situ expression of HK2 in the OVCA tissue section was promoted (Fig. 5E). Notably, the tissue level of HK2 mRNA was moderately correlated to miR-654-3p and mildly associated with circPDXK in these OVCA subjects both in a linear relation (Fig. 5J and K). In OVCA cell lines, HK2 protein expression was upregulated in SKOV3 and A2780 cells (Fig. 5G), and this upregulation could be declined by circPDXK siRNA (Fig. 5H and I); circPDXK silencing-mediated level of HK2 protein could also be elevated by miR-654-3p inhibitor (Fig. 5H and I). These results manifested that HK2 was targeted by miR-654-3p and its expression could be modulated by circPDXK and miR-654-3p.
The validation of HK2 as the functional target gene for miR-654-3p. A Schematic plot showed the predicted miR-654-3p target sites in HK2 3’UTR WT and their mutations in HK2 3ʹUTR MUT. B, C Dual-luciferase reporter assay determined relative luciferase activity of HK2 3ʹUTR WT and HK2 3’UTR MUT report vectors in the condition of miR-654-3p or miR-NC overexpression in SKOV3 and A2780 cells. D QPCR estimated relative HK2 mRNA expression in tumor tissues and adjacent tissues from OVCA cancer patients (n = 52). E IHC showed the in situ expression of HK2 in one pair of adjacent and tumor tissues from one randomly selected OVCA patient. F Western blotting detected relative protein expression of HK2 in tissue extract of three pairs of adjacent and tumor tissues from three randomly selected OVCA patients. G–I Western blotting detected relative protein expression of HK2 in (G) HOSEpiC, SKOV3 and A2780 cells, and (H, I) SKOV3 and A2780 cells after transfections of si-NC, si-circPDXK#2, and si-circPDXK#2 combined with in-miR-NC or in-miR-654-3p. J, K Linear correlation coefficient (r) between HK2 mRNA level and miR-654-3p or circPDXK level in 52 OVCA tumors was determined by Pearson correlation analysis. **P < 0.01; ***P < 0.001
MiR-654-3p mimic transfection resulted in a downregulation of HK2 protein expression in SKOV3 and A2780 cells (Fig. 6B), and functionally caused overall suppressions of CCK8 cell viability (Fig. 6C and D), colony formation number (Fig. 6E), EdU positive rate (Fig. 6F), and Transwell migration and invasion (Fig. 6G and H), as well as ki67 expression (Fig. 6J–L); in addition, apoptosis rate and expressions of Bax and E-cadherin in SKOV3 and A2780 cells were boosted in the condition of miR-654-3p overexpression (Fig. 6I–L). Expectedly, glucose uptake, lactate production, and intracellular ATP level were totally repressed by restoring miR-654-3p via mimic transfection (Fig. 6M–O). These results demonstrated a suppressive role of miR-654-3p in the progression and glycolysis of OVCA cells. Intriguingly, ectopic expression of HK2 via overexpression vector transfection could overall reverse the biological effects of miR-654-3p mimic in OVCA cells (Fig. 6A–O), suggesting an interaction effect between miR-654-3p and its target gene HK2.
Effects of miR-654-3p and HK2 on progression and glycolysis of OVCA cells. A–O SKOV3 and A2780 cells were harvested after transfections of the control pcDNA vector, HK2 overexpression vector, miR-NC, miR-654-3p, and miR-654-3p combined with the control pcDNA vector or HK2 overexpression vector. A, B Western blotting examined the relative protein expression of HK2. C, D CCK8 assay measured OD values (λ = 450 nm) at different culture durations. E Colony formation assay determined the number of colonies. F EdU assay examined EdU-positive cell rate (%). G, H Transwell assays evaluated numbers of migrated cells and invaded cells per field (100 ×) under a microscope. I FCM method and Annexin V-FITC/PI staining showed apoptosis rate (%). J–L Western blotting detected relative protein expression of ki67, Bax and E-cadherin with normalization to β-actin. M–O Glucose uptake, lactate and ATP assay kits separately examined relative glucose uptake, lactate production and intracellular ATP level. *P < 0.05; **P < 0.01; ***P < 0.001
Inhibiting circPDXK via shRNA delays tumor growth of OVCA cells
In vivo role of circPDXK in OVCA was eventually measured in nude mice xenograft tumors. As consequently, xenograft tumors mediated by A2780 cells with sh-circPDXK transfection were bigger and heavier compared to negative transfection (Fig. 7A–C). Molecularly, tumor tissues in the sh-circPDXK group showed a downregulated expression of circPDXK, HK2, and ki67, and an upregulated miR-654-3p, Bax, and E-cadherin (Fig. 7D and E). More importantly, expression analysis using IHC depicted higher ratios of HK2 and ki67 positive cells in sh-circPDXK group tissues (Fig. 7F). These outcomes showed a tumor growth inhibition of circPDXK interference in OVCA in vivo with dysregulations of miR-654-3p and HK2.
In vivo role of circPDXK dysregulation in OVCA. A–F A2780 cells stably transfected with sh-NC and sh-circPDXK were harvested for nude mice xenograft tumor model experiment (n = 5 mice/group). A Most representative image of xenograft tumors in each group was shown. B, C Tumor volume and weight were shown. D QPCR detected relative RNA expression of circPDXK, miR-654-3p, and HK2, and E western blotting measured relative protein expression of HK2, ki67, Bax, and E-cadherin in tissue extract of xenograft tumors in each group. F IHC showed the in situ expression of HK2 and ki67 in one randomly selected xenograft tumor in each group. *, P < 0.05; **, P < 0.01; ***, P < 0.001
Discussion
It was reported that some functional circRNAs exhibited cell/tissue-specific or tissue developmental stage-special expression models [25]. Here, we demonstrated that circPDHK was one upregulated circRNA in human OVCA tissue and cell samples, and its silencing could dampen OVCA cell growth, motility, and glycolysis. Thus, we propose that circPDXK is a novel OVCA-related circRNA, which may be a molecular mechanism and treatment target for OVCA by integrating glycolytic status and cell malignancy.
Our qPCR data supported the previous microarray data [18] of circPDXK upregulation in OVCA patients’ tumor tissues; besides, we also showed a higher level of circPDXK in 4 human OVCA cell lines, and its resistance to exonuclease (RNase R), as well as the favorable half-life under RNA synthesis inhibitor (actinomycin D) condition. These outcomes together showed a stable expression model of circPDXK in OVCA, further suggesting it might be also detectable and more stable in the blood, urine, and other body fluids in the patients; if so, it might harbor the potential as a mini-invasive molecular indicator in the diagnosis of OVCA. In addition, the host gene PDXK was also recently suggested to be oncogenic and as a new biomarker of chemotherapy response, diagnosis, and prognosis in serous OVCA [26, 27]. It’s worth noting that this present study was the first functional validation of circPDXK role in OVCA cells, and we discovered an overall suppression of circPDXK RNA interference on cell viability, colony formation, DNA synthesis, migration, invasion, tumor formation, and glycolytic process.
In terms of the ceRNA mechanism of circPDXK, we showed two common computational miRNAs depending on three online biological algorithms. Among the two, miR-582-3p seemed unstudied in OVCA to date, even though it had been declared to participate in a few malignant cancers including lung cancer [28], prostate cancer [29], and the gynecological malignancy cervical cancer [30]; the other miRNA was miR-654-3p was commonly downregulated in OVCA cells and their exosomes and was implicated in Wnt signaling pathway and insulin resistance, as indicated by the functional enrichment analysis of its target genes [19]. Furthermore, phosphatidylinositol-3-hydroxykinase (PI3K)/AKT signaling pathway could be suppressed by overexpressing miR-654-3p during its anti-cisplatin resistance role in OVCA cells [31]. Accordingly, we further testified miR-654-3p to be one target for circPDXK and left miR-582-3p alone at least in this study,
As one target gene for miR-654-3p, HK2 could exert an integrated control of tumor cell growth, mitochondrial apoptosis, autophagy and chemoresistance via glycolysis and signaling pathways: Akt/mammalian target of rapamycin (mTOR) [32, 33] and MEK/extracellular signal-regulated kinases (ERK) [6, 7, 34]. And, we found that the link between HK2 and PI3K/Akt/mTOR and Wnt signals in OVCA remained largely undetermined yet. Even so, this issue was not involved in this study, at least temporarily. On the other hand, HK2 was the predominant isoform among the four HK isoforms in insulin-sensitive tissues, such as adipose tissue, skeletal muscle, and heart [35]. HK2 expression was generally upregulated in multiple tumors associated with enhanced aerobic glycolysis [35]. Of note, HK2 was localized within cytosol and mitochondria and at the outer mitochondrial membrane, and it was required for anti-apoptosis by interacting with mitochondria; whereas, the effect of HK2 dysregulation on mitochondrial apoptosis-related proteins, such as Bax and Bak [36].
Clinically, imaging cancer metabolism including 18F-deoxy glucose-positron emission tomography (PET) had been becoming a novel strategy to fully characterize tumor biology [37]. Here, we molecularly considered that targeting the circPDXK/miR-654-3p axis might be a therapeutic effect in OVCA cell malignant behaviors by inhibiting HK2 and suppressing glucose metabolism transformation to glycolysis.
In conclusion, this study presented that silencing circPDXK and restoring miR-654-3p could counteract the progression and glycolysis of OVCA via fine-tuning the expression of HK2. The circPDXK/miR-654-3p/HK2 ceRNA axis may be an underlying pathological basis for the malignant development of OVCA and thus may provide a potential interfering approach.
Availability of data and materials
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.
References
Siegel RL, Miller KD, Jemal A (2019) Cancer statistics, 2019. CA Cancer J Clin 69(1):7–34. https://doi.org/10.3322/caac.21551
Karnezis AN, Cho KR, Gilks CB, Pearce CL, Huntsman DG (2017) The disparate origins of ovarian cancers: pathogenesis and prevention strategies. Nat Rev Cancer 17(1):65–74. https://doi.org/10.1038/nrc.2016.113
Nezhat FR, Apostol R, Nezhat C, Pejovic T (2015) New insights in the pathophysiology of ovarian cancer and implications for screening and prevention. Am J Obstet Gynecol 213(3):262–267. https://doi.org/10.1016/j.ajog.2015.03.044
Xie W, Sun H, Li X, Lin F, Wang Z, Wang X (2021) Ovarian cancer: epigenetics, drug resistance, and progression. Cancer Cell Int 21(1):434. https://doi.org/10.1186/s12935-021-02136-y
Abdel-Wahab AF, Mahmoud W, Al-Harizy RM (2019) Targeting glucose metabolism to suppress cancer progression: prospective of anti-glycolytic cancer therapy. Pharmacol Res 150:104511. https://doi.org/10.1016/j.phrs.2019.104511
Siu MKY, Jiang YX, Wang JJ, Leung THY, Han CY, Tsang BK, Cheung ANY, Ngan HYS, Chan KKL (2019) Hexokinase 2 regulates ovarian cancer cell migration, invasion and stemness via FAK/ERK1/2/MMP9/NANOG/SOX9 signaling cascades. Cancers (Basel). https://doi.org/10.3390/cancers11060813
Zhang XY, Zhang M, Cong Q, Zhang MX, Zhang MY, Lu YY, Xu CJ (2018) Hexokinase 2 confers resistance to cisplatin in ovarian cancer cells by enhancing cisplatin-induced autophagy. Int J Biochem Cell Biol 95:9–16. https://doi.org/10.1016/j.biocel.2017.12.010
Mathupala SP, Ko YH, Pedersen PL (2006) Hexokinase II: cancer’s double-edged sword acting as both facilitator and gatekeeper of malignancy when bound to mitochondria. Oncogene 25(34):4777–4786. https://doi.org/10.1038/sj.onc.1209603
Xintaropoulou C, Ward C, Wise A, Queckborner S, Turnbull A, Michie CO, Williams ARW, Rye T, Gourley C, Langdon SP (2018) Expression of glycolytic enzymes in ovarian cancers and evaluation of the glycolytic pathway as a strategy for ovarian cancer treatment. BMC Cancer 18(1):636. https://doi.org/10.1186/s12885-018-4521-4
Zhang M, Liu Q, Zhang M, Cao C, Liu X, Zhang M, Li G, Xu C, Zhang X (2020) Enhanced antitumor effects of follicle-stimulating hormone receptor-mediated hexokinase-2 depletion on ovarian cancer mediated by a shift in glucose metabolism. J Nanobiotechnology 18(1):161. https://doi.org/10.1186/s12951-020-00720-4
Tang X, Ren H, Guo M, Qian J, Yang Y, Gu C (2021) Review on circular RNAs and new insights into their roles in cancer. Comput Struct Biotechnol J 19:910–928. https://doi.org/10.1016/j.csbj.2021.01.018
Hua X, Sun Y, Chen J, Wu Y, Sha J, Han S, Zhu X (2019) Circular RNAs in drug resistant tumors. Biomed Pharmacother 118:109233. https://doi.org/10.1016/j.biopha.2019.109233
Shang Q, Yang Z, Jia R, Ge S (2019) The novel roles of circRNAs in human cancer. Mol Cancer 18(1):6. https://doi.org/10.1186/s12943-018-0934-6
Zhang Q, Wang W, Zhou Q, Chen C, Yuan W, Liu J, Li X, Sun Z (2020) Roles of circRNAs in the tumour microenvironment. Mol Cancer 19(1):14. https://doi.org/10.1186/s12943-019-1125-9
Sheng R, Li X, Wang Z, Wang X (2020) Circular RNAs and their emerging roles as diagnostic and prognostic biomarkers in ovarian cancer. Cancer Lett 473:139–147. https://doi.org/10.1016/j.canlet.2019.12.043
Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong F, Ren D, Ye X, Li C, Wang Y et al (2018) Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer 17(1):79. https://doi.org/10.1186/s12943-018-0827-8
Galluzzi L, Vacchelli E, Michels J, Garcia P, Kepp O, Senovilla L, Vitale I, Kroemer G (2013) Effects of vitamin B6 metabolism on oncogenesis, tumor progression and therapeutic responses. Oncogene 32(42):4995–5004. https://doi.org/10.1038/onc.2012.623
Gong J, Xu X, Zhang X, Zhou Y (2020) Circular RNA-9119 suppresses in ovarian cancer cell viability via targeting the microRNA-21-5p-PTEN-Akt pathway. Aging (Albany NY) 12(14):14314–14328. https://doi.org/10.18632/aging.103470
Zhang S, Zhang X, Fu X, Li W, Xing S, Yang Y (2018) Identification of common differentially-expressed miRNAs in ovarian cancer cells and their exosomes compared with normal ovarian surface epithelial cell cells. Oncol Lett 16(2):2391–2401. https://doi.org/10.3892/ol.2018.8954
Formosa A, Markert EK, Lena AM, Italiano D, Finazzi-Agro E, Levine AJ, Bernardini S, Garabadgiu AV, Melino G, Candi E (2014) MicroRNAs, miR-154, miR-299-5p, miR-376a, miR-376c, miR-377, miR-381, miR-487b, miR-485-3p, miR-495 and miR-654-3p, mapped to the 14q32.31 locus, regulate proliferation, apoptosis, migration and invasion in metastatic prostate cancer cells. Oncogene 33(44):5173–5182. https://doi.org/10.1038/onc.2013.451
Pu JT, Hu Z, Zhang DG, Zhang T, He KM, Dai TY (2020) MiR-654-3p suppresses non-small cell lung cancer tumourigenesis by Inhibiting PLK4. Onco Targets Ther. 13:7997–8008. https://doi.org/10.2147/OTT.S258616
Yang J, Zhang Z, Chen S, Dou W, Xie R, Gao J (2020) miR-654-3p predicts the prognosis of hepatocellular carcinoma and inhibits the proliferation, migration, and invasion of cancer cells. Cancer Biomark 28(1):73–79. https://doi.org/10.3233/CBM-191084
Duan M, Fang M, Wang C, Wang H, Li M (2020) LncRNA EMX2OS induces proliferation, invasion and sphere formation of ovarian cancer cells via regulating the miR-654–3p/AKT3/PD-L1 Axis. Cancer Manag Res 12:2141–2154. https://doi.org/10.2147/CMAR.S229013
Wang T, Hao D, Yang S, Ma J, Yang W, Zhu Y, Weng M, An X, Wang X, Li Y et al (2019) miR-211 facilitates platinum chemosensitivity by blocking the DNA damage response (DDR) in ovarian cancer. Cell Death Dis 10(7):495. https://doi.org/10.1038/s41419-019-1715-x
Chaichian S, Shafabakhsh R, Mirhashemi SM, Moazzami B, Asemi Z (2020) Circular RNAs: a novel biomarker for cervical cancer. J Cell Physiol 235(2):718–724. https://doi.org/10.1002/jcp.29009
Fekete JT, Osz A, Pete I, Nagy GR, Vereczkey I, Gyorffy B (2020) Predictive biomarkers of platinum and taxane resistance using the transcriptomic data of 1816 ovarian cancer patients. Gynecol Oncol 156(3):654–661. https://doi.org/10.1016/j.ygyno.2020.01.006
Tan W, Liu B, Ling H (2020) Pyridoxal kinase (PDXK) promotes the proliferation of serous ovarian cancer cells and is associated with poor prognosis. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 36(6):542–548
Fang L, Cai J, Chen B, Wu S, Li R, Xu X, Yang Y, Guan H, Zhu X, Zhang L et al (2015) Aberrantly expressed miR-582–3p maintains lung cancer stem cell-like traits by activating Wnt/beta-catenin signalling. Nat Commun 6:8640. https://doi.org/10.1038/ncomms9640
Huang S, Zou C, Tang Y, Wa Q, Peng X, Chen X, Yang C, Ren D, Huang Y, Liao Z et al (2019) miR-582-3p and miR-582-5p suppress prostate cancer metastasis to bone by repressing TGF-beta signaling. Mol Ther Nucleic Acids 16:91–104. https://doi.org/10.1016/j.omtn.2019.01.004
Xu J, Zhang Y, Huang Y, Dong X, Xiang Z, Zou J, Wu L, Lu W (2020) circEYA1 functions as a sponge of miR-582–3p to suppress cervical adenocarcinoma tumorigenesis via upregulating CXCL14. Mol Ther Nucleic Acids. 22:1176–1190. https://doi.org/10.1016/j.omtn.2020.10.026
Niu YC, Tong J, Shi XF, Zhang T (2020) MicroRNA-654-3p enhances cisplatin sensitivity by targeting QPRT and inhibiting the PI3K/AKT signaling pathway in ovarian cancer cells. Exp Ther Med 20(2):1467–1479. https://doi.org/10.3892/etm.2020.8878
Wu H, Pan L, Gao C, Xu H, Li Y, Zhang L, Ma L, Meng L, Sun X, Qin H (2019) Quercetin inhibits the proliferation of glycolysis-addicted HCC cells by reducing hexokinase 2 and Akt-mTOR pathway. Molecules. https://doi.org/10.3390/molecules24101993
Zhang K, Zhang M, Jiang H, Liu F, Liu H, Li Y (2018) Down-regulation of miR-214 inhibits proliferation and glycolysis in non-small-cell lung cancer cells via down-regulating the expression of hexokinase 2 and pyruvate kinase isozyme M2. Biomed Pharmacother 105:545–552. https://doi.org/10.1016/j.biopha.2018.06.009
Cui N, Li L, Feng Q, Ma HM, Lei D, Zheng PS (2020) Hexokinase 2 promotes cell growth and tumor formation through the Raf/MEK/ERK signaling pathway in cervical cancer. Front Oncol 10:581208. https://doi.org/10.3389/fonc.2020.581208
Roberts DJ, Miyamoto S (2015) Hexokinase II integrates energy metabolism and cellular protection: Akting on mitochondria and TORCing to autophagy. Cell Death Differ 22(2):248–257. https://doi.org/10.1038/cdd.2014.173
Majewski N, Nogueira V, Bhaskar P, Coy PE, Skeen JE, Gottlob K, Chandel NS, Thompson CB, Robey RB, Hay N (2004) Hexokinase-mitochondria interaction mediated by Akt is required to inhibit apoptosis in the presence or absence of Bax and Bak. Mol Cell 16(5):819–830. https://doi.org/10.1016/j.molcel.2004.11.014
Pantel AR, Ackerman D, Lee SC, Mankoff DA, Gade TP (2018) Imaging cancer metabolism: underlying biology and emerging strategies. J Nucl Med 59(9):1340–1349. https://doi.org/10.2967/jnumed.117.199869
Acknowledgements
Not applicable.
Funding
Chengdu Municipal Health Commission Project (No: 2019107).
Author information
Authors and Affiliations
Contributions
Conceptualization and Methodology: WW and JZ; Formal analysis and Data curation: JZ and HZ; Validation and Investigation: LH and WW; Writing - original draft preparation and Writing - review and editing: LH, WW and JZ. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The present study was approved by the ethical review committee of Beijing Tongren Hospital, Capital Medical University. Written informed consent was obtained from all enrolled patients.
Consent for publication
Patients agree to participate in this work.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hou, L., Wang, W., Zhai, J. et al. Circular RNA pyridoxal kinase (circPDXK) involves in the progression of ovarian cancer and glycolysis via regulating miR-654-3p and hexokinase II. Appl Biol Chem 65, 81 (2022). https://doi.org/10.1186/s13765-022-00747-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13765-022-00747-1