Abstract
Background
Clostridioides (Clostridium) difficile is an important pathogen of healthcare- associated diarrhea, however, an increase in the occurrence of C. difficile infection (CDI) outside hospital settings has been reported. The accumulation of antimicrobial resistance in C. difficile can increase the risk of CDI development and/or its spread. The limited number of antimicrobials for the treatment of CDI is matter of some concern.
Objectives
In order to summarize the data on antimicrobial resistance to C. difficile derived from humans, a systematic review and meta-analysis were performed.
Methods
We searched five bibliographic databases: (MEDLINE [PubMed], Scopus, Embase, Cochrane Library and Web of Science) for studies that focused on antimicrobial susceptibility testing in C. difficile and were published between 1992 and 2019. The weighted pooled resistance (WPR) for each antimicrobial agent was calculated using a random- effects model.
Results
A total of 111 studies were included. The WPR for metronidazole and vancomycin was 1.0% (95% CI 0–3%) and 1% (95% CI 0–2%) for the breakpoint > 2 mg/L and 0% (95% CI 0%) for breakpoint ≥32 μg/ml. Rifampin and tigecycline had a WPRs of 37.0% (95% CI 18–58%) and 1% (95% CI 0–3%), respectively. The WPRs for the other antimicrobials were as follows: ciprofloxacin 95% (95% CI 85–100%), moxifloxacin 32% (95% CI 25–40%), clindamycin 59% (95% CI 53–65%), amoxicillin/clavulanate 0% (0–0%), piperacillin/tazobactam 0% (0–0%) and ceftriaxone 47% (95% CI 29–65%). Tetracycline had a WPR 20% (95% CI 14–27%) and meropenem showed 0% (95% CI 0–1%); resistance to fidaxomicin was reported in one isolate (0.08%).
Conclusion
Resistance to metronidazole, vancomycin, fidaxomicin, meropenem and piperacillin/tazobactam is reported rarely. From the alternative CDI drug treatments, tigecycline had a lower resistance rate than rifampin. The high-risk antimicrobials for CDI development showed a high level of resistance, the highest was seen in the second generation of fluoroquinolones and clindamycin; amoxicillin/clavulanate showed almost no resistance. Tetracycline resistance was present in one fifth of human clinical C. difficile isolates.
Similar content being viewed by others
Introduction
Clostridium difficile, recently reclassified as Clostridioides difficile [1], is an important pathogen of healthcare-associated diarrhea [2]. Recently, however, an increase in the occurrence of CDI outside hospital settings has been reported [3, 4].
Previous antibiotic use was recognized as one of the risk factors for developing CDI through an alteration of gut microbiota. The accumulation of antimicrobial resistance mechanisms may provide an advantage to C. difficile as it is not affected by antimicrobials present in the gut [5].
An antibiotic stewardship intervention, that limited the use of the fluoroquinolones, clindamycin, amoxicillin/clavulanate, and cephalosporins, was shown to be effective in reducing the occurrence of multidrug-resistant epidemic ribotypes, e.g. 001 and 027 [6].
Currently, three antimicrobial agents, metronidazole, vancomycin and fidaxomicin are recommended for the treatment of CDI [7,8,9] and several new anti-CDI drugs are being tested in clinical trials [9]. The new data suggest tigecycline is effective in treating patients with a severe course of CDI [10], and rifaximin might be beneficial in preventing a CDI relapse [11].
In addition to humans, C. difficile has been cultured from livestock, food and the environment [12]. Tetracycline is one of the most commonly used antimicrobials in agriculture providing antimicrobial selective pressure in this sphere. This is supported by observations of a high prevalence of the tetracycline resistance gene tetM in livestock-associated C. difficile ribotype 078 isolates [13]. Moreover, the zoonotic transmission of C. difficile between farm animals and humans has been demonstrated [14].
Carbapenems are antimicrobials used for the treatment of infections caused by multidrug-resistant gram-negative pathogens. However, some carbapenem resistance mechanisms are transferable to other bacterial species [15]. Hence, the monitoring of carbapenem resistance in C. difficile is justified.
We aimed to review the data on the resistance of antimicrobials to C. difficile that have been recommended for CDI treatment; alternative drugs for CDI treatment; high-risk antimicrobials associated with CDI development; agriculture-related antimicrobials; and antimicrobials reserved for the treatment of multidrug pathogens.
Methods
Search strategy and study selection
Five bibliographic databases, including international databases (MEDLINE [PubMed], Scopus, Embase, Cochrane Library and Web of Science) were searched for relevant articles (Until October 2019) using the following keywords: (“Clostridium difficile” OR “Clostridioides difficile” OR C. difficile) AND (“Antimicrobial-Drug Resistance” OR “drug resistance” OR “antibiotic resistance” OR “aminoglycosides” OR “beta-lactams” OR “cephalosporins” OR “clindamycin” OR “tetracyclines” OR “fluoroquinolones” OR “macrolides” OR “vancomycin” OR “metronidazole” OR “fidaxomicin” OR “carbapenems”) in the Title/Abstract/Keywords fields. No limitation was used while searching the databases, but for the study to be included in our analysis, the available abstract had to be written in English. The recorded hits were merged, and any duplicates were removed using EndNote X7 (Thomson Reuters, New York, NY, USA).
Selection criteria and data extraction
All selected studies were reviewed by three authors independently: Ebrahim Kouhsari, Behnam Ahmadzadeh and Abbas Maleki. Studies were excluded if they met the following conditions: (1) C. difficile antibiotic resistance was not presented; (2) resistance rates were not clearly reported; (3) no human clinical C. difficile strain was tested; (4) it was a meta-analysis and systematic review or a review article or not an original research article; (5) a duplicated report using the same database; (6) a conference abstract and article without the full text upon request from the author; (7) less than 5 isolates were tested. Any discrepancies and inconsistencies with the selection of an article were resolved through discussion, and a fourth author (Nourkhoda Sadeghifard) acted as arbiter.
The information extracted from each included study was: (1) author; (2) publication year; (3) study period; (4) number of C. difficile isolates; (5) antimicrobial susceptibility methods; (6) interpretation of resistance; (7) resistance rates (Supplementary Data 1).
Quality assessment
A quality evaluation of the included studies was performed independently (Behnam Ahmadzadeh, Ebrahim Kouhsari), using an adapted version of the tool proposed by the Newcastle-Ottawa assessment scale adapted for cross-sectional studies [16] (Supplementary Table 1). A score ranging from 0 to 8 points was attributed to each study (≥ 5 points: high quality, 4–3 points: Moderate quality, ≤ 2 points: low quality). A higher score indicated a higher study quality. A third reviewer (Leila Molaeipour) adjudicated in any cases where there was a disagreement.
Definition of resistance
In individual studies, resistance was defined according to either the European Committee on Antimicrobial Susceptibility Testing (EUCAST) [17] or the Clinical & Laboratory Standards Institute (CLSI) [18] minimal inhibitory concentration (MIC) interpretative breakpoints. The individual MICs were as follows: vancomycin ≥32 mg/L; metronidazole ≥32 mg/L; clindamycin ≥8 mg/L; tetracycline ≥16 mg/L; ciprofloxacin ≥8 mg/L; moxifloxacin ≥8 mg/L; meropenem ≥16 mg/L; piperacillin/tazobactam ≥128/4 mg/L, amoxicillin/clavulanate ≥16/8 mg/L and ceftriaxone 64 mg/L according to the (CLSI) [18]. The MIC interpretive breakpoints for vancomycin >2 mg/L, metronidazole >2 mg/L, rifampin >0.004 mg/L, moxifloxacin >4 mg/L and tigecycline >0.25 mg/L were based on the epidemiological cut-off values (ECOFFs) defined by EUCAST [17].
Statistical analysis
Studies presenting raw data on antimicrobial resistance were included in the meta-analysis which was performed by computing the pooled prevalence of resistance for each antimicrobial agent using a random- effects model with Stata/SE software, v.14.1 (StataCorp, College Station, TX). The inconsistency across studies was examined by the forest plot as well as the I2 statistic. Values of I2 (25, 50 and 75%) were interpreted as the presence of low, medium or high heterogeneity, respectively and the random effects models were used [19]. Subgroup analyses were then employed by assuming continents, year, antimicrobial susceptibility testing, and the quality of studies as sources of variation. All statistical interpretations were reported on a 95% confidence interval (CI) basis.
Study outcomes
The main outcome of interest was the weighted pooled resistance rate (WPR) of strains resistant to specific antimicrobial agents according to the CLSI and/or EUCAST guidelines, respectively. A subgroup analysis was performed (1) for geographical regions (Asia, Europe, Africa, Oceania, South and North America); (2) publication date (1992–2014, and 2015–2019, 3) antimicrobial susceptibility testing method (agar dilution, Etest, and microbroth dilution); and (4) the quality of the studies (high quality, moderate quality, low quality). Subgroup analyses were not performed when the number of studies in the category was lower than five.
Results
Search results
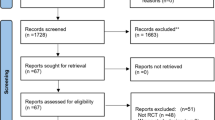
We evaluated six electronic databases and categorized 14,582 articles published up to October 2019 (Fig. 1). From these, after an initial screening of the title and abstract, 11,204 articles were excluded, due to their irrelevance and duplication, but the full text of the remaining 335 articles was reviewed (Fig. 1). From the 335 articles, 224 were excluded again for the following reasons: review, not original research, conference abstract and article without full text (n = 162), no human clinical C. difficile strains (n = 24), no data for susceptibility testing or used disk diffusion method or no resistance data (n = 27), and data using the same isolates or low number of isolates (n = 11). Finally, 111 studies were included in this systematic review and meta-analysis (Supplementary Data 1). The studies included in the meta-analysis assessed antibiotic resistance to metronidazole, clindamycin, tetracycline, moxifloxacin and ciprofloxacin, meropenem, piperacillin/tazobactam, amoxicillin/clavulanate, vancomycin, rifampin and tigecycline.
Characteristics of the included studies
The 111 included studies [20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130] were performed in 35 countries and investigated 19,733 C. difficile isolates. The majority of the studies originated in Asia (n = 42), followed by Europe (n = 37).
Epsilometer (E-test) strips were the most frequent antimicrobial susceptibility testing method used (n = 58), followed by agar dilution (n = 49). All studies had a cross-sectional design, and the mean Newcastle-Ottawa score was 4.5. The quality was high in 62 (55.8%) studies, medium in 46 (41.4%) studies, and low in 3 (2.7%) studies (Supplementary Data 1). Most of the studies (93.69%) included in the meta-analysis had determined the resistance to metronidazole.
The WPR rates for each antimicrobial are shown in Table 1 and Fig. 2. The forest plots that show the analyses for resistance to individual antimicrobials and subgroups are displayed in the Supplementary Figure 1. Data on the resistance of each antimicrobial and the subgroup analyses by year, continent, quality and method of susceptibility testing are shown in the Supplementary Table 2.
Resistance to metronidazole
The susceptibility to metronidazole was determined in 104 studies and included 19,645 C. difficile isolates.
The EUCAST breakpoint (2 mg/L) was applied in 32 studies (5900 C. difficile isolates) and 190 C. difficile isolates were found to be resistant (3.2%); the WPR (to metronidazole) was 1% (95% CI 0–3%) with substantial heterogeneity (I2 = 91.97%).
The subgroup analysis that compared the data from 1992 to 2014 (WPR 0%; 95% CI 0–1%) and 2015–2019 (WPR 2%; 95% CI 0–4%) indicated an increase in the resistance rate. However, this difference was not statistically significant (P = 0.06). Based on the quality of the studies, the resistance rates did not differ between the groups (p = 0.998), T\he highest resistance rate was reported in Asia, followed by North America and Europe (4, 95% CI 0–12%; 3, 95% CI 0–8%; 1, 95% CI 0–2%). No statistical difference was found in the method used for antimicrobial susceptibility testing (AST), (p = 0.523).
The CLSI breakpoint (32 mg/L) was applied to 69 studies and 129 of the 13,207 C. difficile isolates investigated were found to be resistant (1.0%); the WPR was 0% (95% CI 0–0%), with substantial heterogeneity (I2 = 81.4%). No statistical significant difference was found between data from 1992 to 2014 and 2015–2019 (p = 0.280). Subgroup analyses by continent showed a significant difference between the groups (p = 0.038); the highest resistance was in Europe (1%; 95% CI 0–2%). No statistical difference was found in the method used for AST (P = 0.281).
Resistance to vancomycin
A susceptibility to vancomycin was determined in 94 studies where 15,515 C. difficile isolates were tested.
Eighteen studies used the breakpoint of 32 mg/L and 13 C. difficile isolates of the 2307 isolates tested were resistant (0.6%). Another nine studies used the breakpoint of 16 mg/L and ten C. difficile isolates of 2296 tested were resistant (0.4%). Eight studies used the breakpoint of 4 mg/L and seven isolates of the 1107 isolates tested were resistant (0.6%). Overall the WPRs were, based on the breakpoints of 32, 16 and 4 mg/L, 0% (95% CI 0–0%), 0% (95% CI 0–0%), and 0% (95% CI 0–1%) with a heterogeneity of ≤50.1%. The subgroup analyses were not performed.
Based on the EUCAST breakpoint of 2 mg/L, a sensitivity to vancomycin was investigated in 58 studies and of the 11,188 C. difficile isolates tested, 416 isolates were found to be resistant (3.7%). The WPR was 1% (95% CI 0–2%), with substantial heterogeneity (I2 = 93.89%, P = 0.00).
The subgroup analysis, that compared data from 1992 to 2014 (WPR 1%; 95% CI 0–2%) and from 2015 to 2019 (WPR 1%; 95% CI 0–3%), indicated an increase in the resistance rate, however, this difference was not statistically significant (P = 0.48). In the continental subgroup analyses, a significant difference was found (P = 0.000) and the highest resistance rates were reported in South America followed by North America (53, 95% CI 38–68 and 4%, 95% CI 1–9%).
The resistance rates differ significantly when comparing the quality of studies (P = 0.01). In the low quality articles, the WPR was 6% (95% CI 2–11%) higher than in the moderate and high quality articles with a WPR of 2% (95% CI 0–2%). No statistical difference was found in the method used for AST (P = 0.47).
Resistance to moxifloxacin
A susceptibility to moxifloxacin was determined in 78 studies and from those studies 14,383 isolates were investigated.
Using the CLSI breakpoint of 8 mg/L, 11484 C. difficile isolates were investigated and 3912 isolates were found to be resistant (34.1%); the WPR to moxifloxacin was 32% (95% CI, 25–40%) with a substantial heterogeneity (I2 = 93.89%, P = 0.00).
The subgroup analysis, that compared data from 1992 to 2014 and 2015–2019, did not find any significant difference between these groups (p = 0.508). In the continental categorisation, the difference between the groups was significant (P = 0.000); the highest WPR was in Africa, followed by North America and Asia (94, 95% CI 85–98%; 44, 95% CI 33–55 and 33%, 95% CI 25–40%), respectively. In a subgroup analysis on the quality of articles, the results showed a significant difference (P = 0.014); the low, moderate and high quality reports gave WPRs of 54% (95% CI 39–68%), 35% (95% CI 22–49%), and 30% (95% CI 22–38%). No statistical difference was found in the method used for AST (P = 0.543).
Using the EUCAST breakpoint of 4 mg/L, 11484 C. difficile isolates were investigated and 3912 isolates were found to be resistant; 34.1%); the WPR was 49% (95% CI 0.34–0.65) with a substantial heterogeneity (I2 = 98.4%, P = 0.00). The subgroup analyses were not performed.
Resistance to ciprofloxacin
The susceptibility to ciprofloxacin was determined in 28 studies investigating 4339 C. difficile isolates and used a breakpoint of 8 mg/L. From them, 3356 isolates were found to be resistant (77%); the WPR to ciprofloxacin was 95% (95% CI 85–100%) with a substantial heterogeneity (I2 = 99.12%, P = 0.00).
A subgroup analysis, that compared the data from 1992 to 2014 and 2015–2019, showed a significant difference (P = 0.001), 100% (95% CI 100%) versus 79% (95% CI 54–97%). The difference in continental categorisation was also significant (P = 0.000); the highest WPR was in South America (100, 95% CI 40–100%) followed by Asia (96, 95% CI 89–100% and North America (94, 95% CI 40–100%). No statistical difference was found in the method used for AST (P = 0.495).
Resistance to clindamycin
The susceptibility to clindamycin was determined in 64 studies investigating 19,645 C. difficile isolates and, using the CLSI breakpoint (8 mg/l), 6685 C. difficile isolates were reported to be resistant (34.0%).
The overall WPR to clindamycin was 59% (95% CI, 53–65%), with a substantial heterogeneity (I2 = 97.50%, P = 0.00); there was no significant difference in the time categories (P = 0.96). The groups differed in continental categorization (p = 0.000) with the highest rates in Asia and South America (72, 95% CI 65–78 and 59%, 95% CI 19–94%, respectively). Also, in the subgroup analysis on the quality of articles, the results showed a significant difference (P = 0.000); the low, moderate and high quality reports reported resistance rates of 17% (95%CI 9–27%), 57% (95% CI 46–68%) and 63% (95% CI 55–70%), respectively. There was statistical significance between the methods used for AST (p = 0.020).
Resistance to tetracycline
The susceptibility to tetracycline was determined in 31 studies investigating 4861 C. difficile isolates and from those 886 isolates (18.2%) were found to be resistant using the breakpoint of 16 mg/L. The WPR was 20% (95% CI, 14–27%), with substantial heterogeneity (I2 = 97.04%, P = 0.00).
There was no difference between the data from 1992 to 2014 and 2015–2019 (p = 0.26). A statistically significant difference was found in the continental categorization (P = 0.000); the highest resistances were 34% (95% CI, 26–43%), 26% (95% CI, 17–35%), and 16% (95% CI, 5–31%) in Oceania, Asia, and Europe, respectively. In a subgroup analysis on the quality of articles, the results showed a significant difference (P = 0.01); the low, moderate and high quality reports gave resistance rates of 40% (95%CI 29–52%), 16% (95% CI 7–28%) and 22% (95% CI 13–32%), respectively. No statistical difference was found in the method used for AST (P = 0.216).
Meropenem
The susceptibility to meropenem was determined in 17 studies using the breakpoint (≥16 mg/L mg/L) and 2756 C. difficile isolates were investigated; 20 isolates found to be resistant (0.7%). The overall WPR was 0% (95% CI, 0%-%1) with moderate heterogeneity (I2 = 71.49%, P = 0.00). No statistical difference was found between the data from 1992 to 2014 and 2015–2019 (p = 0.106). The continental, quality and methods subgroup differences were not analysed.
Amoxicillin/Clavulanate
The susceptibility to co-amoxicillin was investigated in 10 studies using the breakpoint of ≥16/8 mg/L. A total of 2803 C. difficile isolates were investigated and 4 isolates were reported as resistant (0.1%); the WPR was 0% (95% CI, 0–0%), with low heterogeneity (I2 = 45.4%, P = 0.06). No subgroup analyses were performed.
Piperacillin/Tazobactam
The susceptibility to piperacillin/tazobactam was investigated in 17 studies applying the breakpoint of ≥128/4 mg/L mg/L and included 3041 C. difficile isolates. Eight isolates were found to be resistant (0.3%); the WPR to this antibiotic was 0% with (95% CI, 0–0%). No subgroup analyses were performed.
Ceftriaxone
The susceptibility to ceftriaxone was investigated in 13 studies. Of the 3476 C. difficile isolates investigated, 1289 isolates were found to be resistant (37.1%) using the breakpoint of ≥64 mg/L. The WPR for ceftriaxone was 47% (95% CI, 29–65%), with substantial heterogeneity (I2 = 99.05%, P = 0.00). No subgroup analyses were performed.
Rifampin
The susceptibility to rifampin was investigated in 10 studies on 1861 of C. difficile isolates. Using the breakpoint of 0.004 mg/L, 787 isolates were reported to be resistant (42.3%), the WPR was 37% (95% CI, 18–58%) with substantial heterogeneity (I2 = 97.69%, P = 0.00). No subgroup analyses were performed.
Tigecycline
The susceptibility to tigecycline was investigated in 10 studies in 2375 C. difficile isolates. Thirty-nine isolates were reported to be resistant (1.6%) based on the breakpoint of 0.25 mg/L; the WPR was 1% (95% CI 0–3%) with substantial heterogeneity (I2 = 83.53%, P = 0.00). No subgroup analyses were performed.
Fidaxomicin
The susceptibility to fidaxomicin was investigated in 1184 isolates from six studies. One isolate found to be resistant (0.08%) based on the breakpoint of ≥8 mg/L. The analyses were not performed because of the absence of a recommended breakpoint and the low number of studies.
Discussion
Due to the limited number of antimicrobials that can be used for the treatment [7,8,9] of CDI, it is important to obtain information about the resistance profiles of circulating C. difficile strains. Moreover, the accumulation of resistance mechanisms gives C. difficile an advantage since CDI can develop after the use of antimicrobials due to an alteration in gut microbiota [5].
Several methods can be used to determine the MIC in antimicrobial susceptibility testing. In our study, the Etest was the most used method followed by agar dilution. The agar dilution method is suitable for AST when there is high number of isolates since there is a need to prepare fresh testing plates for each experiment; however the commercially available Etest can be used independently for individual isolates.
Three antimicrobials are recommended for the treatment of CDI; metronidazole, vancomycin and fidaxomicin. For AST, there is still no MIC breakpoint available for fidaxomicin; for vancomycin and metronidazole two values exist but with a wide range: EUCAST 2 mg/L and CLSI 32 mg/L, The difference between the resistance rates, according to the breakpoint used, was also noted in our study. For metronidazole, the WPR was 1% (95% CI, 0–3%) using EUCAST but using CLSI, the WPR was 0% (95% CI, 0–0%). A similar pattern was also observed for vancomycin where using the EUCAST breakpoint, the WPR was higher (1% (95% CI 0–2%) than for the CLSI breakpoint 0% (95% CI, 0–0%).
Recently, a systematic review and meta-analysis [131] of metronidazole and vancomycin resistance in C. difficile showed higher WPRs than observed in our study; 1.9% (95% CI, 0.5–3.6%) for metronidazole and 2.1% (95% CI, 0–5.1%) for vancomycin. The analyses differed in the date of publication for data collection, (1982–2017) vs (1992–2019), and in the origin of the isolates since, in our analyses, the data on the C. difficile isolates of animal origin were not included.
The data on the susceptibility testing for metronidazole, vancomycin and moxifloxacin were included in the “enhanced level” of a CDI surveillance protocol published by the European Centre for Disease Prevention and Control (ECDC) [2]. Moxifloxacin, a fluoroquinolone, is not considered as a drug for CDI treatment but moxifloxacin resistance in C. difficile strains was shown to be an important marker for the spread of C. difficile in a healthcare setting [132]. Two representatives of fluoroquinolones were analysed in our study: ciprofloxacin and moxifloxacin. From all the antimicrobials in our study, ciprofloxacin showed the highest level of resistance (WPR 95%) and the resistance to moxifloxacin was 32 and 49% according to the CLSI and EUCAST breakpoints, respectively.
In addition to fluoroquinolones, clindamycin, amoxicillin/clavulanate and cephalosporins are indicated for limited use in hospital settings in order to reduce CDI rates [6]. From these four classes of antimicrobials, three classes exhibited high rates of resistance; however with amoxicillin/clavulanate, only 4 isolates out of 2803 isolates were investigated.
Rifaximin has been suggested as an alternative to existing CDI therapies, especially in CDI recurrences and their prevention [133, 134]. Data on rifampin resistance, which correlate with rifaximin [135], showed a high level of resistance in investigated C. difficile isolates (787/1861) and suggest more risk to treatment failure due to C. difficile strain resistance compared to recommended CDI treatments.
The effectiveness of tigecycline use in the treatment of CDI was evaluated in several studies [10]. According to the reported resistance rates in our study, treatment failure is less likely with tigecycline than with rifaximin. However, recently, the emergence of mobile tigecycline-resistance genes, tet(X3) and tet(X4) that inactivate all tetracyclines, including tigecycline, was reported recently in gram-negative bacteria [136]. Moreover, Tet proteins have, in vitro, the potential to acquire mutations leading to an increased MICs for tigecycline [137]. From the available data, the tet classes of ribosomal protection genes are the most common molecular mechanism for tetracycline resistance in C. difficile [13]. The spread of newly detected tet(X) genes or mutations in present tet classes genes (e.g tetM or tetW) could increase the prevalence of resistance to tigecycline.
Conclusion
A resistance to metronidazole, vancomycin, fidaxomicin, meropenem and piperacillin-tazobactam is reported rarely. From alternative CDI treatment drugs, tigecycline had a lower resistance rate than rifampicin. The high-risk antimicrobials for CDI development showed a high level of resistance, the highest was seen in the second generation of fluoroquinolones and clindamycin; amoxicillin/clavulanate showed almost no resistance. Tetracycline resistance was present in one fifth of human clinical C. difficile isolates.
Availability of data and materials
All data were included.
Abbreviations
- WPR:
-
Weighted Pooled Resistance
- CDI:
-
Clostridium difficile infection
- EUCAST:
-
European Committee on Antimicrobial Susceptibility Testing
- CLSI:
-
Clinical & Laboratory Standards Institute
- MIC:
-
Minimal inhibitory concentration
- ECOFFs:
-
Epidemiological cut-off values
- AST:
-
Antimicrobial susceptibility testing
- ECDC:
-
European Centre for Disease Prevention and Control
References
Lawson PA, Citron DM, Tyrrell KL, Finegold SM. Reclassification of clostridium difficile as clostridioides difficile (Hall and O’Toole 1935) Prévot 1938. Anaerobe. 2016;40:95–9.
Krutova M, Kinross P, Barbut F, Hajdu A, Wilcox M, Kuijper E, et al. How to: surveillance of Clostridium difficile infections. Clin Microbiol Infect. 2018;24(5):469–75.
Fawley WN, Davies KA, Morris T, Parnell P, Howe R, Wilcox MH. Enhanced surveillance of Clostridium difficile infection occurring outside hospital, England, 2011 to 2013. Eurosurveillance. 2016;21(29):30295.
Barbut F, Day N, Bouée S, Youssouf A, Grandvoinnet L, Lalande V, et al. Toxigenic Clostridium difficile carriage in general practice: results of a laboratory-based cohort study. Clin Microbiol Infect. 2019;25(5):588–94.
Rupnik M, Wilcox MH, Gerding DN. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol. 2009;7(7):526–36.
Lawes T, Lopez-Lozano J-M, Nebot CA, Macartney G, Subbarao-Sharma R, Wares KD, et al. Effect of a national 4C antibiotic stewardship intervention on the clinical and molecular epidemiology of Clostridium difficile infections in a region of Scotland: a non-linear time-series analysis. Lancet Infect Dis. 2017;17(2):194–206.
Debast SB, Bauer MP, Kuijper EJ. Committee. European Society of Clinical Microbiology and Infectious Diseases: update of the treatment guidance document for Clostridium difficile infection. Clin Microbiol Infect. 2014;20:1–26.
McDonald LC, Gerding DN, Johnson S, Bakken JS, Carroll KC, Coffin SE, et al. Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin Infect Dis. 2018;66(7):e1–e48.
Ooijevaar R, Van Beurden Y, Terveer E, Goorhuis A, Bauer M, Keller J, et al. Update of treatment algorithms for Clostridium difficile infection. Clin Microbiol Infect. 2018;24(5):452–62.
Kechagias KS, Chorepsima S, Triarides NA, Falagas ME. Tigecycline for the treatment of patients with Clostridium difficile infection: an update of the clinical evidence. Eur J Clin Microbiol Infect Dis. 2020;39:1053–8.
Garey KW, Ghantoji SS, Shah DN, Habib M, Arora V, Jiang Z-D, et al. A randomized, double-blind, placebo-controlled pilot study to assess the ability of rifaximin to prevent recurrent diarrhoea in patients with Clostridium difficile infection. J Antimicrob Chemother. 2011;66(12):2850–5.
Stabler RA, Dawson LF, Valiente E, Cairns MD, Martin MJ, Donahue EH, et al. Macro and micro diversity of Clostridium difficile isolates from diverse sources and geographical locations. PLoS One. 2012;7(3):e31559.
Dingle KE, Didelot X, Quan TP, Eyre DW, Stoesser N, Marwick CA, et al. A role for tetracycline selection in recent evolution of agriculture-associated Clostridium difficile PCR Ribotype 078. MBio. 2019;10(2):e02790–18.
Lim S-C, Riley TV, Knight DR. One health: the global challenge of Clostridium difficile infection. Microbiology Australia. 2020;41(1):23–7.
Elshamy AA, Aboshanab KM. A review on bacterial resistance to carbapenems: epidemiology, detection and treatment options. Future Science OA. 2020;6(3):FSO438.
Modesti PA, Reboldi G, Cappuccio FP, Agyemang C, Remuzzi G, Rapi S, et al. Panethnic differences in blood pressure in Europe: a systematic review and meta-analysis. PLoS One. 2016;11(1):e0147601.
Eucast T. European committee on antimicrobial susceptibility testing, breakpoint tables for interpretation of MICs and zone diameters. In: European Society of Clinical Microbiology and Infectious Diseases Basel; 2018.
Wayne P. Clinical and Laboratory Standards Institute (CLSI) performance standards for antimicrobial disk diffusion susceptibility tests 28th ed. approved standard. CLSI doc M100-S28. 2009;29(2011):M100–S128.
DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–88.
Ackermann G, Degner A, Cohen SH, Silva J Jr, Rodloff AC. Prevalence and association of macrolide–lincosamide–streptogramin B (MLSB) resistance with resistance to moxifloxacin in Clostridium difficile. J Antimicrob Chemother. 2003;51(3):599–603.
Ackermann G, Löffler B, Adler D, Rodloff AC. In vitro activity of OPT-80 against Clostridium difficile. Antimicrob Agents Chemother. 2004;48(6):2280–2.
Álvarez-Pérez S, Blanco JL, Harmanus C, Kuijper E, García ME. Subtyping and antimicrobial susceptibility of Clostridium difficile PCR ribotype 078/126 isolates of human and animal origin. Vet Microbiol. 2017;199:15–22.
Androga GO, Knight DR, Lim S-C, Foster NF, Riley TV. Antimicrobial resistance in large clostridial toxin-negative, binary toxin-positive Clostridium difficile ribotypes. Anaerobe. 2018;54:55–60.
Aoki K, Takeda S, Miki T, Ishii Y, Tateda K. Antimicrobial susceptibility and molecular characterization using whole-genome sequencing of Clostridioides difficile collected in 82 hospitals in Japan between 2014 and 2016. Antimicrob Agents Chemother. 2019;63(12):e01259–19.
Aptekorz M, Szczegielniak A, Wiechuła B, Harmanus C, Kuijper E, Martirosian G. Occurrence of Clostridium difficile ribotype 027 in hospitals of Silesia, Poland. Anaerobe. 2017;45:106–13.
Arca-Suárez J, Galán-Sánchez F, Cano-Cano F, García-Santos G, Rodríguez-Iglesias M. Antimicrobial susceptibility and molecular typing of toxigenic clinical isolates of Clostridium difficile causing infections in the south of Spain. Anaerobe. 2018;54:146–50.
Baghani A, Ghourchian S, Aliramezani A, Yaseri M, Mesdaghinia A, Douraghi M. Highly antibiotic-resistant Clostridium difficile isolates from Iranian patients. J Appl Microbiol. 2018;125(5):1518–25.
Balassiano IT, dos Santos-Filho J, Vital-Brazil JM, Nouér SA, Souza CR, Brazier JS, et al. Detection of cross-infection associated to a Brazilian PCR-ribotype of Clostridium difficile in a university hospital in Rio de Janeiro, Brazil. Antonie Van Leeuwenhoek. 2011;99(2):249–55.
Beran V, Chmelar D, Vobejdova J, Konigova A, Nemec J, Tvrdik J. Sensitivity to antibiotics of Clostridium difficile toxigenic nosocomial strains. Folia Microbiol. 2014;59(3):209–15.
Beran V, Kuijper E, Harmanus C, Sanders I, van Dorp S, Knetsch C, et al. Molecular typing and antimicrobial susceptibility testing to six antimicrobials of Clostridium difficile isolates from three Czech hospitals in eastern Bohemia in 2011–2012. Folia Microbiol. 2017;62(5):445–51.
Berger FK, Rasheed SS, Araj GF, Mahfouz R, Rimmani HH, Karaoui WR, et al. Molecular characterization, toxin detection and resistance testing of human clinical Clostridium difficile isolates from Lebanon. Int J Med Microbiol. 2018;308(3):358–63.
Bourgault A-M, Lamothe F, Loo VG, Poirier L. Group C-Cs. In vitro susceptibility of Clostridium difficile clinical isolates from a multi-institutional outbreak in southern Quebec, Canada. Antimicrob Agents Chemother. 2006;50(10):3473–5.
Büchler AC, Rampini SK, Stelling S, Ledergerber B, Peter S, Schweiger A, et al. Antibiotic susceptibility of Clostridium difficile is similar worldwide over two decades despite widespread use of broad-spectrum antibiotics: an analysis done at the University Hospital of Zurich. BMC Infect Dis. 2014;14(1):607.
Byun J-H, Kim H, Kim JL, Kim D, Jeong SH, Shin JH, et al. A nationwide study of molecular epidemiology and antimicrobial susceptibility of Clostridioides difficile in South Korea. Anaerobe. 2019;60:102106.
Chatedaki C, Voulgaridi I, Kachrimanidou M, Hrabak J, Papagiannitsis C, Petinaki E. Antimicrobial susceptibility and mechanisms of resistance of Greek Clostridium difficile clinical isolates. J Global Antimicrob Resist. 2019;16:53–8.
Chen Y-B, Gu S-L, Shen P, Lv T, Fang Y-H, Tang L-L, et al. Molecular epidemiology and antimicrobial susceptibility of Clostridium difficile isolated from hospitals during a 4-year period in China. J Med Microbiol. 2018;67(1):52–9.
Cheng J-W, Yang Q-W, Xiao M, Yu S-Y, Zhou M-L, Kudinha T, et al. High in vitro activity of fidaxomicin against Clostridium difficile isolates from a university teaching hospital in China. J Microbiol Immunol Infect. 2018;51(3):411–6.
Chia J-H, Lai H-C, Su L-H, Kuo A-J, Wu T-L. Molecular epidemiology of Clostridium difficile at a medical center in Taiwan: persistence of genetically clustering of A− B+ isolates and increase of A+ B+ isolates. PLoS One. 2013;8(10):e75471.
Chow VC, Kwong TN, So EW, Ho YI, Wong SH, Lai RW, et al. Surveillance of antibiotic resistance among common Clostridium difficile ribotypes in Hong Kong. Sci Rep. 2017;7(1):1–6.
Costa CL, de Carvalho CBM, González RH, Gifoni MAC, de Albuquerque RR, Quesada-Gómez C, et al. Molecular epidemiology of Clostridium difficile infection in a Brazilian cancer hospital. Anaerobe. 2017;48:232–6.
Dong D, Zhang L, Chen X, Jiang C, Yu B, Wang X, et al. Antimicrobial susceptibility and resistance mechanisms of clinical Clostridium difficile from a Chinese tertiary hospital. Int J Antimicrob Agents. 2013;41(1):80–4.
Dong D, Peng Y, Zhang L, Jiang C, Wang X, Mao E. Clinical and microbiological characterization ofClostridium difficileinfection in a tertiary care hospital in Shanghai, China. Chin Med J. 2014;127(9):1601–7.
Ebrahim-Saraie HS, Heidari H, Amanati A, Bazargani A, Taghavi SA, Nikokar I, et al. A multicenter-based study on epidemiology, antibiotic susceptibility and risk factors of toxigenic Clostridium difficile in hospitalized patients in southwestern Iran. Infez Med. 2018;26(4):308–15.
Eckert C, Coignard B, Hebert M, Tarnaud C, Tessier C, Lemire A, et al. Clinical and microbiological features of Clostridium difficile infections in France: the ICD-RAISIN 2009 national survey. Med Mal Infect. 2013;43(2):67–74.
Eitel Z, Terhes G, Sóki J, Nagy E, Urbán E. Investigation of the MICs of fidaxomicin and other antibiotics against Hungarian Clostridium difficile isolates. Anaerobe. 2015;31:47–9.
Fenner L, Frei R, Gregory M, Dangel M, Stranden A, Widmer A. Epidemiology of Clostridium difficile-associated disease at university hospital Basel including molecular characterisation of the isolates 2006–2007. Eur J Clin Microbiol Infect Dis. 2008;27(12):1201.
Fraga EG, Nicodemo AC, Sampaio JLM. Antimicrobial susceptibility of Brazilian Clostridium difficile strains determined by agar dilution and disk diffusion. Braz J Infect Dis. 2016;20(5):476–81.
Freeman J, Vernon J, Morris K, Nicholson S, Todhunter S, Longshaw C, et al. Pan-European longitudinal surveillance of antibiotic resistance among prevalent Clostridium difficile ribotypes. Clin Microbiol Infect. 2015;21(3):248. e249–16.
Gao Q, Wu S, Huang H, Ni Y, Chen Y, Hu Y, et al. Toxin profiles, PCR ribotypes and resistance patterns of Clostridium difficile: a multicentre study in China, 2012–2013. Int J Antimicrob Agents. 2016;48(6):736–9.
Giufrè M, Accogli M, Ricchizzi E, Barbanti F, Farina C, Fazii P, et al. Multidrug-resistant infections in long-term care facilities: extended-spectrum β-lactamase–producing Enterobacteriaceae and hypervirulent antibiotic resistant Clostridium difficile. Diagn Microbiol Infect Dis. 2018;91(3):275–81.
Goudarzi M, Goudarzi H, Alebouyeh M, Rad MA, Mehr FSS, Zali MR, et al. Antimicrobial susceptibility of Clostridium difficile clinical isolates in Iran. Iran Red Crescent Med J. 2013;15(8):704.
Harvala H, Alm E, Åkerlund T, Rizzardi K. Emergence and spread of moxifloxacin-resistant Clostridium difficile ribotype 231 in Sweden between 2006 and 2015. New Microbes New Infect. 2016;14:58–66.
Hastey CJ, Dale SE, Nary J, Citron D, Law JH, Roe-Carpenter DE, et al. Comparison of Clostridium difficile minimum inhibitory concentrations obtained using agar dilution vs broth microdilution methods. Anaerobe. 2017;44:73–7.
Hecht DW, Galang MA, Sambol SP, Osmolski JR, Johnson S, Gerding DN. In vitro activities of 15 antimicrobial agents against 110 toxigenic Clostridium difficile clinical isolates collected from 1983 to 2004. Antimicrob Agents Chemother. 2007;51(8):2716–9.
Hidalgo-Villeda F, Tzoc E, Torres L, Bu E, Rodríguez C, Quesada-Gómez C. Diversity of multidrug-resistant epidemic Clostridium difficile NAP1/RT027/ST01 strains in tertiary hospitals from Honduras. Anaerobe. 2018;52:75–8.
Huang H, Wu S, Wang M, Zhang Y, Fang H, Palmgren A-C, et al. Clostridium difficile infections in a Shanghai hospital: antimicrobial resistance, toxin profiles and ribotypes. Int J Antimicrob Agents. 2009;33(4):339–42.
Huang H, Weintraub A, Fang H, Wu S, Zhang Y, Nord CE. Antimicrobial susceptibility and heteroresistance in Chinese Clostridium difficile strains. Anaerobe. 2010;16(6):633–5.
Hung Y-P, Tsai P-J, Lee Y-T, Tang H-J, Lin H-J, Liu H-C, et al. Nationwide surveillance of ribotypes and antimicrobial susceptibilities of toxigenic Clostridium difficile isolates with an emphasis on reduced doxycycline and tigecycline susceptibilities among ribotype 078 lineage isolates in Taiwan. Infect Drug Resist. 2018;11:1197.
Jamal WY, Mokaddas EM, Verghese TL, Rotimi V. In vitro activity of 15 antimicrobial agents against clinical isolates of Clostridium difficile in Kuwait. Int J Antimicrob Agents. 2002;20(4):270–4.
Jamal WY, Rotimi VO. Surveillance of antibiotic resistance among hospital-and community-acquired toxigenic Clostridium difficile isolates over 5-year period in Kuwait. PLoS One. 2016;11(8):e0161411.
Jiang Z, DuPont H, La Rocco M, Garey K. In vitro susceptibility of Clostridium difficile to rifaximin and rifampin in 359 consecutive isolates at a university hospital in Houston, Texas. J Clin Pathol. 2010;63(4):355–8.
Jiménez A, Araya R, Paniagua D, Camacho-Mora Z, Du T, Golding G, et al. Molecular epidemiology and antimicrobial resistance of Clostridium difficile in a national geriatric hospital in Costa Rica. J Hosp Infect. 2018;99(4):475–80.
Jin D, Luo Y, Huang C, Cai J, Ye J, Zheng Y, et al. Molecular epidemiology of Clostridium difficile infection in hospitalized patients in eastern China. J Clin Microbiol. 2017;55(3):801–10.
John R, Brazier J. Antimicrobial susceptibility of polymerase chain reaction ribotypes of Clostridium difficile commonly isolated from symptomatic hospital patients in the UK. J Hosp Infect. 2005;61(1):11–4.
Karlowsky JA, Zhanel GG, Hammond GW, Rubinstein E, Wylie J, Du T, et al. Multidrug-resistant north American pulsotype 2 Clostridium difficile was the predominant toxigenic hospital-acquired strain in the province of Manitoba, Canada, in 2006–2007. J Med Microbiol. 2012;61(5):693–700.
Karlowsky JA, Adam HJ, Kosowan T, Baxter MR, Nichol KA, Laing NM, et al. PCR ribotyping and antimicrobial susceptibility testing of isolates of Clostridium difficile cultured from toxin-positive diarrheal stools of patients receiving medical care in Canadian hospitals: the Canadian Clostridium difficile surveillance study (CAN-DIFF) 2013–2015. Diagn Microbiol Infect Dis. 2018;91(2):105–11.
Kim J, Kang JO, Pai H, Choi TY. Association between PCR ribotypes and antimicrobial susceptibility among Clostridium difficile isolates from healthcare-associated infections in South Korea. Int J Antimicrob Agents. 2012;40(1):24–9.
Knight DR, Giglio S, Huntington PG, Korman TM, Kotsanas D, Moore CV, et al. Surveillance for antimicrobial resistance in Australian isolates of Clostridium difficile, 2013–14. J Antimicrob Chemother. 2015;70(11):2992–9.
Knight DR, Riley TV. Clostridium difficile clade 5 in Australia: antimicrobial susceptibility profiling of PCR ribotypes of human and animal origin. J Antimicrob Chemother. 2016;71(8):2213–7.
Kociolek LK, Gerding DN, Osmolski JR, Patel SJ, Snydman DR, McDermott LA, et al. Differences in the molecular epidemiology and antibiotic susceptibility of Clostridium difficile isolates in pediatric and adult patients. Antimicrob Agents Chemother. 2016;60(8):4896–900.
Kouhsari E, Douraghi M, Krutova M, Yaseri HF, Talebi M, Baseri Z, et al. The emergence of metronidazole and vancomycin reduced susceptibility in Clostridium difficile isolates in Iran. J Global Antimicrob Resist. 2019;18:28–33.
Krutova M, Matejkova J, Tkadlec J, Nyc O. Antibiotic profiling of Clostridium difficile ribotype 176–a multidrug resistant relative to C. difficile ribotype 027. Anaerobe. 2015;36:88–90.
Kullin BR, Reid S, Abratt V. Clostridium difficile in patients attending tuberculosis hospitals in Cape Town, South Africa, 2014-2015. Afr J Lab Med. 2018;7(2):1–9.
Kuwata Y, Tanimoto S, Sawabe E, Shima M, Takahashi Y, Ushizawa H, et al. Molecular epidemiology and antimicrobial susceptibility of Clostridium difficile isolated from a university teaching hospital in Japan. Eur J Clin Microbiol Infect Dis. 2015;34(4):763–72.
Lachowicz D, Pituch H, Obuch-Woszczatyński P. Antimicrobial susceptibility patterns of Clostridium difficile strains belonging to different polymerase chain reaction ribotypes isolated in Poland in 2012. Anaerobe. 2015;31:37–41.
Li H, Li W-G, Zhang W-Z, Yu S-B, Liu Z-J, Zhang X, et al. Antibiotic resistance of clinical isolates of Clostridioides difficile in China and its association with geographical regions and patient age. Anaerobe. 2019;60:102094.
Liao C-H, Ko W-C, Lu J-J, Hsueh P-R. Characterizations of clinical isolates of Clostridium difficile by toxin genotypes and by susceptibility to 12 antimicrobial agents, including fidaxomicin (OPT-80) and rifaximin: a multicenter study in Taiwan. Antimicrob Agents Chemother. 2012;56(7):3943–9.
Lidan C, Linhai L, Yang L, Zhaohui S, Xiaoyan H, Yuling S. Molecular characterization and antimicrobial susceptibility of tcdA-negative Clostridium difficile isolates from Guangzhou, China. Diagn Microbiol Infect Dis. 2016;84(4):361–5.
López-Ureña D, Quesada-Gómez C, Miranda E, Fonseca M, Rodríguez-Cavallini E. Spread of epidemic Clostridium difficile NAP1/027 in Latin America: case reports in Panama. J Med Microbiol. 2014;63(2):322–4.
López-Ureña D, Quesada-Gómez C, Montoya-Ramírez M, del Mar G-CM, Somogyi T, Rodríguez C, et al. Predominance and high antibiotic resistance of the emerging Clostridium difficile genotypes NAPCR1 and NAP9 in a Costa Rican hospital over a 2-year period without outbreaks. Emerg Microbes Infect. 2016;5(1):1–5.
Luo Y, Zhang W, Cheng J-W, Xiao M, Sun G-R, Guo C-J, et al. Molecular epidemiology of Clostridium difficile in two tertiary care hospitals in Shandong Province, China. Infect Drug Resist. 2018;11:489.
Mutlu E, Wroe AJ, Sanchez-Hurtado K, Brazier JS, Poxton IR. Molecular characterization and antimicrobial susceptibility patterns of Clostridium difficile strains isolated from hospitals in south-East Scotland. J Med Microbiol. 2007;56(7):921–9.
Nasereddin LM, Bakri FG, Shehabi AA. Clostridium difficile infections among Jordanian adult hospitalized patients. Am J Infect Control. 2009;37(10):864–6.
Ngamskulrungroj P, Sanmee S, Pusathit P, Piewngam P, Elliott B, Riley TV, et al. Molecular epidemiology of Clostridium difficile infection in a large teaching hospital in Thailand. PLoS One. 2015;10(5):e0127026.
Niyogi SK. Antimicrobial susceptibility of Clostridium difficile strains isolated from hospitalised patients with acute diarrhoea. J Diarrhoeal Dis Res. 1992;10:156–8.
Novak A, Spigaglia P, Barbanti F, Goic-Barisic I, Tonkic M. First clinical and microbiological characterization of Clostridium difficile infection in a Croatian University hospital. Anaerobe. 2014;30:18–23.
Obuch-Woszczatyński P, Dubiel G, Harmanus C, Kuijper E, Duda U, Wultańska D, et al. Emergence of Clostridium difficile infection in tuberculosis patients due to a highly rifampicin-resistant PCR ribotype 046 clone in Poland. Eur J Clin Microbiol Infect Dis. 2013;32(8):1027–30.
Oka K, Osaki T, Hanawa T, Kurata S, Okazaki M, Manzoku T, et al. Molecular and microbiological characterization of Clostridium difficile isolates from single, relapse, and reinfection cases. J Clin Microbiol. 2012;50(3):915–21.
Peng Z, Addisu A, Alrabaa S, Sun X. Antibiotic resistance and toxin production of Clostridium difficile isolates from the hospitalized patients in a large hospital in Florida. Front Microbiol. 2017;8:2584.
Peretz A, Tkhawkho L, Pastukh N, Brodsky D, Halevi CN, Nitzan O. Correlation between fecal calprotectin levels, disease severity and the hypervirulent ribotype 027 strain in patients with Clostridium difficile infection. BMC Infect Dis. 2016;16(1):309.
Piepenbrock E, Stelzer Y, Berger F, Jazmati N. Changes in Clostridium (Clostridioides) difficile PCR-Ribotype distribution and antimicrobial resistance in a German tertiary care hospital over the last 10 years. Curr Microbiol. 2019;76(4):520–6.
Pinto LJ, Alcides AP, Ferreira EO, Avelar KE, Sabrá A, Domingues RM, et al. Incidence and importance of Clostridium difficile in paediatric diarrhoea in Brazil. J Med Microbiol. 2003;52(12):1095–9.
Pirš T, Avberšek J, Zdovc I, Krt B, Andlovic A, Lejko-Zupanc T, et al. Antimicrobial susceptibility of animal and human isolates of Clostridium difficile by broth microdilution. J Med Microbiol. 2013;62(9):1478–85.
Putsathit P, Maneerattanaporn M, Piewngam P, Knight DR, Kiratisin P, Riley TV. Antimicrobial susceptibility of Clostridium difficile isolated in Thailand. Antimicrob Resist Infect Control. 2017;6(1):58.
Ramírez-Vargas G, Quesada-Gómez C, Acuña-Amador L, López-Ureña D, Murillo T, del Mar G-CM, et al. A Clostridium difficile lineage endemic to Costa Rican hospitals is multidrug resistant by acquisition of chromosomal mutations and novel mobile genetic elements. Antimicrob Agents Chemother. 2017;61(4):e02054–16.
Reil M, Hensgens M, Kuijper E, Jakobiak T, Gruber H, Kist M, et al. Seasonality of Clostridium difficile infections in southern Germany. Epidemiol Infect. 2012;140(10):1787–93.
Rodriguez C, Avesani V, Taminiau B, Van Broeck J, Brévers B, Delmée M, et al. Investigation of Clostridium difficile interspecies relatedness using multilocus sequence typing, multilocus variable-number tandem-repeat analysis and antimicrobial susceptibility testing. Vet J. 2015;206(3):349–55.
Russello G, Russo A, Sisto F, Scaltrito MM, Farina C. Laboratory diagnosis of Clostridium difficile associated diarrhoea and molecular characterization of clinical isolates. New Microbiol. 2012;35(3):307–16.
Saatian B, Banerjee C, Carroll KC, Ross TL, Kamangar F. Characterization of clostridium difficile infection and analysis of recovered isolates in a community hospital population in Baltimore, Maryland. Infect Dis Clin Pract. 2010;18(6):383–8.
Samonis G, Vardakas K, Tansarli G, Dimopoulou D, Papadimitriou G, Kofteridis D, et al. Clostridium difficile in Crete, Greece: epidemiology, microbiology and clinical disease. Epidemiol Infect. 2016;144(1):161–70.
Sandell S, Rashid M-U, Jorup-Rönström C, Ellström K, Nord CE, Weintraub A. Clostridium difficile recurrences in Stockholm. Anaerobe. 2016;38:97–102.
Santos A, Isidro J, Silva C, Boaventura L, Diogo J, Faustino A, et al. Molecular and epidemiologic study of Clostridium difficile reveals unusual heterogeneity in clinical strains circulating in different regions in Portugal. Clin Microbiol Infect. 2016;22(8):695–700.
Pelaez T, Alcala L, Alonso R, Rodriguez-Creixems M, Garcia-Lechuz J, Bouza E. Reassessment of Clostridium difficile susceptibility to metronidazole and vancomycin. Antimicrob Agents Chemother. 2002;46(6):1647–50.
Roberts S, Heffernan H, Al Anbuky N, Pope C, Paviour S, Camp T, et al. Molecular epidemiology and susceptibility profiles of Clostridium difficile in New Zealand, 2009. NZ Med J. 2011;124(1332):45–51.
Secco DA, Balassiano IT, Boente RF, Miranda KR, Brazier J, Hall V, et al. Clostridium difficile infection among immunocompromised patients in Rio de Janeiro, Brazil and detection of moxifloxacin resistance in a ribotype 014 strain. Anaerobe. 2014;28:85–9.
Seo M-R, Kim J, Lee Y, Lim D-G, Pai H. Prevalence, genetic relatedness and antibiotic resistance of hospital-acquired Clostridium difficile PCR ribotype 018 strains. Int J Antimicrob Agents. 2018;51(5):762–7.
Seugendo M, Mshana S, Hokororo A, Okamo B, Mirambo M, von Müller L, et al. Clostridium difficile infections among adults and children in Mwanza/Tanzania: is it an underappreciated pathogen among immunocompromised patients in sub-Saharan Africa? New Microbes New Infect. 2015;8:99–102.
Shayganmehr F-S, Alebouyeh M, Azimirad M, Aslani MM, Zali MR. Association of tcdA+/tcdB+ Clostridium difficile genotype with emergence of multidrug-resistant strains conferring metronidazole resistant phenotype. Iran Biomed J. 2015;19(3):143.
Shoaei P, Shojaei H, Khorvash F, Hosseini SM, Ataei B, Tavakoli H, et al. Molecular epidemiology of Clostridium difficile infection in Iranian hospitals. Antimicrob Resist Infect Control. 2019;8(1):12.
Snydman D, McDermott L, Jacobus N, Thorpe C, Stone S, Jenkins S, et al. US-based national sentinel surveillance study for the epidemiology of Clostridium difficile-associated diarrheal isolates and their susceptibility to fidaxomicin. Antimicrob Agents Chemother. 2015;59(10):6437–43.
Snydman DR, McDermott LA, Thorpe CM, Chang J, Wick J, Walk ST, et al. Antimicrobial susceptibility and ribotypes of Clostridium difficile isolates from a phase 2 clinical trial of ridinilazole (SMT19969) and vancomycin. J Antimicrob Chemother. 2018;73(8):2078–84.
Spigaglia P, Barbanti F, Mastrantonio P, Brazier JS, Barbut F, Delmée M, et al. Fluoroquinolone resistance in Clostridium difficile isolates from a prospective study of C. difficile infections in Europe. J Med Microbiol. 2008;57(6):784–9.
Spigaglia P, Barbanti F, Mastrantonio P. difficile ESGoC, Ackermann G, Balmelli C, et al. Multidrug resistance in European Clostridium difficile clinical isolates. J Antimicrob Chemother. 2011;66(10):2227–34.
Spigaglia P, Barbanti F, Castagnola E, Diana MC, Pescetto L, Bandettini R. Clostridium difficile causing pediatric infections: new findings from a hospital-based study in Italy. Anaerobe. 2017;48:262–8.
Taori SK, Hall V, Poxton IR. Changes in antibiotic susceptibility and ribotypes in Clostridium difficile isolates from southern Scotland, 1979–2004. J Med Microbiol. 2010;59(3):338–44.
Tenover FC, Tickler IA, Persing DH. Antimicrobial-resistant strains of Clostridium difficile from North America. Antimicrob Agents Chemother. 2012;56(6):2929–32.
T-t T, J-h Z, Yang J, C-x Q, Z-r L, Chen J, et al. Molecular characterization of Clostridium difficile isolates from human subjects and the environment. PLoS One. 2016;11(3):e0151964.
Tickler IA, Goering RV, Whitmore JD, Lynn AN, Persing DH, Tenover FC. Strain types and antimicrobial resistance patterns of Clostridium difficile isolates from the United States, 2011 to 2013. Antimicrob Agents Chemother. 2014;58(7):4214–8.
Tickler IA, Obradovich AE, Goering RV, Fang FC, Tenover FC, Consortium H. Changes in molecular epidemiology and antimicrobial resistance profiles of Clostridioides (Clostridium) difficile strains in the United States between 2011 and 2017. Anaerobe. 2019;60:102050.
Tkhawkho L, Nitzan O, Pastukh N, Brodsky D, Jackson K, Peretz A. Antimicrobial susceptibility of Clostridium difficile isolates in Israel. J Global Antimicrobial Resist. 2017;10:161–4.
Tokimatsu I, Shigemura K, Osawa K, Kinugawa S, Kitagawa K, Nakanishi N, et al. Molecular epidemiologic study of Clostridium difficile infections in university hospitals: results of a nationwide study in Japan. J Infect Chemother. 2018;24(8):641–7.
Venugopal AA, Riederer K, Patel SM, Szpunar S, Jahamy H, Valenti S, et al. Lack of association of outcomes with treatment duration and microbiologic susceptibility data in Clostridium difficile infections in a non-NAP1/BI/027 setting. Scand J Infect Dis. 2012;44(4):243–9.
Wang B, Lv Z, Zhang P, Su J. Molecular epidemiology and antimicrobial susceptibility of human Clostridium difficile isolates from a single institution in Northern China. Medicine. 2018;97(25):e11219.
Wieczorkiewicz JT, Lopansri BK, Cheknis A, Osmolski JR, Hecht DW, Gerding DN, et al. Fluoroquinolone and macrolide exposure predict Clostridium difficile infection with the highly fluoroquinolone-and macrolide-resistant epidemic C. difficile strain BI/NAP1/027. Antimicrob Agents Chemother. 2016;60(1):418–23.
Wolfe C, Pagano P, Pillar CM, Shinabarger DL, Boulos RA. Comparison of the in vitro antibacterial activity of Ramizol, fidaxomicin, vancomycin, and metronidazole against 100 clinical isolates of Clostridium difficile by broth microdilution. Diagn Microbiol Infect Dis. 2018;92(3):250–2.
Wultańska D, Banaszkiewicz A, Radzikowski A, Obuch-Woszczatyński P, Młynarczyk G, Brazier J, et al. Clostridium difficile infection in polish pediatric outpatients with inflammatory bowel disease. Eur J Clin Microbiol Infect Dis. 2010;29(10):1265–70.
Yang J, Zhang X, Liu X, Cai L, Feng P, Wang X, et al. Antimicrobial susceptibility of Clostridium difficile isolates from ICU colonized patients revealed alert to ST-37 (RT 017) isolates. Diagn Microbiol Infect Dis. 2017;89(2):161–3.
Zhou Y, Burnham C-AD, Hink T, Chen L, Shaikh N, Wollam A, et al. Phenotypic and genotypic analysis of Clostridium difficile isolates: a single-center study. J Clin Microbiol. 2014;52(12):4260–6.
Zhou Y, Mao L, Yu J, Lin Q, Luo Y, Zhu X, et al. Epidemiology of Clostridium difficile infection in hospitalized adults and the first isolation of C. difficile PCR ribotype 027 in central China. BMC Infect Dis. 2019;19(1):232.
Wang R, Suo L, Chen HX, Song LJ, Shen YY, Luo YP. Molecular epidemiology and antimicrobial susceptibility of Clostridium difficile isolated from the Chinese People’s liberation Army general Hospital in China. Int J Infect Dis. 2018;67:86–91.
Saha S, Kapoor S, Tariq R, Schuetz AN, Tosh PK, Pardi DS, et al. Increasing antibiotic resistance in Clostridioides difficile: a systematic review and meta-analysis. Anaerobe. 2019;58:35–46.
Dingle KE, Didelot X, Quan TP, Eyre DW, Stoesser N, Golubchik T, et al. Effects of control interventions on Clostridium difficile infection in England: an observational study. Lancet Infect Dis. 2017;17(4):411–21.
Major G, Bradshaw L, Boota N, Sprange K, Diggle M, Montgomery A, et al. Follow-on RifAximin for the prevention of recurrence following standard treatment of infection with Clostridium Difficile (RAPID): a randomised placebo controlled trial. Gut. 2019;68(7):1224–31.
Ng QX, Loke W, Foo NX, Mo Y, Yeo W-S, Soh AYS. A systematic review of the use of rifaximin for Clostridium difficile infections. Anaerobe. 2019;55:35–9.
Miller MA, Blanchette R, Spigaglia P, Barbanti F, Mastrantonio P. Divergent rifamycin susceptibilities of Clostridium difficile strains in Canada and Italy and predictive accuracy of rifampin Etest for rifamycin resistance. J Clin Microbiol. 2011;49(12):4319–21.
He T, Wang R, Liu D, Walsh TR, Zhang R, Lv Y, et al. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat Microbiol. 2019;4(9):1450–6.
Linkevicius M, Sandegren L, Andersson DI. Potential of tetracycline resistance proteins to evolve tigecycline resistance. Antimicrob Agents Chemother. 2016;60(2):789–96.
Acknowledgements
We are grateful for the useful comments and suggestions from anonymous referees.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors. We thank the Ilam University of Medical Sciences (A-10-2575-2) for financial support.
Author information
Authors and Affiliations
Contributions
Mohammad Sholeh, Ebrahim Kouhsari: Conceptualization, Methodology, Software. Mehdi Forouzesh, Sergey Mironov, Nourkhoda Sadeghifard: Data curation, Writing- Original draft preparation. Leila Molaeipour, Abbas Maleki: Visualization, Investigation. Marcela Krutova: Supervision, Validation: Ebrahim Kouhsari, Marcela Krutova: Writing- Reviewing and Editing. The author(s) read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Ilam University of Medical Sciences (http://ethics.research.ac.ir/IR.MEDILAM.REC.1398.136).
Consent for publication
Not applicable.
Competing interests
The authors report no conflicts of interest in this work.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Sholeh, M., Krutova, M., Forouzesh, M. et al. Antimicrobial resistance in Clostridioides (Clostridium) difficile derived from humans: a systematic review and meta-analysis. Antimicrob Resist Infect Control 9, 158 (2020). https://doi.org/10.1186/s13756-020-00815-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13756-020-00815-5