Abstract
Ray-finned fishes (Actinopterygii) represent more than 50 % of extant vertebrates and are of great evolutionary, ecologic and economic significance, but they are relatively underrepresented in ‘omics studies. Increased availability of transcriptome data for these species will allow researchers to better understand changes in gene expression, and to carry out functional analyses. An international project known as the “Transcriptomes of 1,000 Fishes” (Fish-T1K) project has been established to generate RNA-seq transcriptome sequences for 1,000 diverse species of ray-finned fishes. The first phase of this project has produced transcriptomes from more than 180 ray-finned fishes, representing 142 species and covering 51 orders and 109 families. Here we provide an overview of the goals of this project and the work done so far.
Similar content being viewed by others
Background
Ray-finned fishes (Actinopterygii) are the most diverse and abundant group of extant vertebrates. Thus far, approximately 32,900 fish species are recorded in FishBase [1]. Fishes encompass enormous variation in morphology, physiology and ecology. They are of great economic and medical significance as a primary source of protein for people worldwide, as a novel source of active ingredients in pharmaceuticals [2], and as evolutionary models for specific human diseases and conditions [3].
However, genomic resources for fishes are relatively underrepresented and published genetic data represent only a small fraction of extant fish species. So far, the whole genomes of only 38 fish species have been published (Additional file 1) and, although the number is growing (Additional file 2), searching the National Center for Biotechnology Information (NCBI)’s Sequence Read Archive (SRA) database for “fish AND transcriptome” yields 16,975 transcriptomes of only 242 fish species (Table 1). A lack of genomic resources for most fish species motivated us to generate large-scale fish transcriptome data and establish a database that may be used by scientists around the world. To this end, we initiated the “Transcriptomes of 1,000 Fishes” (Fish-T1K) project, an effort devoted to sequencing the transcriptomes of 1,000 different species of ray-finned fishes.
Fish-T1K
Fish-T1K is an international, collaborative and non-profit initiative officially launched by BGI and the China National Genebank (CNGB) in November 2013. The objective is to generate RNA-seq transcriptome sequences for 1,000 diverse fish species to help scientists unravel the mysteries of fish evolution, and pursue innovative approaches and strategies for addressing challenges in fish breeding, disease control and prevention, seafood safety, and biodiversity conservation.
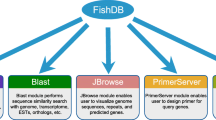
Through this project, an integrated biobank will be established, incorporating a high-level bio-repository and a large-scale transcriptome database. The biobank will collect and store fish genetic resources including vouchers and frozen tissues, DNA and RNA nucleotides, together with related sample information documented according to standard operating procedures (SOPs). A companion database, committed to being the world’s largest database of fish transcriptomes, has already been established and provides access to the sequences via BLAST search.
The Fish-T1K consortium
More than 40 scientists from 25 institutions across seven countries are active members of the Fish-T1K project (Fig. 1; Additional file 3). The Steering Committee consists of six core consortium members who are recognized experts in ichthyology, taxonomy, bioinformatics, phylogenetics, and evolution. In addition to the head office at BGI in Shenzhen, China, we have also established a hub at the Smithsonian National Museum of Natural History (NMNH) in Washington DC, USA, to facilitate quality sample collection from North America.
Distribution of Fish-T1K Consortium members. See detailed information of these numbered institutions in Additional file 3
Species selection
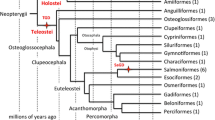
Fish-T1K proposes to sequence 1,000 different ray-finned fish species representing all the orders and major families [4], and filling important gaps in the phylogenetic tree. Species that are endangered, of great economic and medical significance, or exhibit extreme phenotypes will also be targeted. Candidate species will be decided based on their importance and availability, while the target number will be a compromise between scientific needs and practical limitations such as financial constraints and availability of specimens.
Subprojects
To maximize usage of these transcripts, Fish-T1K has launched several subprojects to address specific questions in fish evolution. The major research goal of Fish-T1K is to reconstruct a comprehensive molecular phylogeny of ray-finned fishes to further resolve and test existing phylogenetic hypotheses. Additional subprojects include analysis of the evolutionary genomics of fish venoms, evolution of the annual life cycle in killifishes, and adaptations related to marine-to-freshwater transitions/migration.
SOPs and best practices
In the past two years, the Fish-T1K Team has established a series of SOPs, approved by BGI’s Institutional Review Board on Bioethics and Biosafety (No. BGI-IRB 15139), to ensure high quality sampling is achieved. Adhering to these SOPs means that all of our genetic resources, data and associated metadata are appropriately obtained, documented, and stored, which is helpful in establishing and optimizing standards common to large-scale transcriptome and genome sequencing projects.
Transcriptome data from multiple tissues of five fishes were generated as a pilot quality control test (Additional file 4). Accordingly, total RNA is now routinely extracted from gills and other tissues of interest, and approximately 3.5 Gb of raw data are generated for each sample. Clean reads are assembled de novo into contigs with SOAPdenovo-Trans (v1.3) [5], and the final assembled transcripts are used for annotation, ortholog prediction and other analyses.
Current RNA sequencing progress
The Fish-T1K team has established a collaborative global network for collecting specimens. As of January 2016, 7,000 high quality fish samples were collected from Australia, the Caribbean, Denmark, Singapore, the UK, USA, and many places in China such as the Tibetan Plateau, Sanya, and the Yellow Sea. From these 7,000 samples, RNA samples were extracted from 142 ray-finned species covering 51 orders and 109 families, and around 180 transcriptomes have been produced (Table 1; Additional file 5). Meanwhile, more RNA samples from other species are being isolated and sequenced.
Website and database
The official Fish-T1k website [6] is equipped with a database for BLAST search. The website provides detailed information about the Fish-T1K project, and particular sample information (RNA quality, sample provider, etc.) and data quality (raw data size, scaffold size and number, etc.) are presented in the database. Users can access the BLAST tool and download sequences of interest. Data will be uploaded periodically as sample collection and transcriptome sequencing progresses.
Data sharing policy and data availability
All sequences generated from Fish-T1K will be deposited in NCBI and GigaDB in addition to the Fish-T1K database, following the Fort Lauderdale rules [7] and Toronto International Data Release Workshop guidelines [8], and will be released at least in the time of publication of any resulting papers. We plan to peer review and publish the SOP and method papers, and publications for some of the ongoing subprojects are also expected in one the coming year.
Fish-T1K membership
All are welcome to participate in Fish-T1K and to propose new subprojects; these should address a major question in fish evolution and lead to (a) significant publication(s). Interested researchers can email fisht1k@genomics.cn with a brief proposal. The significance, question(s) to be addressed and fishes/tissues to be sequenced and analyzed should be included. On acceptance of a proposal, the lead scientist(s) will be asked to collect any fish tissues that are not in our list, and to be in charge of analyzing and publishing the generated data.
Conclusions
Similar initiatives already exist to sequence the transcriptomes of large numbers of plants (1KP [9]) and insects (1KITE [10]). They have been well received and have been useful in establishing Fish-T1K. Although some progress has already been made, Fish-T1K is at an early stage. We will continue to expand the scope of the project: in the first phase we aim to cover all orders, and all families in the second phase. More species will be added as required by subprojects. As the world’s first large-scale transcriptome database exclusively for fish, Fish-T1K will greatly enhance the study of fish biology, and eventually contribute efforts towards global fish biodiversity conservation and the sustainable utilization of natural fish resources.
Abbreviations
- 1KITE:
-
1 K Insect Transcriptome Evolution
- 1KP:
-
1000 Plants
- Fish-T1K:
-
transcriptomes of 1,000 Fishes
- NGS:
-
next-generation sequencing
- RNA-seq:
-
RNA sequencing
- SOPs:
-
standard operating procedures. SRA: Sequence Read Archive.
- SRA:
-
Sequence Read Archive
References
Froese R, Pauly D. FishBase. 2015. Available at: http://www.fishbase.org/. Accessed 12 Apr 2016.
Han S, Sun X, Ritzenthaler JD, et al. Fish oil inhibits human lung carcinoma cell growth by suppressing ILK. Mol Cancer Res. 2009;7(1):108–17.
Albertson RC, Cresko W, Detrich HW, et al. Evolutionary mutant models for human disease. Trends Genet. 2009;25(2):74–81.
Betancur-R R, Wiley Ed, Bailly N, et al. Phylogenetic classification of bony fishes (version 3). DeepFin. 2015; Available at: http://deepfin.bio.ou.edu/. Accessed 12 Apr 2016.
Luo R, Liu B, Xie Y, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 2012;1:18.
Transcriptomes of 1,000 fishes. Available at: www.fisht1k.org. Accessed 12 Apr 2016.
The Wellcome Trust (Fort Lauderdale January 14–15, 2003). Sharing data from large-scale biological research projects: a system of tripartite responsibility. 2003; Available at: https://www.genome.gov/Pages/Research/WellcomeReport03.03.pdf. Accessed 12 Apr 2016.
Birney E, Hudson TJ, Green ED, et al. Prepublication data sharing. Nature. 2009;461(7261):168–70.
Matasci N, Hung LH, Yan ZX, et al. Data access for the 1,000 Plants (1KP) project. GigaScience. 2014;3:17.
Misof B, Liu S, Meusemann K, et al. Phylogenomics resolves the timing and pattern of insect evolution. Science. 2014;346(6210):763–7.
Acknowledgements
We wish to acknowledge the Fish-T1K Consortium members, partners, advisors and supporters who have made Fish-T1K possible. We would also like to thank Junxing Yang from the Kunming Institute of Zoology of the Chinese Academy of Sciences, Qiang Lin from South China Sea Institute of Oceanology of the Chinese Academy of Sciences, Carol Stepien from the University of Toledo, Luiz Rocha from the California Academy of Sciences, Donald Stewart from the State University of New York College of Environmental Science and Forestry, and Andrew Thompson from The George Washington University for their tremendous support with sample collection.
Funding
This work was supported by China 863 Projects (No. 2012AA10A407 & 2014AA093501), Shenzhen and Hong Kong Innovation Circle Project (No. SGLH20131010105856414), Shenzhen Special Program for Future Industrial Development (No. JSGG20141020113728803), Shenzhen Special Program for Bio-industry Development (Nos. HY20130205008 and NYSW20130326010014) and Special Project on the Integration of Industry, Education and Research of Guangdong Province (No. 2013B090800017). A grant from the Smithsonian Institution (Biogenomics/GGI) to C. Baldwin, G. Orti, and R. Betancur provides partial funding for RNA extractions at the NMNH (Washington, DC, USA).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
YS drafted the original text with detailed input from YH, XL, GO, BV and QS. All authors have read and approved the final manuscript and participated in Fish-T1K.
Additional files
Additional file 1:
List of fishes with published genome data. (DOCX 30 kb)
Additional file 2:
Number of fish species with newly published transcriptomes in SRA of the NCBI from 2009 to 2015. (TIF 4045 kb)
Additional file 3:
List of the current Fish-T1K Consortium members. (DOCX 19 kb)
Additional file 4:
Transcriptome data of five species for quality control. (DOCX 20 kb)
Additional file 5:
List of fishes with transcriptome data generated by Fish-T1K. (XLSX 87 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Sun, Y., Huang, Y., Li, X. et al. Fish-T1K (Transcriptomes of 1,000 Fishes) Project: large-scale transcriptome data for fish evolution studies. GigaSci 5, 18 (2016). https://doi.org/10.1186/s13742-016-0124-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13742-016-0124-7